Abstract
Apyrases hydrolyze nucleoside triphosphates and diphosphates and are found in all eukaryotes and a few prokaryotes. Although their enzymatic properties have been well characterized, relatively little is known regarding their subcellular localization and physiological function in plants. In this study, we used reverse genetic and biochemical approaches to investigate the role of potato (Solanum tuberosum)-specific apyrase. Silencing of the apyrase gene family with RNA interference constructs under the control of the constitutive 35S promoter led to a strong decrease in apyrase activity to below 10% of the wild-type level. This decreased activity led to phenotypic changes in the transgenic lines, including a general retardation in growth, an increase in tuber number per plant, and differences in tuber morphology. Silencing of apyrase under the control of a tuber-specific promoter led to similar changes in tuber morphology; however, there were no direct effects of apyrase inhibition on tuber metabolism. DNA microarrays revealed that decreased expression of apyrase leads to increased levels of transcripts coding for cell wall proteins involved in growth and genes involved in energy transfer and starch synthesis. To place these results in context, we determined the subcellular localization of the potato-specific apyrase. Using a combination of approaches, we were able to demonstrate that this enzyme is localized to the apoplast. We describe the evidence that underlies both this fact and that potato-specific apyrase has a crucial role in regulating growth and development.
ATP-diphosphohydrolases, or apyrases, are enzymes that hydrolyze both the γ- and β-phosphates of ATP and ADP. They are distinct from other phosphohydrolases with respect to their specific activity, nucleotide substrate specificity, divalent cation requirement, and sensitivity to inhibitors (Plesner, 1995; Handa and Guidotti, 1996). Apyrases are ubiquitously expressed in eukaryotes and have additionally been found in some prokaryotes, indicating a general role for these enzymes across species. Furthermore, apyrase function has been intensively studied in mammalian systems, in which the role of apyrase is to hydrolyze extracellular adenosine nucleoside triphosphates and diphosphates, which is important both in the inactivation of synaptic ATP as a neurotransmitter following nerve stimulation (Todorov et al., 1997) and in the inhibition of ADP-induced platelet aggregation to prevent thrombosis (Marcus et al., 1997). Adenosine deriving from the concerted action of apyrase and ecto-5′-nucleotidase is subsequently transported into mammalian cells via a sodium/adenosine cotransporter (Che et al., 1992).
In contrast to the situation described above for mammalian systems, relatively little is known about the function of apyrase in plants. The enzyme has been characterized at the biochemical level in potato (Solanum tuberosum; Kalckar, 1944; Molnar and Lorand, 1961; Kettlun et al., 1982) and various legumes, such as Glycine max, Medicago truncatula, and Pisum sativum (Day et al., 2000; Hsieh et al., 2000; Cohn et al., 2001). Potato apyrase has additionally been speculated to be involved in starch synthesis (Fanta et al., 1988). The reasoning behind this hypothesis was the fact that many steps of the starch biosynthetic pathway are regulated by the levels of ATP, ADP, or inorganic phosphate (Pi) and that potato tuber, which accumulates vast quantities of starch (Geigenberger and Fernie, 2006), contains a specific apyrase distinct from those found in other plants (Handa and Guidotti, 1996; Roberts et al., 1999). A special feature of potato-specific apyrase is that it is soluble and does not appear to be strongly bound to membranes, as is the case for all other plant apyrases characterized to date. On the basis of phylogenetic analysis and a DNA hybridization experiment reported by Handa and Guidotti (1996), it has been postulated that potato additionally contains apyrases that more closely resemble general plant apyrases (Roberts et al., 1999). The deduced amino acid sequence of the potato-specific cDNA contains a putative N-terminal signal peptide for the secretory pathway (Supplemental Fig. S1). In M. truncatula, four apyrase cDNAs were isolated and two of them showed increased transcript levels in response to nodulation (Cohn et al., 2001). Nodulation-promoting factors were also shown to bind to a lectin domain of an apyrase found in roots of Dolichos biflorus, indicating a role of this enzyme during nodulation (Etzler et al., 1999). However, this lectin domain appears to be conserved, also being present in the potato apyrase and the apyrases AtAPY1 and AtAPY2 from Arabidopsis (Arabidopsis thaliana; Steinebrunner et al., 2000). In addition to sequence analysis, some information is available regarding the subcellular localization of these proteins, with the legume-specific apyrase DbLNP from D. biflorus demonstrated to have a cell membrane localization (Etzler et al., 1999). Plasma membrane localization was also found for the G. max apyrase GS52 (Day et al., 2000; Kalsi and Etzler, 2000), and a second, structurally similar apyrase has been documented as being localized to the Golgi apparatus (Day et al., 2000). While GS52-specific antibodies blocked nodulation in G. max (Day et al., 2000), overexpression of GS52 in Lotus japonicus enhanced nodulation (McAlvin and Stacey, 2005). These studies point to a specific function of apyrase in nodulation processes of legumes. Furthermore, studies of apyrase function in nonlegumes have been performed in Arabidopsis. These studies include the analysis of transgenic plants overexpressing an apyrase from P. sativum and T-DNA knockout mutants of two apyrase genes (AtAPY1 and AtAPY2) identified in Arabidopsis. Interestingly, expression of the heterologous apyrase led to increased scavenging of extracellular phosphate (Thomas et al., 1999) and to higher resistance to xenobiotics (Thomas et al., 2000) and herbicides (Windsor et al., 2003). In contrast, single knockout mutants of AtAPY1 or AtAPY2 lack a discernible phenotype, while the corresponding double mutant exhibited a complete inhibition of pollen germination in Arabidopsis, indicating a role of apyrase in sexual reproduction of plants (Steinebrunner et al., 2003).
Although AtAPY1 and AtAPY2 are thought to be ecto-apyrases, only indirect evidence supporting an apoplastic localization for these enzymes has been presented to date (Wu et al., 2007). Apyrase activity, occasionally termed “latent UDPase,” has additionally been identified in Golgi vesicles from P. sativum, wherein it appears to use UDP as a substrate (Orellana et al., 1997; Neckelmann and Orellana, 1998). It is believed that this activity hydrolyzes UDP to UMP, which is subsequently translocated into the symplast in exchange for a nucleotide sugar. The sugar moiety of this nucleotide sugar is subsequently transferred to a nascent cell wall polysaccharide or protein glycosyl residue yielding UDP. It seems likely that the elimination of UDP is required for continued operation of the cycle, since it is an inhibitor of the glucosyltransferase-catalyzed reactions that constitute a portion of the cycle (Neckelmann and Orellana, 1998). On the basis of their proteomic analyses of Golgi preparations, Dunkley et al. (2004) suggested a Golgi localization of AtAPY2. While these previous studies revealed a number of possible functions of apyrase, the general importance of this enzyme in plants has not been fully resolved. Moreover, the subcellular localizations of plant apyrases in general, and the potato-specific apyrase in particular, have not been unambiguously demonstrated.
In this study, we cloned two new potato-specific apyrases closely related to the one identified by Handa and Guidotti (1996). Using database searches, we were able to find tentative consensus sequences of potato apyrases that in fact more closely resemble general plant apyrases, supporting the assumption that potato contains additional apyrases with special function in addition to general plant apyrases (Roberts et al., 1999). We used a reverse genetic approach to investigate the importance and role of potato-specific apyrase. Transgenic analyses were performed to study the effect of altered apyrase expression on growth, development, metabolism, and gene expression. In addition, the subcellular localization of the potato apyrase was investigated using nonaqueous fractionation, GFP fusion constructs, apoplastic washings, and tuber slice incubation experiments. The combined results of these studies indicate that potato apyrase is an apoplastic enzyme of major importance for growth and development.
RESULTS
Potato Contains at Least Three Different Potato-Specific Apyrases with High Expression in Sink Tissues
Using primers based on the 5′ and 3′ untranslated region sequences of the previously cloned potato apyrase (Handa and Guidotti, 1996), two apyrases with very high identity to this apyrase were cloned. These novel clones were thus termed StAPY2 and StAPY3 (with StAPY1 being the apyrase cloned previously). StAPY3 showed 98.7% identity to StAPY1, and StAPY2 still showed 92.6% identity to StAPY1 at the protein level (alignment included in Supplemental Fig. S1 and Supplemental Table S1). A comprehensive database search revealed tentative consensus (TC) sequences generated from ESTs for all three apyrases in the potato gene indices database provided by the Computational Biology and Functional Genomics Laboratory (Supplemental Table S1). StAPY2 was assigned to TC134863, and both StAPY1 and StAPY3 were assigned to TC137054 alone, but the single nucleotide polymorphism report for this TC clearly verifies the existence of both very similar isoforms. In addition to this, TCs putatively coding for other apyrases were identified in this database (TC142822 and TC144101; Supplemental Table S1). Complete or partial protein sequences were derived from these TC sequences and analyzed with regard to identity toward StAPY1 to StAPY3 and AtAPY1 and AtAPY2. While comparison at the nucleotide level was relatively unclear, the protein sequences of these TCs were markedly more similar to the protein sequences of the Arabidopsis apyrases (59%–66%) than to StAPY1 to StAPY3 (49%–52%). This comparison thus provides support for the assumption by Roberts et al. (1999) that in addition to potato-specific apyrase, additional apyrases with high similarity to general plant apyrases, such as AtAPY1 and AtAPY2, exist in the potato genome.
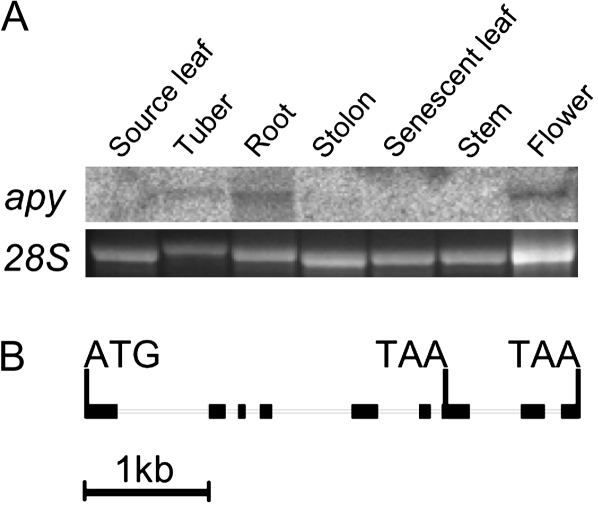
The cDNA of StAPY2 has a comparable length to StAPY1 and StAPY3. However, in addition to the TAA stop codon found in the other apyrases at positions 1,363 to 1,365, it has an additional stop codon at positions 850 to 852. The protein sequences of both isoforms contain all four apyrase conserved regions identified by Handa and Guidotti (1996; Supplemental Fig. S1). The anterior stop codon, however, was not found in several ESTs from the StAPY2 cDNA cloned from the cultivar Shepody, and it is hence possible that this cDNA is not translated into a functional apyrase protein in the cultivar Desiree. Due to the high identity between the different potato-specific apyrases (>92%; Supplemental Table S1) and their lower identity with other detected apyrases at the nucleotide level, it was possible to determine the combined transcript levels of all three potato-specific isoforms in different tissues using a probe generated from the cDNA sequence of StAPY3. Expression was found to occur to a considerable extent in sink tissues such as root, tuber, and flower and to a much lesser extent in stolon tissue (Fig. 1A). In contrast, virtually no transcript was detectable in source leaves, senescent leaves, or stem tissue.
Figure 1.
Gene expression pattern of potato apyrase and gene structure of the potato apyrase 2. A, Fifty micrograms of RNA from different tissues was separated, and transcript levels of StAPY1 to StAPY3 were quantified with a probe consisting of StAPY3. The detected band had a size of 1.5 kb. As a loading control, the band of the 28S ribosomal RNA (28S) on the agarose gel is also shown. B, The complete gene of the truncated potato apyrase StAPY2 consisting of nine exons (black blocks) and eight introns is shown, including the start codon and the stop codon leading to truncation in exon 7 as well as the stop codon found in StAPY1 to StAPY13 in the last exon.
Isolation of a genomic clone of StAPY2 confirmed that the truncation found in the cDNA was not an artifact of the cloning procedure. The 4-kb sequence obtained contained the 3,909-bp DNA sequence from the start to the posterior stop codon identified in the cDNA (Fig. 1B). In total agreement with the anterior stop codon found in the cDNA of StAPY2, the genomic sequence contained the corresponding stop codon in exon 7 at positions 2,855 to 2,857. The gene consists of nine exons and eight introns, exactly like the Arabidopsis apyrases AtAPY1 and AtAPY2, and some of the splicing sites are conserved between the potato and Arabidopsis apyrases. It thus seems likely that the general plant apyrases and the potato-specific apyrases originated from a common ancestral protein.
Generation of Transgenic Potato Plants with Altered Expression of Apyrase
Potato plants with reduced/elevated apyrase expression were generated by Agrobacterium tumefaciens-mediated transformation of Desiree potato plants, as described by Rocha-Sosa et al. (1989). The full-length sequence of StAPY3 was cloned into pART7/pART27 (Gleave, 1992) to create a plasmid overexpressing StAPY3 under the control of the ubiquitous cauliflower mosaic virus 35S promoter. For the production of the 35S-RNA interference (RNAi) plasmid, the first 500 nucleotides of StAPY3 were cloned in the sense-intron-antisense orientation into pHANNIBAL/pART27 (Wesley et al., 2001; for full cloning details, see Supplemental Fig. S2, A–C). Due to the high identity of StAPY1 to StAPY3 at the RNA level, it seems probable that these RNAi constructs would affect the transcript levels of all three potato-specific apyrases but not of those with higher identity to AtAPY1 and AtAPY2. Furthermore, a database search in the potato gene indices of the Computational Biology and Functional Genomics Laboratory using the 500-bp RNAi fragment as a query suggested that it was highly unlikely that this construct would affect any other unintended target. A third plasmid for tuber-specific reduction of apyrase was produced by replacement of the 35S promoter with the B33 promoter. Following regeneration, transformed plant material was clonally propagated, cuttings were cultivated in tissue culture, and explants were grown under controlled conditions in order to generate plant material for biochemical analysis and phenotypic characterization.
Lines with Altered Expression of Apyrase under the Control of the Constitutive 35S Promoter Showed Marked Changes in Apyrase mRNA and Activity
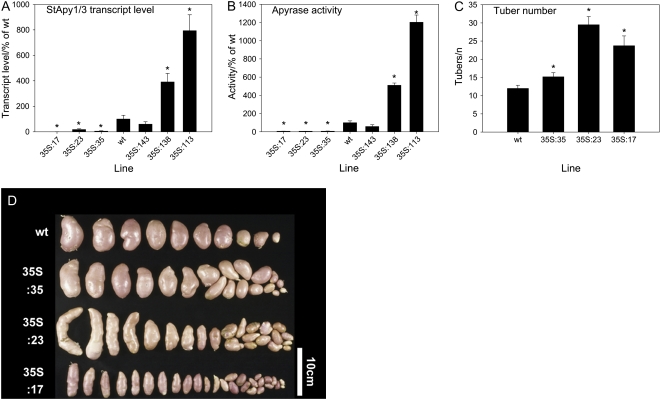
To investigate the expression of apyrase in the transgenic lines, both apyrase mRNA and activity were determined. The primers used for the real-time PCR were specific for isoforms StApy1 and StApy3 but would not allow amplification of StApy2. Differentiation between StApy1 and StApy3 was virtually impossible, due to the high similarity at the nucleotide level downstream of the sequence used for RNAi induction. Apyrase activity was measured in the soluble fraction of enzyme extracts using ATP as substrate and in the absence of phosphatase inhibitors, since potato apyrase is the predominant ATP-hydrolyzing phosphatase in enzyme extracts of potato tubers (see below). As shown in Figure 2, A and B, both the apyrase mRNA and activity of all 35S-RNAi lines were significantly reduced, whereas the apyrase mRNA and activity of two of the 35S-overexpressing lines were significantly higher than those in wild-type tubers. While line 35S:17 was clearly most strongly reduced in StAPY1/3 transcript levels (0.13% of the wild type) compared with lines 35S:35 (5.3%) and 35S:23 (18%), apyrase activity was similarly reduced in all three RNAi lines. Line 35S:23 appeared to be the most affected, followed by lines 35S:17 and 35S:35 at the level of activity. However, these lines were not very different from one another, with activities ranging from 6.0% to 7.1% of the total wild-type activity. From this, we conclude that neither transcript levels nor phosphatase activities allow unimpeachable conclusions regarding the degree of reduction of apyrase in these lines. That said, they clearly demonstrate that apyrase is markedly reduced in all of them.
Figure 2.
Altered apyrase transcript and activity under the control of the 35S promoter affects tuber development. A, Combined transcript levels of StApy1 and StApy3 in tuber tissue of RNAi lines (two-digit code) and overexpressors (three-digit code) were determined from cDNA by real-time PCR and expressed as percentages of the wild type. B, Apyrase activity was analyzed using an assay containing 1 mm ATP as substrate, and the activities are expressed as percentages of the wild type. C, Lines with decreased apyrase expression grown for 10 weeks in a phytotron produced a higher number of smaller tubers showing a rod-like shape (see D). Results are means ± se, n = 6 to 8 (A and B) and 10 to 13 (C). Significant differences from the wild type according to Student's t test are indicated with asterisks (P < 0.05). D shows typical examples.
Using specific primers for StAPY2, we found that this isoform is expressed to a lower extent when compared with StAPY1 and StAPY3 and that its expression is also reduced in two of the 35S-RNAi lines (Supplemental Fig. S3, A–C). As observed for the wild type, apyrase activities of the transgenics were similar when UTP, ADP, and UDP were provided as substrate (see Fig. 7H below). However, it should be noted that the turnover of the monophosphates was markedly lower, suggesting that ATP was hydrolyzed by apyrase, not by any phosphatase. The relative turnover of the triphosphates and diphosphates tested was invariant in the overexpressing lines.
Figure 7.
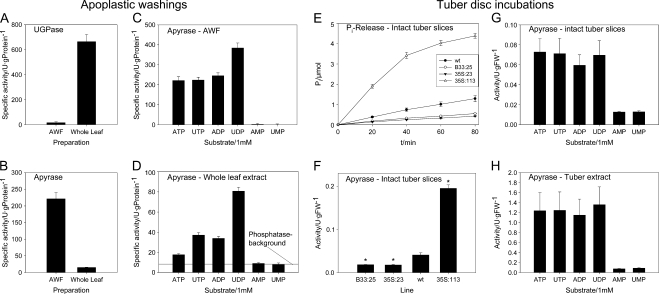
Apyrase activity in apoplastic washings and in tuber slices in incubation experiments. A to D, To investigate the subcellular localization of apyrase, the specific activities of apyrase (B; 1 mm ATP as substrate) and of the cytosolic marker enzyme UGPase (A) were determined in AWF obtained by centrifugation of infiltrated wild-type potato leaves and compared with the activities assayed in whole leaf extract obtained from the same plants. The substrate specificity of apyrase/5′-nucleotidase was assayed in both AWF (C) and whole leaf extract (D) using 1 mm ATP, UTP, ADP, UDP, AMP, or UMP. The horizontal line indicates the level of nonspecific phosphatase activity on the basis of nucleoside monophosphate hydrolysis. E to H, Experiment with tuber slices. Freshly prepared and washed tuber slices of the wild type, B33-RNAi line B33:25, 35S-RNAi line 35S:23, and 35S overexpressor line 35S:113 were incubated with 3 μmol of ATP in 3 mL of buffer in a final concentration of 1 mm to determine apoplastic apyrase activity as release of Pi from ATP in a time course (E) or in absolute activities per gram fresh weight (FW; F). The substrate specificity of apyrase/5′-nucleotidase in intact tuber slices (G) and in total tuber enzyme extracts produced from wild-type plants using substrate concentrations of 1 mm (H) are also shown. Results are means ± se (n = 6–8). In F, significant differences from the wild type according to Student's t test are indicated with asterisks (P < 0.05).
All of the lines exhibited a resting ATP-hydrolyzing activity of approximately 6% of that found in the wild type. It seems unlikely that all of this activity corresponds to residual potato-specific apyrase, since apyrase protein was not detectable on a western blot in the strongest RNAi line and consistently increased in the strongest overexpressing line (Supplemental Fig. S3D). This result suggests that the remaining ATP hydrolysis activity is catalyzed either by apyrase isoforms that do not cross-react with the antibody or by less specific phosphatases. Irrespective of what is responsible for this residual activity, the data presented here clearly demonstrate that potato-specific apyrase isoforms contribute to the vast majority of apyrase activity measured in potato tubers.
Lines with Decreased Expression of Apyrase under the Control of the Constitutive 35S Promoter Showed Changes in Growth and Development
When grown in a greenhouse, potato plants with decreased constitutive expression of apyrase showed developmental alterations compared with the wild type. There was a decrease in shoot growth; however, this was not strictly dependent on the decrease in apyrase activity (Supplemental Fig. S4, A and B). Lines with decreased apyrase, however, produced a larger number of tubers (Fig. 2C), which were considerably smaller in size (Fig. 2D) and developed a more longitudinal shape than tubers of the wild type (Fig. 2D). In contrast to this, lines with increased expression of apyrase under control of the 35S promoter had no visible alterations in overall plant growth or tuber development in comparison with the wild type (data not shown).
Lines with Decreased Expression of Apyrase under the Control of the Tuber-Specific B33 Promoter Showed Increased Longitudinal Tuber Growth
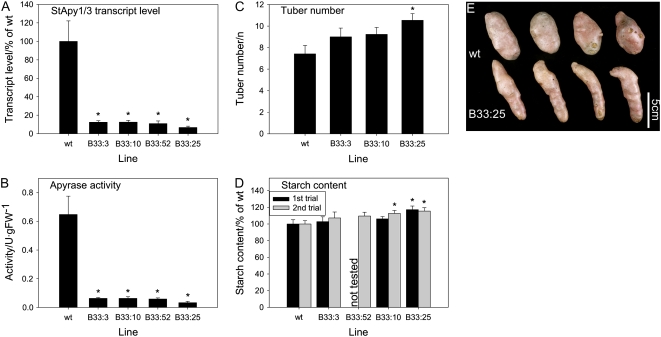
In order to investigate whether tuber development was specifically affected by a decrease in apyrase activity in the growing tuber, as opposed to pleiotropic changes as a consequence of its reduced expression in other plant tissues, further transgenic lines were generated that expressed the apyrase-RNAi chimera under the control of the tuber-specific B33 patatin promoter. Following screening of the resultant transformants, four lines were selected that displayed a strong decrease in both tuber apyrase mRNA and activity with respect to the wild type (Fig. 3, A and B). As would perhaps be expected, there was no substantial decrease in shoot growth when apyrase expression was decreased using the B33 promoter (data not shown), since this promoter has been demonstrated to confer tuber-specific expression (Liu et al., 1990). However, tuber morphoplogy was altered in the tissue-specific lines in a similar manner to that described above for the lines exhibiting constitutive repression of apyrase expression (compare Fig. 3E with Fig. 2D). When taken together, these data provide strong evidence that the changes in tuber morphology are directly linked to alterations in the tuber apyrase activity rather than consequences of pleiotropic effects of the modulation of apyrase activity in other parts of the plant.
Figure 3.
Altered apyrase transcript and activity under the control of the tuber-specific B33 promoter affects tuber phenotype and starch content. A, Combined transcript levels of StApy1 and StApy3 in tubers of 8-week-old plants were determined from cDNA by real-time PCR and expressed as percentages of the wild type. B, Apyrase activity was analyzed using an assay containing 1 mm ATP as substrate and displayed in units per gram fresh weight (FW). C and D, Tuber number (C) and starch content (D) in two independent trials were determined in tubers from 13-week-old plants. There were characteristic changes in tuber morphology (see E), which were similar to those observed in lines in which apyrase was decreased using the constitutive 35S promoter (see Fig. 2D). Results are means ± se, n = 8 (A and B) and 12 to 15 (C and D). Significant differences from the wild type according to Student's t test are indicated with asterisks (P < 0.05). E shows typical examples.
The total tuber number per plant was only slightly increased in the B33 lines in comparison with the wild type, and this was only significant in a single line (Fig. 3C). These results contrast with those of the 35S lines, which exhibited a very clear increase in tuber number (Fig. 2C). This discrepancy, however, could be due to several reasons, including differential promoter strength and the fact that the results of the 35S expression could be complicated by modulating apyrase expression in a broad range of tissue types.
Decreased Expression of Apyrase under the Control of the Tuber-Specific B33 Promoter Did Not Lead to Substantial Alterations in Metabolite Levels
We further investigated whether decreased expression of apyrase via the tuber-specific B33 promoter affects the composition of the tubers. Starch, which represents the major carbohydrate in potato tubers, was only slightly elevated in response to the repression of apyrase, the increase being significant only in two of the lines showing a strong decrease in apyrase activity (Fig. 3D). The reduction of apyrase activity did not substantially affect the relative contents of total protein (data not shown) or of cell wall components such as cellulose or uronic acids (Supplemental Fig. S5, A and B). Moreover, the composition of cell wall matrix polysaccharides was unaltered in the transgenic lines, providing evidence that cell wall matrix architecture was not affected by the change in apyrase activity (Supplemental Fig. S5C). Given that uronic acids and cell wall matrix polysaccharides are produced in the Golgi, the lack of changes in these components argues against the possibility that potato-specific apyrases act as latent UDPases functionally linked to the production of such cell wall structures in the Golgi. As such, these results oppose the hypothesis postulated for the apyrase of P. sativum (Orellana et al., 1997; Neckelmann and Orellana, 1998); however, it should be borne in mind that apyrases likely play different roles in diverse species.
In order to investigate whether the changes in tuber development and starch content in the transformants were attributable to changes in metabolite levels, we next assessed these using HPLC and gas chromatography-mass spectrometry (GC-MS). There were no significant changes in the levels of adenylate pools or adenylate-related parameters for cellular energy charge in the different genotypes, with the exception of the concentration of AMP in line B33:3 (Supplemental Fig. S6). The other uridylates and guanylates (UTP/UDP/UMP/GTP/GDP/GMP) measured by HPLC were also unaltered in comparison with the wild type (data not shown).
Although steady-state levels of nucleotides were unchanged, we postulated that reduction of apyrase may have led to a decline in the turnover of nucleotides. Such a scenario would be anticipated to result in lowered respiration rates, since it would entail lower demand for respiratory ATP production. For this reason, oxygen consumption of tubers was determined by transferring freshly cut tuber slices into a Clarke-type electrode. However, this parameter was also largely unaltered in the transformants (Supplemental Fig. S7), suggesting that modification of apyrase did not greatly affect overall cellular ATP turnover. Systematic profiling of metabolites using a GC-MS-based method also failed to reveal major changes in metabolite levels in the transformants (Supplemental Table S2). Furthermore, even minor changes were not statistically significant following correction of the P values returned using the false discovery rate (FDR) method (Benjamini and Hochberg, 1995) implemented in the R software (n = 8). This observation has important implications, since it suggests that the changes in growth caused by reduction of apyrase are not primarily due to an effect of altered apyrase activity upon metabolism.
Decreased Expression of Apyrase under the Control of the Tuber-Specific B33 Promoter Leads to Changes in Transcript Profiles
To investigate whether the changes in tuber development and starch content in response to the decreased apyrase activity are attributable to broader changes in gene expression, transcript profiles were analyzed in growing tubers using The Institute for Genomic Research (TIGR) 10k EST potato array. This array contains 12,091 cDNAs in replicates and covers approximately 25% of the 46,000 potato genes found so far according to the Canadian Potato Genome Project. Four arrays were analyzed that were hybridized with two independent tuber samples from line B33:10 versus wild-type samples and two independent tuber samples from line B33:25 versus wild-type samples. The P values obtained after normalization were corrected for multiple testing using the FDR algorithm implemented in the R software (the FDR corrected P values are subsequently referred to as q values). In some cases, several clones of one gene were spotted on the array. After elimination of the redundant clones, 11,375 individual genes derived from unique tentative consensus sequences remained. Of these, 540 genes (4.7% of all genes) were differentially regulated. Only 58 of them were down-regulated compared with 481 genes that were up-regulated, showing that reduced apyrase activity leads to a preferential activation of genes. Of the up-regulated genes, 48 showed a greater than 2-fold induction; however, no repression of corresponding magnitude was observed in the down-regulated genes. Using a recently developed MapMan mapping file (Rotter et al., 2007), all genes were sorted into bins arranged according to functional ontologies. This also allowed visualization of the genes involved in general metabolism using the MapMan software (Thimm et al., 2004; Fig. 4). In addition, all of the genes exhibiting a greater than 2-fold change were manually explored for tentative functions using BLASTN (Altschul et al., 1997) in an attempt to identify homologs. This facilitated the association of several previously unassigned genes to a functional ontology. However, 18 such genes remained unidentified and are thus listed as coding for hypothetical/expressed proteins. Three genes had contradictory 5′ and 3′ EST sequences derived from the clone spotted on the array. The inconsistency between the 5′ and 3′ ends of a clone is probably caused by chimeras consisting of more than one cDNA in one plasmid. The remaining 27 genes are listed in Table I.
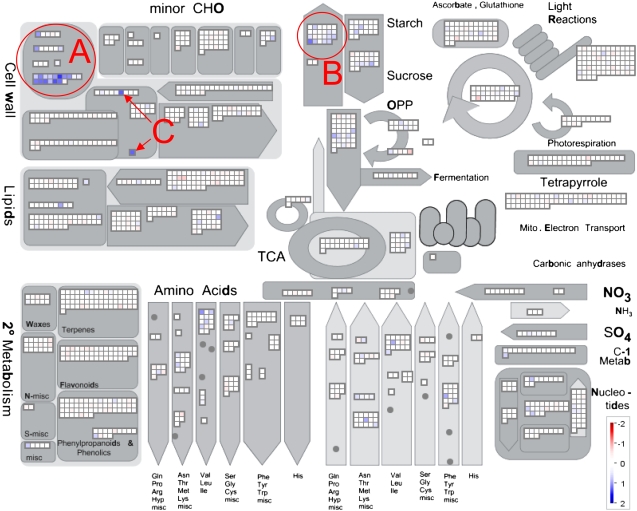
Figure 4.
Tubers with decreased apyrase under the control of the tuber-specific B33 promoter show specific changes in their transcript profiles compared with the wild type. RNA was extracted from growing tubers of separate plants from the wild type and lines B33:10 and B33:25 and converted into dye-labeled second-strand cDNA. The second-strand DNA was hybridized together with complementary labeled wild-type second-strand cDNA on the TIGR 10k potato array, giving four hybridized arrays in total. Within the two replicate arrays for one of the two lines, a dye swap was performed. Data are shown as a snapshot of the metabolic overview slide of MapMan (Thimm et al., 2004) using a recently developed mapping file (Rotter et al., 2007). Changes in transcript levels coding for extensins and related cell wall proteins (A), genes involved in starch synthesis (B), and glycosyltransferases (C) are highlighted.
Table I.
Summary of differentially regulated transcripts in tubers exhibiting decreased apyrase expression under the control of the tuber-specific B33 promoter in comparison with the wild type
Only genes with tentative function that passed the t test corrected for multiple comparisons using the FDR and that are at least 2-fold changed compared with the wild type are listed. Genes were assigned into bins as suggested by the MapMan mapping file for the TIGR 10k potato array developed by Rotter et al. (2007). Transcript changes are given in log2 scale, and the q value corresponds to the P value corrected for multiple comparison. Sequence information for the spotted clones can be retrieved using the identifier from GenBank or the TIGR Solanaceae Genomics Resource. If significant induction was observed on alternative spots on the array coding for the same gene, the identifier is given in parentheses.
| Bin | Identifier/Clone | Log2 | q Value | Description |
|---|---|---|---|---|
| Amino acid metabolism.synthesis.branched chain group.common | STMHX23 | 1.06 | 0.0010 | Homolog to UP|Q9SM58_PEA (Q9SM58) acetohydroxy acid isomeroreductase precursor, partial (36%) |
| Cell wall.cell wall proteins | STMIJ81 | 1.23 | 0.0016 | Similar to UP|Q02021_LYCES (Q02021) Gly-rich protein, partial (97%) |
| Cell wall.cell wall proteins.AGPs | STMCJ47 | 1.02 | 0.0071 | Homolog to UP|Q52QJ6_LYCES (Q52QJ6) cell wall-attached arabinogalactan/Pro-rich protein, partial (98%) |
| Cell wall.cell wall proteins.HRGP | STMJK30 (STMHU57) | 1.02 | 0.0118 | Similar to TIGR_Ath1|At1g53645.1 68414.m06102 Hyp-rich glycoprotein family protein, partial (47%) |
| Cell wall.cell wall proteins.Pro-rich proteins | STMGX29 | 1.05 | 0.0063 | Similar to UP|Q40402 (Q40402) Nicotiana plumbaginifolia extensin precursor, partial (28%) |
| STMIA95 | 1.08 | 0.0033 | Homolog to UP|Q40402 (Q40402) N. plumbaginifolia extensin precursor, partial (33%) | |
| STMHV77 | 1.17 | 0.0033 | Similar to UP|Q6QNA3 (Q6QNA3) Pro-rich protein 1 (extensin), partial (92%) | |
| STMID50 (STMIK90) | 1.21 | 0.0016 | Homolog to UP|Q41707 (Q41707) extensin class 1 protein precursor, partial (56%) | |
| STMJI39 | 1.12 | 0.0029 | Homolog to UP|O82066 (O82066) Pro-rich protein (extensin), partial (91%) | |
| STMJN80 | 1.21 | 0.0016 | Homolog to Solanum lycopersicum extensin (class I) gene, partial (76%) | |
| STMHZ32 | 1.45 | 0.0010 | Homolog to UP|Q09084 (Q09084) extensin (class II) precursor, partial (26%) | |
| STMIR05 | 1.33 | 0.0011 | Potato mRNA for extensin | |
| STMIG94 (STMIH94) | 1.25 | 0.0012 | Homolog to UP|Q43504 (Q43504) extensin-like protein Dif10 precursor (fragment), partial (24%) | |
| STMIK16 | 1.02 | 0.0026 | Similar to UP|Q43504 (Q43504) extensin-like protein Dif10 precursor (fragment), partial (21%) | |
| Cell wall.pectin synthesis | STMHQ60 | 1.26 | 0.0041 | Similar to TIGR_Ath1|At3g25140.1 68416.m03139 glycosyltransferase family 8 protein, partial (88%) |
| Miscellaneous.UDP glucosyltransferase and glucoronyltransferase | STMHU42 (STMJB73) | 1.19 | 0.0062 | Similar to TIGR_Ath1|At5g60700.1 68418.m07617 glycosyltransferase family protein 2, partial (16%) |
| STMHQ80 | 1.25 | 0.0101 | Similar to TIGR_Ath1|At1g18580.1 68414.m02317 glycosyltransferase family protein 8 | |
| Major CHO metabolism.synthesis.starch.transporter/transport.metabolite transporters at the mitochondrial membrane | STMDR46 (STMDI11) (STMGA51) (STMGJ88) (STMHK79) | 1.20 | 0.0010 | UP|ADT2_SOLTU (P27081) ADP/ATP carrier protein 2, mitochondrial precursor (ADP/ATP translocase 2) adenine nucleotide translocator 2 (ANT2; fragment), complete |
| Redox.thioredoxin | STMHQ72 | 1.21 | 0.0010 | Homolog to plastidial thioredoxin reductase (cNTR) AT2G41680 |
| Protein.degradation.ubiquitin.E3.RING | STMGI84 | 1.03 | 0.0122 | Similar to TIGR_Ath1|At1g68070.1 68414.m07776 zinc finger (C3HC4-type RING finger) family protein |
| Protein.synthesis.miscellaneous ribosomal protein | STMJN92 (STMDU75) | 1.02 | 0.0016 | Homolog to UP|RS18_ARATH (P34788) 40S ribosomal protein S18, complete |
| RNA.processing.splicing | STMGI77 | 1.08 | 0.0190 | Similar to TIGR_Ath1|At5g51300.1 68418.m06359 splicing factor-related, partial (26%) |
| RNA.RNA binding | STMIY63 | 1.42 | 0.0071 | Similar to TIGR_Ath1|At3g13224.2 68416.m01658 RNA recognition motif-containing protein, partial (59%) |
| Signaling.G proteins | STMEF69 | 1.07 | 0.0067 | Similar to TIGR_Ath1|At5g27540.1 68418.m03297 GTP-binding protein-related, partial (39%) |
| Signaling.unspecified | STMIM39 | 1.27 | 0.0024 | TIP41-like family protein, contains Pfam PF04176: TIP41-like family; identical to cDNA putative cytoskeletal protein mRNA, partial cds GI:5031529 |
| Stress.abiotic.cold | STMIJ15 | 1.10 | 0.0016 | Similar to UP|GRP2_NICSY (P27484) Gly-rich protein 2, partial (95%) |
| Stress.abiotic.heat | STMJJ69 | 1.22 | 0.0067 | Similar to TIGR_Ath1|At1g74250.1 68414.m08599 DNAJ heat shock N-terminal domain-containing protein, partial (97%) |
Almost half of the genes displaying a 2-fold increase in transcript levels code for apoplastic proteins belonging to the class of extensins or related proteins (Table I; Fig. 4A). Extensins are Hyp-rich proteins that are highly O-glycosylated at the Hyp residues. They are structural components of the cell wall with signaling function and have been implicated in many developmental processes and various biotic and abiotic stress responses (Baumberger et al., 2001; Wu et al., 2001; Hall and Cannon, 2002; Merkouropoulos and Shirsat, 2003).
Decreased apyrase led to increased levels of transcripts involved in the provision of carbon and energy for starch synthesis in the amyloplast (Fig. 4B). Both Suc synthase genes 2 (STMDV33, log2 = 0.55, q = 0.0279) and 4 (STMHF04, log2 = 0.85, q = 0.0018) were induced in response to the decreased expression of apyrase. Also, the genes coding for the mitochondrial ATP/ADP translocator 2 (ANT2; Table I) and ANT1 were significantly up-regulated (STMDW96, log2 = 0.76, q = 0.0010) as well as the plastidic ATP/ADP translocator 1 (AANT1; STMGG40, log2 = 0.60, q = 0.0399), as were the Glc-6-P translocator 1 (STMJF75, log2 = 0.66, q = 0.0066) and 2 (STMEC08, log2 = 0.59, q = 0.0429). These changes are in agreement with previous studies showing that the activities of Suc synthase, the plastidic ATP/ADP translocator, and the plastidic Glc-6-P transporter positively correlate with starch content in potato tubers (Tjaden et al., 1998; Geigenberger, 2003; Zhang et al., 2006). The nucleus-encoded subunit of the key enzyme of starch synthesis, ADP-Glc pyrophosphorylase (AGPase), which has a high control coefficient over starch synthesis in potato tubers (Sweetlove et al., 1999; Geigenberger et al., 2004), was significantly induced (STMDW17, log2 = 0.71, q = 0.0422). There was also a strong increase in transcripts coding for a plastidial NADPH-dependent thioredoxin reductase (cNTR; Table I) in response to reduced apyrase. Recently, the posttranslational redox regulation of AGPase was found to be a powerful mechanism in the regulation of starch synthesis in the plastid (Tiessen et al., 2002; Kolbe et al., 2005; Oliver et al., 2008).
Although no influence of altered apyrase activity was found on the composition or amount of cell wall matrix sugars or uronic acids (Supplemental Fig. S5, B and C), two genes coding for glycosyltransferases were strongly up-regulated in response to the decrease in apyrase expression (Table I; Fig. 4C).
Investigation of the Subcellular Localization of Potato Apyrase
The lack of significant changes in nucleotide pool levels upon reduction of apyrase indicates that the potato apyrase resides in a compartment that is physically separated from the major nucleotide pools of the cell: those found in the cytosol, mitochondria, and plastid. Unfortunately, the subcellular localization of apyrases in plants is still a matter of debate. The apyrases GS52 from G. max and DbLNP from the legume D. biflorus were found to be localized to the cell membrane (Day et al., 2000; Kalsi and Etzler, 2000), while evidence has been provided that another, structurally similar, apyrase termed GS50 is localized in the Golgi apparatus of G. max (Day et al., 2000). In Arabidopsis, apyrases have been identified by mass spectrometry only in membrane fractions containing Golgi proteins (Neckelmann and Orellana, 1998; Dunkley et al., 2004). Controversially, Dunkley et al. (2004) have suggested that AtAPY2 is Golgi localized, while Roux and coworkers suggested it to be localized in the cell membrane with the active site facing the apoplast (Steinebrunner et al., 2003). In contrast to the apyrases from other plant species, potato-specific apyrases are soluble when extracted under native conditions (Figs. 2B and 3B). This solubility is probably not an artifact of proteolytic cleavage, because the protein migrates at a predicted size independent of whether extracted under native conditions (Supplemental Fig. S3D, lane 1) or with Lämmli buffer containing a high concentration of ionic detergent (Supplemental Fig. S3D, lanes 2–4). However, given the crucial importance of this question, we decided to investigate the subcellular localization of potato apyrase by following several different approaches in parallel.
Determination of Apyrase in Fractionated Tuber Tissue
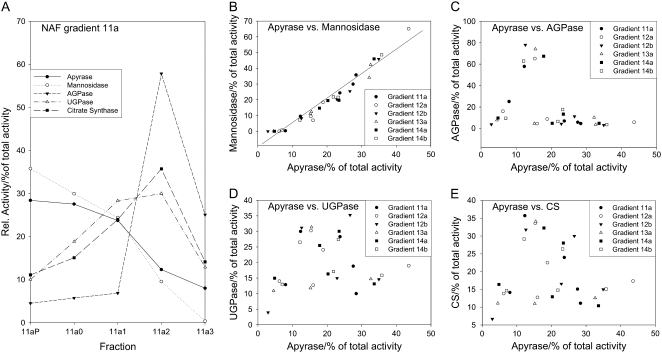
Potato tuber tissue can be fractionated using a nonaqueous fractionation technique to analyze the subcellular localization of a specific protein or metabolite using marker enzymes (Farre et al., 2001; Tiessen et al., 2002). To investigate the distribution of apyrase, six gradients consisting of five different fractions each were analyzed, and the distribution of apyrase activity was compared with the distribution of the activities of several marker enzymes in these fractions, such as UGPase (cytosol), AGPase (plastid), citrate synthase (mitochondria), and mannosidase (vacuole + apoplast).
The pattern of apyrase activity was substantially different from the pattern of the cytosolic, plastidial, and mitochondrial marker activities (Fig. 5). Thus, it is unlikely that apyrase is localized in the cytosol, plastids, or mitochondria. The high recovery rate of apyrase activity (100% ± 20% for all gradients; data not shown) also excludes that ATP hydrolysis was carried out by another phosphatase than apyrase. The vacuolar/apoplastic marker mannosidase shows a similar pattern to apyrase and correlated highly significantly with apyrase in its distribution in the representative gradient 11a (Fig. 5A) and all five gradients tested additionally (r = 0.977, P < 0.0001, n = 30; Fig. 5B). At the moment, it is not possible to distinguish between vacuole and apoplast in the nonaqueous gradients due to cofractionation of apoplastic and vacuolar markers (Fettke et al., 2005). Therefore, it cannot be determined with this method whether apyrase is localized in the apoplast or in the vacuole.
Figure 5.
Comparison of apyrase and marker enzymes in nonaqueous fractions (NAF) of lyophilized wild-type tuber tissue. A, The activities of subcellular marker enzymes and apyrase are shown along a typical gradient consisting of five fractions with increasing density from right to left. B to E, Correlations between apyrase and the markers for vacuole and apoplast (mannosidase; B), plastid (AGPase; C), cytosol (UGPase; D) and mitochondria (citrate synthase [CS]; E) were calculated from activities obtained from six individual gradients. The same nonaqueous gradients were used that were characterized previously (Tiessen et al., 2002).
Transient Expression of Apyrase-GFP in Leaf Epidermis Cells
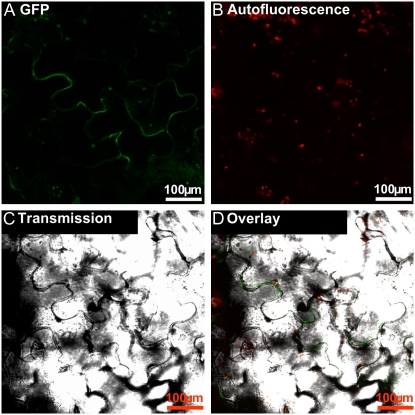
The data obtained from the nonaqueous fractionation gradients suggest vacuolar or apoplastic localization for apyrase. To distinguish between these possibilities, localization was further investigated by the use of an apyrase-GFP fusion protein. Potato apyrase StAPY3 was cloned into pA7-GFP (Sohlenkamp et al., 2002; Voelker et al., 2006; Supplemental Fig. S2D). The resultant plasmid coded for the potato apyrase C-terminally fused to enhanced GFP. A preparation of the plasmid coding for the apyrase-GFP fusion protein was loaded onto gold particles and shot into Arabidopsis leaves with a biolistic gun. After 2 d of incubation at 25°C, the leaves were examined with a confocal microscope and images of cells temporarily expressing apyrase-GFP were recorded. As shown in Figure 6, A, C, and D, GFP accumulates in a layer that surrounds particular epidermis cells of the bombarded leaves. The signal is diffusely spread in the apoplastic space and can be clearly distinguished from the autofluorescence of the chloroplasts recorded at a higher wavelength (Fig. 6B). This shows that apyrase is localized in the apoplast rather than the vacuole. Another control plasmid coding for the membrane-localized ammonium transporter AtAMT2 fused to GFP (Sohlenkamp et al., 2002) was also shot into leaves; however, the fluorescence pattern of this transporter was clearly different from the diffusive signal of StApy3-GFP and restricted to the plasma membrane of a single cell instead (Supplemental Fig. S8).
Figure 6.
Localization of a temporally expressed StAPY3-GFP fusion protein. Arabidopsis leaf epidermal cells were bombarded with a plasmid coding for the StAPY3 C-terminally fused to GFP. A, Fluorescence of GFP recorded between 500 and 525 nm. B, Autofluorescence of the chloroplasts recorded between 630 and 690 nm. C, Bright-field image of the section. D, An overlay of all three images (given in A–C). Typical examples are shown.
Determination of Apyrase Activity in Apoplastic Washing Fluid from Leaves of Potato Wild-Type Plants
If apyrase is a soluble apoplastic protein or easily detachable from an apoplastic component, it should be possible to detect its activity in apoplastic washing fluid (AWF). This method has already been used not only for the determination of apoplastic metabolites but also for the extraction of apoplastic proteins (Boudart et al., 2005; Fecht-Christoffers et al., 2006). Pools of AWF from leaves from eight 6-week-old wild-type potato plants with reasonably low activity of UGPase, a soluble cytosolic enzyme with high activity and an almost identical molecular mass to that of apyrase, were prepared to analyze apyrase activity. In parallel, total cellular enzyme extracts from leaves of all eight plants were also prepared for comparison. The AWF contained less than 3% of specific UGPase activity compared with that contained in whole leaf extracts (Fig. 7A), which is indicative of a relatively low level of contamination of the AWF with cytosolic proteins. In contrast, the AWF contained 13 times higher values of specific apyrase activity than the whole leaf extracts (Fig. 7B). These data give clear evidence for the existence of apoplastic apyrase in potato leaves. This apyrase turns over ATP, UTP, and ADP with the same speed, and UDP is hydrolyzed almost twice as fast as the other nucleotides. Hydrolysis of AMP and UMP was almost undetectable (Fig. 7C). The highly specific turnover of triphosphates and diphosphates, but not of the monophosphate in the AWF, indicates that the phosphatase activity measured was carried out by an apyrase and not by an unspecific phosphatase. Apyrase measured in the whole leaf preparations displayed different kinetic properties than the enzyme in the AWF. UTP and ADP were hydrolyzed twice as fast as ATP, and UDP was hydrolyzed even four times as fast (Fig. 7D). There was a substantial hydrolysis of AMP and UMP, indicating apyrase-independent turnover of nucleotides by unspecific phosphatases in the whole leaf extract. When the turnover of unspecific nucleoside monophosphate hydrolysis was subtracted from the hydrolysis of triphosphates and diphosphates to provide a more specific measure of apyrase activity, the differences between the turnover rates for the respective nucleoside triphosphates and diphosphates were even more pronounced.
Although it is possible that the enzyme might have been modified in one of the fractions or that the presence of effectors led to the differences in kinetic properties, these results suggest the existence of apyrases with different kinetic properties localized in different subcellular compartments in potato leaves. The leaf apoplastic apyrase possesses comparable relative turnover rates for the different purine and pyrimidine nucleotides tested. These kinetic properties also apply to the apyrases changed in activity in this study; therefore, it is highly likely that they are identical. A second leaf apyrase shows preference to the pyrimidine nucleotide UDP. From the experiment performed, no clear conclusions can be drawn concerning its localization with the exception of the fact that we can rule out that it is not a soluble apoplastic protein. Indeed, its substrate preference suggests that it is more likely to be the Golgi-localized latent UDPase, the apyrase potentially involved in the nucleotide sugar transfer (Orellana et al., 1997; Neckelmann and Orellana, 1998).
Determination of Apyrase Activity in Intact Tuber Slices of Potato Wild-Type Plants and Lines with Altered Apyrase Activity
Since apoplastic washings cannot be performed with potato tubers, we used a different experimental approach to measure apoplastic apyrase activity using tuber slices. Tuber slices consist of compactly packed cells without physical barriers such as cuticles and can be kept in buffer for several hours without loss of cell membrane integrity. In contrast to tuber cells in culture, a freshly prepared tuber slice represents a much more physiologically relevant state of this tissue, since it harbors naturally organized cell walls. For this reason, tubers from 8-week-old wild-type and transgenic plants were sliced into discs. The slices were washed five times with buffer to withdraw intracellular contaminations and finally incubated in buffer. To determine apoplastic apyrase activity, ATP was added to the slices. After different time intervals, aliquots were collected from the external medium and analyzed for apyrase activity by determining the conversion of the external ATP to Pi. By this method, the apyrase activities of tuber slices of the strongest B33-RNAi line, the second strongest 35S-RNAi line (the strongest was incapable of producing suitable tuber material for analysis), and the strongest 35S overexpressor were determined. As shown in Figure 7E, apyrase activity could be measured as Pi formation in the medium containing the wild-type tuber slices after the addition of ATP. This release is linear with time, indicating an enzymatic turnover of ATP at saturating substrate concentration rather than a leakage of Pi from damaged cells. The absolute activity determined approximates to 0.05 to 0.08 units g−1 fresh weight for intact wild-type slices (Fig. 7, F and G), which corresponds to roughly 10% of total cellular activity (Figs. 3B and 7H). In comparison with this, UGPase activity measured in washed intact slices did not exceed 0.3% of the activity in total enzyme extracts (data not shown).
The apyrase activities of intact tuber slices from the strongest B33-RNAi and the second strongest 35S-RNAi line (B33:25 and 35S:23) were significantly reduced to a level of 45% and 44% of the wild-type activity, respectively (Fig. 7F). By contrast, the apyrase activity was strongly elevated in the strongest 35S overexpressor (Fig. 7E). The rate of hydrolysis of adenosine triphosphates and diphosphates is apparently much higher than the rate of hydrolysis of the monophosphate in wild-type tuber slices (Fig. 7G). This finding, when taken together with the fact that apyrase activity in the discs of the different lines was altered according to the respective apyrase expression level, provides clear evidence that hydrolysis was carried out by an apyrase rather than an unspecific phosphatase. Furthermore, the maximal activities for the substrates ATP, UTP, ADP, and UDP were identical, as was the case for apyrase characterized in tuber extracts from the wild type (Fig. 7H) and also in tuber extracts from the strongest StAPY3 overexpressor, 35S:113 (data not shown), and they were similar to those determined in the AWF from wild-type potato leaves (Fig. 7C).
DISCUSSION
Potato-Specific Apyrases May Constitute a Small Gene Family
The cloning of three very similar potato-specific apyrases, one of them truncated, may suggest that these apyrases constitute a small gene family. Whether these different isoforms are encoded on different loci or are merely different alleles of the same gene cannot currently be stated confidently, due to the high degree of heterozygosity and the tetraploid genome of potato. The fact that StAPY2 and StAPY3 were cloned using primers that bind to the untranslated region, which generally exhibits low conservation between different genes, would favor the one gene/different allele scenario. However, the existence of all three isoforms in the cultivar Bintje and a Southern-blot analysis showing several bands when probed with StAPY3 (data not shown) provide strong support for the assumption of a small gene family. Additionally, two more apyrases were identified from TC sequences (Supplemental Table S1) that resemble more closely general plant apyrases like AtAPY1 and AtAPY2, but their total number is probably higher because Arabidopsis already contains seven predicted apyrases (The Arabidopsis Information Resource). Potato-specific apyrases are expressed predominantly in sink tissues, like root, flower, stolon, and tuber, but to a much lower extent, if at all, in leaf or stem tissue (Fig. 1A). When expression of the potato-specific apyrase was altered in transgenic plants, strong morphological alterations were found in sink tissues such as tubers. The RNAi approach led to the reduction of nearly all of the apyrase activity in growing tubers, indicating that these potato-specific apyrases encode for most of the apyrase activity in this tissue (Figs. 2B and 3B).
Potato Apyrase Is Confined to the Apoplast
To date, most plant apyrases have been demonstrated or postulated to be localized at the plasma membrane or in the Golgi (Orellana et al., 1997; Neckelmann and Orellana, 1998; Etzler et al., 1999; Thomas et al., 1999; Day et al., 2000; Wu et al., 2007). Here, we provide several independent lines of evidence indicating that the potato-specific apyrases characterized in this study are localized in the apoplast without strong interaction with structural components of the cell. Furthermore, our studies revealed that the potato-specific apyrases account for the majority of apyrase activity in this compartment. First, apyrase activity correlated strongly with the distribution of a vacuolar/apoplastic marker in tuber fractions separated on nonaqueous gradients (Fig. 5). When taken in the context of the high contribution of the potato-specific apyrases to the total tuber activity, this suggests that, of the major cellular compartments, the potato apyrases may be localized in either the vacuole or apoplast, but not in the cytosol, mitochondria, or plastids. Second, the StAPY3-GFP fusion protein was targeted to the apoplast when transiently expressed in Arabidopsis leaves (Fig. 6). While we cannot fully exclude mistargeting of apyrase within the secretory pathway leading to apoplastic accumulation of the StAPY3-GFP fusion protein, it is very unlikely that the fusion protein was entering this path due to C-terminal fusion of GFP to apyrase, since targeting sequences for this path are located at the N terminus and were predicted for all three potato apyrases. Moreover, the strong sequence identity makes it very likely that all three potato apyrases will have the same topogenesis. Third, apyrase activity was detected in the apoplast of tuber slices, showing identical substrate specificity as StAPY3 and being altered in response to StAPY3 or general potato-specific apyrase expression in a corresponding manner (Fig. 7). Fourth, apyrase activity was also demonstrated in apoplastic leaf washings, having almost identical substrate specificity to the apyrase analyzed in the apoplast of tubers but displaying different substrate specificity to the apyrase activity found in the total tissue extracts of potato leaves (Fig. 7). The latter shows high activity when UDP is supplied as substrate, making it quite likely that it corresponds to a latent UDPase that has been shown to be localized to the Golgi (Orellana et al., 1997). In combination, these independent experiments provide evidence that the potato-specific apyrases are apoplastically localized and suggest that they have no strong association to membranes of the cell. Moreover, since the size of the extracted apyrase as analyzed in western blots (approximately 52 kD; Supplemental Fig. S3D) matches the predicted size, it is rather unlikely that the protein has been proteolytically cleaved from a membrane-bound form.
In order to determine whether potato-specific apyrase directly participates in Golgi sugar transfer, an analysis of the cell wall with particular focus on the structural components that assemble in the Golgi was conducted (Supplemental Fig. S5). This analysis revealed that neither the absolute amounts of uronic acids nor the composition of cell wall matrix sugars varied in the transgenic lines. Given that these compounds are likely the largest source of sugars transferred via the Golgi sugar transfer system, it follows that apyrase is not a component of this pathway.
Potato Apyrase Affects Tuber Development
Our results revealed strong effects of apyrase on specific aspects of tuber development. Tubers displaying either constitutive or tuber-specific decreases in the expression of apyrase developed a rod-like shape, indicating stimulation of longitudinal growth of the tubers, in comparison with the wild type (Figs. 2D and 3E). The 35S lines also exhibited a significant decrease in tuber size and an increase in tuber number in response to a reduction of apyrase. Furthermore, the 35S-RNAi lines additionally displayed a decrease in overall plant growth. This was not strictly dependent on the reduction of apyrase (Supplemental Fig. S4). It does, however, at least partially confirm recent studies in Arabidopsis suggesting that plant growth is strongly dependent on apyrase expression (Wolf et al., 2007; Wu et al., 2007). That said, as in our current study, a direct relationship between the extent of apyrase expression and plant growth effects was difficult to establish.
Potato Apyrase Does Not Directly Perturb Major Metabolic Processes but Leads to Characteristic Changes in the Expression Pattern of Nuclear Genes
Although changes in apyrase expression did not affect tuber metabolite profiles (Supplemental Table S2), they led to characteristic changes in the expression pattern of nuclear genes (Fig. 4; Table I). By far the most striking change was the large induction of transcripts coding for cell wall proteins belonging to the class of extensins and related proteins, which made up 13 of the 27 genes displaying a considerable up-regulation at the transcript level. We can rule out that this strong up-regulation is caused by one or a few dominant extensin cDNAs hybridizing to the spots coding for the other extensin genes, since all of the differentially regulated extensins are fairly divergent in nucleotide sequence (data not shown).
Extensins represent a multigene family coding for cell wall proteins with structural and signaling functions in the apoplast. Strong correlative evidence exists that extensins are involved in wounding, biotic and abiotic stress responses, and developmental processes such as pollen recognition and fertilization, embryo development, cell division, differentiation, abscission, and senescence (Rumeau et al., 1990; Roberts and Shirsat, 2006). Unfortunately, to date, relatively little insight into the function of extensins has been obtained from more direct approaches, such as the use of mutants or transgenic plants with altered extensin expression. Two studies using Arabidopsis null mutants or overexpressors of the extensins LRX and Atext1 showed that these manipulations produced only weak effects on the growth of inflorescences and root hair morphogenesis (Baumberger et al., 2001; Roberts and Shirsat, 2006). A third study showed that the knockout of an Arabidopsis extensin encoded by the RSH gene was embryo lethal (Hall and Cannon, 2002). Therefore, it seems reasonable to speculate that the pronounced morphological changes in growing tubers upon reduction in apyrase are due to the increased expression of several extensin genes.
The majority of transcripts encoding genes associated with central metabolism were not affected by the reduction of apyrase. This observation was in strong accordance with the lack of changes in metabolite levels of intermediates of these pathways. Interestingly, many transcripts encoding proteins related to starch metabolism were altered in a manner that would be anticipated to promote starch synthesis. These included both structural enzymes of the Suc-to-starch conversion pathway, such as Suc synthase, the plastidial Glc-6-P translocator, and AGPase (Geigenberger, 2003), and enzymes esoteric to the pathway, such as intercompartmental adenylate transporters and the plastidial adenylate kinase (Geigenberger et al., 2004; Oliver et al., 2008). Interestingly, a plastidial form of a NADP thioredoxin reductase containing a thioredoxin domain (Serrato et al., 2004) was also found to be up-regulated, which could be important for posttranslational redox regulation of starch synthesis (Tiessen et al., 2002; Kolbe et al., 2005). Although it was not investigated whether these changes in transcript abundance lead to subsequent changes in the protein levels or activities, the sum of these changes would be in favor of an increase in starch production by transcriptional regulation at several sites in the pathway. Indeed, strong reduction of apyrase using a tuber-specific promoter resulted in a slight increase in starch accumulation in two of the lines (Fig. 3D). Recent results by Wolf et al. (2007) also document the influence of apyrase on starch accumulation, although this point was not discussed directly by the authors. Their study revealed that a conditional double knockout of AtAPY1 and AtAPY2 had larger starch granules in the chloroplasts of Arabidopsis leaves. In the context of the lack of changes in metabolite levels, the increase in starch levels suggests regulation of the biosynthesis of starch at the transcriptional rather than the posttranscriptional level.
A Possible Link between Potato Apyrase and Apoplastic Signaling Processes
The results of this study demonstrate that decreased expression of potato apyrase isoforms via RNAi leads to changes in tuber development, resulting in a stimulation of longitudinal growth of the tubers. Changes in apyrase activity do not directly perturb major metabolic processes; rather, they lead to specific changes in the expression of nuclear genes encoding extensins that may explain the developmental alterations, since they are suggested to play a role in polar growth (Baumberger et al., 2001; Hall and Cannon, 2002). Since the potato-specific apyrases were found to be located in the apoplast, the effects of changes in apyrase activity are likely to be caused by alterations of apoplastic signaling processes, specifically those involving extracellular ATP. Moreover, the specific effects of altered apyrase activity on gene expression rather than metabolism fit into existing models linking apyrase activity to apoplastic signaling in a manner analogous to that described for mammalian systems (Chivasa et al., 2005; Kim et al., 2006; Song et al., 2006; Wu et al., 2007). These conclusions confirm previous studies that provided evidence for a link between apoplastic ATP and growth. In planta visualization studies revealed that the highest level of eATP is found in actively growing regions, a fact that supports the theory that it plays a major role in the support of growth (Kim et al., 2006). In studies by Chivasa et al. (2005), increasing apoplastic ATP by the addition of external ATP increased the viability of the cell, while the removal of external ATP by external application of apyrase led to the opposite effect. Our study postulates that changes in the expression of the potato-specific apyrase affect development via a perturbation in the relay of signals emanating from extracellular ATP. Future studies in this subject area are required both to provide direct evidence for this hypothesis and to identify components of the putative signaling pathway.
MATERIALS AND METHODS
Cloning Work
cDNA of StAPY2 and StAPY3
The coding sequences of StAPY2 and StAPY3 were amplified by PCR from potato (Solanum tuberosum) tuber cDNA using the oligonucleotides flaypyf (5′-TCTTGGATCCGGGGCAAAATGTTGAACCAAAATAG-3′) and flapyr (5′-ATTGGAATTCCAACACATTAAGATGATGCAACTC-3′), both of them flanked with restriction sites. After digestion with BamHI and EcoRI, the PCR products were ligated into pBluescript SK+ (Stratagene). Each isoform was cloned three times in three independent PCRs out of two different cDNA preparations and sequenced (AGOWA).
Genomic DNA of StAPY2
The genomic sequence of StAPY2 was amplified by PCR from potato genomic DNA using the oligonucleotides allapyf (5′-CTCTTCAAATAGGGGCAAAATGTTGAACC-3′) and apy2UTR (5′-CAAGTTTCCCACCAATACAAGTACAAGATTT-3′) and ligated into pCR2.1 (Invitrogen). The fragment was cloned only once, since exons did not diverge from the sequence of the cDNA of StAPY2 according to the sequencing results.
35S Overexpressor
The full-length sequence of StAPY3 in pBluescript SK+ was amplified using the oligonucleotides RNAis2f (5′-TCTTCTCGAGGGGGCAAAATGTTGAACCAAAATAG-3′) and flapyr (see above), both of them flanked with restriction sites. After digestion with EcoRI and XhoI, the fragment was ligated into pART7 (Gleave, 1992). The cassette containing the 35S promoter, full length StAPY3, and the OCS terminator was excised using NotI and ligated into pART27 (Gleave, 1992).
35S-RNAi
The partial sequence of StAPY3 from nucleotides −8 to 489 was amplified from StAPY3 in pBluescript SK+ using the oligonucleotides RNAis2f (see above) and RNAis2r (5′-ATCAGGTACCGTGACCCATTGATCTTTGCTATGG-3′), both flanked with restriction sites. After digestion with XhoI and KpnI, the fragment was ligated in the sense orientation into pHannibal (Wesley et al., 2001). The identical partial sequence was also amplified using the oligonucleotides RNAiaf (5′-AACAGGATCCCTTAGGATGTTAAAAGGGGATGCAGC-3′) and RNAia2r (5′-ATCAATCGATGTGACCCATTGATCTTTGCTATGG-3′), both flanked with restriction sites. After digestion with BamHI and ClaI, the fragment was ligated in the antisense orientation into pART7 containing the sense fragment. The cassette containing the 35S promoter, the RNAi construct, and the OCS terminator was excised using NotI and ligated into pART27.
B33-RNAi
Using BamHI and XhoI, the RNAi construct consisting of the StAPY3 sense and antisense fragments comprising the pdk intron was excised from pHannibal (see above) and ligated into pART33, a derivative of pART7 with a B33 promoter instead of the 35S promoter. The cassette containing the B33 promoter, the RNAi construct, and the OCS terminator was excised and ligated into pART27 using NotI.
StAPY3-GFP
The sequence of StAPY3 was amplified from StAPY3 in pBluescript SK+ using the oligonucleotides RNAis2f (see above) and alIapy1r (5′-CAACGTCGACAGATGATGCAACTCTAATTTTG-3′), both flanked with restriction sites. After digestion with XhoI and SalI, the full-length fragment without stop codon was ligated into dephosphorylated pA7-GFP. The coding sequence of StAPY3 of this plasmid was sequenced to guarantee that it was accurate.
Sequence Analysis
Alignments were produced using the ClustalW algorithm embedded in the MegAlign software (DNAstar). This software was also used for sequence identity calculations using identity weight matrix. For homology searches, the GenBank database was explored using BLASTN (Altschul et al., 1997).
Generation of Transgenic Potato Plants
The plasmids were transformed into Agrobacterium tumefaciens strain pGV2260 using a gene pulser electroporator (Bio-Rad) as described by Mattanovich et al. (1989). The recombinant agrobacteria were used to transform sterile, freshly injured leaves of Desiree potato by the method established by Rocha-Sosa et al. (1989).
Plant Growth Conditions
Desiree potato plants were cultivated in tissue culture for long-term storage and propagation. Cuttings were transferred to soil and grown in a phytotron for 2 weeks. Then the plants were transferred to pots with a diameter of 18 cm and grown in the phytotron (soil, 60%/75% humidity [day/night], 22°C/16°C, 400 μmol s−1 m2 light intensity, 16-h days and 18-h nights), a conditioned glasshouse (soil, 60% humidity, 22°C/16°C, 350 μmol s−1 m2 light intensity, 16-h days and 18-h nights), or in an uncontrolled greenhouse.
Northern-Blot Analysis
After separation of 20 μg of RNA derived from different tissues on a denaturing agarose gel and transfer on a membrane (Hybond-XL; APBiotech), the membrane was probed with the full-length fragment of StAPY3 labeled with 32P. Hybridization was detected using a BAS-1800II phosphoimager (Fuji).
Western-Blot Analysis
After separation of 5 μg of tuber protein using PAGE and transfer to a polyvinylidene difluoride membrane (Roche), the membrane was probed with a serum directed against potato apyrase kindly provided by Dr. Pal Nyren (Royal Institute of Technology, Stockholm). Binding was detected enzymatically using a secondary antibody conjugated to horseradish peroxidase (Bio-Rad).
Real-Time PCR
RNA was extracted from 60 mg fresh weight using the RNeasy Plant Mini Kit (Qiagen), and DNA was digested as suggested by the supplier. RNA (500 ng) was used to produce cDNA in a final volume of 100 μL using reverse transcriptase and RNase inhibitor provided by Invitrogen. For cDNA quantification, 2 μL was used as template in real-time PCR mixed with 10 μL of Power SYBR Green (Applied Bioscience) and 10 μL of primer mix (forward and reverse primers, each 0.5 μm). The following primer mixes were used for quantification: StAPY1 and StAPY3 (forward, 5′-GCTTGTTGATGGATTTGGACTAAA-3′; reverse, 5′-GCCATGCTGCTCCAACTAGATAG-3′), StAPY2 (forward, 5′-GCTTGTTGATGGATTTGGACTAAA-3′; reverse, 5′-GCCATGCTGCTTTAATTTGGTAA-3′), StGAPDH5′ (forward, 5′-AAGGACAAGGCTGCTGCTCAC-3′; reverse, 5′-AACTCTGGCTTGTATTCATTCTCG-3′), StGAPDH3′ (forward, 5′-TTCAACATCATCCCTAGCAGCACT-3′; reverse, 5′-TAAGGTCGACAACAGAAACATCAG-3′), and EF1-α (forward, 5′-CATTGCTTGCTTTCACCCTTGGTG-3′; reverse, 5′-CCTAGCCTTGGAGTACTTGGGGG-3′). The amount of cDNA of the induced apyrase was related to the amount of EF1-α. Only samples with equal threshold cycle values (±1) for the GAPDH cDNA 5′ and 3′ ends were considered for analysis.
Expression Profiling Using the TIGR 10k Potato Array
RNA extraction, cDNA synthesis, labeling, hybridization, and scanning were performed as described previously (Degenkolbe et al., 2005). This method includes quality control of first-strand cDNA synthesis by real-time PCR using 3′ and 5′ primers for the GAPDH transcript (see above). TIGR 10k potato arrays (TIGR Solanaceae Genomics Resource) were hybridized and scanned with a Fuji MAS FLA-8000 microarray scanner. The GeneSpotter software (MicroDiscovery) was used for the grid positioning and signal quantification. The resulting data were analyzed using the LIMMA package (Smyth et al., 2005) for the bioconductor software (Gentleman et al., 2004). Data were normalized using within-array print-tip Loess and between-array quantile normalization. Statistical analysis was performed using a mixed model incorporating the duplicate spots for each transcript. P values were corrected using a FDR correction (Benjamini and Hochberg, 1995). Transcripts were considered to be differentially expressed if they had a FDR P value (q value) of <0.05. Data were visualized with the MapMan software (Thimm et al., 2004) using a mapping file developed by Rotter et al. (2007).
Transient Expression of GFP Fusion Proteins in Arabidopsis
A 50-μL gold particle suspension (1 μm in diameter, 6% [w/v] in 50% glycerol) was mixed with 5 μL of plasmid DNA (0.5 μg/μL), 50 μL of 2.5 m CaCl2, and 20 μL of 0.1 m spermidine. After vortexing, the suspension was shortly centrifuged and washed four times with 100% ethanol. The suspension was then resuspended in 48 μL of 100% ethanol. A 10-μL gold particle suspension was transferred on a 1,100-psi rupture disc (Bio-Rad) and shot on freshly cut Arabidopsis (Arabidopsis thaliana) C24 leaves placed upside down at a distance of 9 cm in a Bio-Rad PDS-1000 microprojectile bombardment system as described previously (Sohlenkamp et al., 2002). The bombarded leaves were incubated for 24 to 48 h at 25°C and then inspected with a Leica DM IRBE microscope equipped with a Leica TCS SPII confocal scanner. The excitation wavelength was 488 nm, the emission window for the GFP channel covered 500 to 525 nm, and autofluorescence was recorded in a window between 630 and 690 nm.
Metabolite Analysis
Nucleotides were quantified from TCA extracts prepared as described by Jelitto et al. (1992) using a Kontron HPLC system (Bio-Tek Instruments) equipped with a Partisil-SAX 10 anion-exchange column and a UV photometer detecting at 254 nm (Geigenberger et al., 1998). Starch was measured as Glc obtained after three digestions of the TCA precipitate with amylase and amyloglucosidase by an enzyme-linked assay described previously (Geigenberger et al., 1998). GC-MS analysis and preceding extraction and derivatization were performed as described by Roessner et al. (2001) using a HP 5980 gas chromatograph (Agilent) coupled to a Pegasus II time-of-flight mass spectrometer (Leco).
Cell Wall Analysis
Determination of Matrix Sugar Composition
The pellet of a TCA extract of 200 mg of potato tissue was washed once with 1.5 mL of methanol:chloroform (1:1) and once with 1.5 mL of water. To dispose starch, the pellets were digested three times overnight with 1.5 mL of amylose-amyloglucosidase solution as described above (Geigenberger et al., 1998) and washed twice again with 1.5 mL of water. The pellet was then mixed with 50 μg of inositol and digested with 250 μL of 2 m trifluoracetic acid for 1 h at 121°C. 2-Propanol (300 μL) was added to the solution, and the mixture was evaporated at 40°C with a N2 stream. Evaporation was repeated three times in total. The pellet was extracted with 400 μL of water using a sonificator. The pellet was dried and used for the determination of crystalline cellulose. Fifty microliters of the watery extract was used for the determination of uronic acids. The remaining 350 μL was dried and used for the determination of matrix sugar composition according to Englyst and Cummings (1984). Analysis was done by GC-MS on a SP-2380 fused silica capillary column (30 m × 0.25 mm, 0.2 μm film thickness; Supelco) in an Agilent 6890 N gas chromatograph coupled to an Agilent 5973 mass selective detector (Agilent Technologies). The temperature program started at 160°C for 2 min, then increased to 200°C at 20°C min−1, was held for 5 min, then increased to 245°C at 20°C min−1, then was held for 12 min. The system was calibrated for Glc, Gal, Ara, Xyl, Rha, Fuc, Man, and inositol.
Determination of Uronic Acids
Uronic acids were determined in a 50-μL aliquot separated after trifluoroacetic acid hydrolysis as described above using the biphenyl assay as described by Filisetti-Cozzi and Carpita (1991) with GalUA as a standard.
Determination of Crystalline Cellulose
The preparation and hydrolysis of crystalline cellulose from the pellet left after trifluoroacetic acid hydrolysis as described above was done as described previously (Updegraff, 1969). The cellulose-derived Glc was determined using the anthrone assay (Dische, 1962).
Enzyme Analysis
Enzyme Extraction
Enzymes were extracted as described previously by Geigenberger and Stitt (1993) with Pefabloc and polyvinylpolypyrrolidone (PVPP) instead of phenylmethylsulfonyl fluoride in the extraction buffer and omitting bovine serum albumin. Enzymes were extracted from 30 to 100 mg of frozen homogenized tissue using 1,500 μL of 50 mm HEPES/KOH, pH 7.4, containing 5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 0.1% Triton X-100, 10% glycerol, 2 mm benzamidine, 2 mm γ-aminocaproic acid, 0.5 mm Pefabloc, 5 mm dithiothreitol, and 0.1% PVPP. Extracts were kept at 4°C prior to measurement.
Preparation of Apoplastic Washings
Thirty-two leaves of a potato plant not older than 6 weeks were vacuum infiltrated with 20 mm MES, pH 6.0, using a 60-mL syringe (Braun). The infiltrated leaves were quickly dried with tissue and rolled into an Eppendorf 5-mL pipet tip, which was plugged with a 1.5-mL Eppendorf cup containing 2 μL of 20 mm benzamidine, 20 mm γ-aminocaproic acid, 5 mm Pefabloc, 50 mm dithiothreitol, and 0.1% PVPP in MES buffer. The washings were then collected by centrifugation at 250g for 4 min. One microliter of each washing was tested for cytosolic UGPase activity to check for cytosolic contamination, as suggested by Farran et al. (2002). Only washings with reasonably low UGPase activity (<2.5% of overall leaf UGPase activity) were pooled for further analysis.
Apyrase/5′-Nucleotidase in Soluble Extracts or AWF
Five microliters of extract or AWF was assayed as described previously (Riewe et al., 2008), using 100 mm MES/KOH, pH 6.0, and 5 mm CaCl2 instead of HEPES/EDTA/EGTA buffer. Enzymatic reaction was started by adding the substrate (ATP/UTP/ADP/UDP/AMP/UMP) to a final concentration of 1 mm. The release of 1 μmol of Pi per minute was defined as 1 unit.
Apyrase/5′-Nucleotidase in Intact Tuber Slices
Five tuber slices were washed five times with buffer containing 20 mm MES/KOH, pH 6.0, and 1 mm CaCl2 and incubated in 3 mL of this buffer. The reaction was started by adding the substrate to a final concentration of 1 mm. After certain time points, aliquots of 80 μL were taken to determine Pi liberation as described for the soluble extracts.
UGPase in Soluble Extracts or AWF
UGPase was assayed in the direction of Glc-1-P as described by Zrenner et al. (1993). For intact tuber slices, five slices (8 mm diameter and 2 mm thick) were washed three times with 20 mm MES/KOH, pH 6.0, and 1 mm CaCl2 twice with 100 mm Tris-HCl containing 2 mm MgCl2. Then, the slices were incubated with UGPase assay mix and the reaction was started with PPi as described above. At several time points, aliquots of 200 μL were subtracted and NADPH production was immediately measured in a photometer. There was no production of NADPH detectable in the absence of PPi.
Analysis of Respiration Rates
Respiration rates of two freshly prepared tuber discs (8 mm diameter and 2 mm thick) were analyzed in an oxygen electrode (Hansatech) filled with 1 mL of 20 mm MES/KOH, pH 6.0, as described previously (Loef et al., 1999).
Sequence data from this article can be found in the GenBank data library under accession numbers AF535135 (StAPY2 mRNA), EU125183 (StAPY3 mRNA), and EU125182 (StAPY2 genomic DNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of potato-specific apyrases cloned to date.
Supplemental Figure S2. Plasmids used for manipulation and localization of apyrase.
Supplemental Figure S3. Potato apyrase transcript and protein abundance.
Supplemental Figure S4. Effect of a decrease in apyrase activity under control of the 35S promoter on overall shoot growth.
Supplemental Figure S5. Cell wall composition of tubers from the B33-RNAi lines.
Supplemental Figure S6. Overall adenylate levels in tubers with decreased apyrase under the control of the tuber-specific B33 promoter.
Supplemental Figure S7. Respiration rates of tuber slices of the B33-RNAi lines.
Supplemental Figure S8. Localization of a temporally expressed plasma membrane-localized GFP fusion protein.
Supplemental Table S1. Sequence identity of potato-specific and general plant apyrases.
Supplemental Table S2. Metabolite profile of tubers from the B33-RNAi lines.
Supplemental Table S3. Transcript profile dataset of tubers from the B33-RNAi lines B33:10 and B33:25.
Supplementary Material
Acknowledgments
We thank Ewa Urbanczyk-Wochniak (Plant Biology Division, The Samuel Roberts Noble Foundation, Ardmore, OK) and Matthew Hannah (Max-Planck Institute of Molecular Plant Physiology) for their support with the microarray analysis. We are grateful to Ulrike Haensel (Bayer CropScience, Frankfurt, Germany) and Peter Immerzeel (Umea Plant Science Center, Umea, Sweden) for their support with the cell wall analysis, to Axel Tiessen (CINVESTAV, Irapuato, Mexico) for providing nonaqueous fractions and marker activities, to Pal Nyren (Royal Institute of Technology, Stockholm, Sweden) for providing the anti-apyrase serum, and to Jens Kossman (Stellenbosch, South Africa) and Jeremy Clark (Max-Planck Institute of Molecular Plant Physiology) for advice in the beginning of the experiments.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Ge 878/1–5).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Peter Geigenberger (geigenberger@mpimp-golm.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Ringli C, Keller B (2001) The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 15 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57 289–300 [Google Scholar]
- Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, Esquerre-Tugaye MT, Pont-Lezica R (2005) Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: identification by mass spectrometry and bioinformatics. Proteomics 5 212–221 [DOI] [PubMed] [Google Scholar]
- Che M, Nishida T, Gatmaitan Z, Arias IM (1992) A nucleoside transporter is functionally linked to ectonucleotidases in rat liver canalicular membrane. J Biol Chem 267 9684–9688 [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Lindsey K, Slabas AR (2005) Extracellular ATP functions as an endogenous external metabolite regulating plant cell viability. Plant Cell 17 3019–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn JR, Uhm T, Ramu S, Nam YW, Kim DJ, Penmetsa RV, Wood TC, Denny RL, Young ND, Cook DR, et al (2001) Differential regulation of a family of apyrase genes from Medicago truncatula. Plant Physiol 125 2104–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RB, McAlvin CB, Loh JT, Denny RL, Wood TC, Young ND, Stacey G (2000) Differential expression of two soybean apyrases, one of which is an early nodulin. Mol Plant Microbe Interact 13 1053–1070 [DOI] [PubMed] [Google Scholar]
- Degenkolbe T, Hannah MA, Freund S, Hincha DK, Heyer AG, Kohl KI (2005) A quality-controlled microarray method for gene expression profiling. Anal Biochem 346 217–224 [DOI] [PubMed] [Google Scholar]
- Dische Z (1962) General color reactions. Methods Carbohydr Chem 1 478–492 [Google Scholar]
- Dunkley TP, Watson R, Griffin JL, Dupree P, Lilley KS (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol Cell Proteomics 3 1128–1134 [DOI] [PubMed] [Google Scholar]
- Englyst HN, Cummings JH (1984) Simplified method for the measurement of total non-starch polysaccharides by gas-liquid chromatography of constituent sugars as alditol acetates. Analyst 109 937–942 [DOI] [PubMed] [Google Scholar]
- Etzler ME, Kalsi G, Ewing NN, Roberts NJ, Day RB, Murphy JB (1999) A nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA 96 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanta N, Anich M, Mancilla M, Kettlum AM, Valenzuela MA, Traverso-Cori A (1988) Starch, adenine nucleotides and apyrase changes during potato tuber development. Arch Biol Med Exp (Santiago) 21 129–133 [Google Scholar]
- Farran I, Sanchez-Serrano JJ, Medina JF, Prieto J, Mingo-Castel AM (2002) Targeted expression of human serum albumin to potato tubers. Transgenic Res 11 337–346 [DOI] [PubMed] [Google Scholar]
- Farre EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L (2001) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiol 127 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecht-Christoffers MM, Fuhrs H, Braun HP, Horst WJ (2006) The role of hydrogen peroxide-producing and hydrogen peroxide-consuming peroxidases in the leaf apoplast of cowpea in manganese tolerance. Plant Physiol 140 1451–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettke J, Poeste S, Eckermann N, Tiessen A, Pauly M, Geigenberger P, Steup M (2005) Analysis of cytosolic heteroglycans from leaves of transgenic potato (Solanum tuberosum L.) plants that under- or overexpress the Pho 2 phosphorylase isozyme. Plant Cell Physiol 46 1987–2004 [DOI] [PubMed] [Google Scholar]
- Filisetti-Cozzi TM, Carpita NC (1991) Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197 157–162 [DOI] [PubMed] [Google Scholar]
- Geigenberger P (2003) Regulation of sucrose to starch conversion in growing potato tubers. J Exp Bot 54 457–465 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR (2006) Starch biosynthesis in the potato tuber. In Food Biochemistry and Food Processing. Blackwell Publishing, Oxford, pp 253–270
- Geigenberger P, Hajirezaei M, Geiger M, Deiting U, Sonnewald U, Stitt M (1998) Overexpression of pyrophosphatase leads to increased sucrose degradation and starch synthesis, increased activities of enzymes for sucrose-starch interconversions, and increased levels of nucleotides in growing potato tubers. Planta 205 428–437 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M (1993) Sucrose synthase catalyzes a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta 189 329–339 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M, Fernie AR (2004) Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ 27 655–673 [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hall Q, Cannon MC (2002) The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa M, Guidotti G (1996) Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum). Biochem Biophys Res Commun 218 916–923 [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Song CJ, Roux SJ (2000) Regulation of a recombinant pea nuclear apyrase by calmodulin and casein kinase II. Biochim Biophys Acta 1494 248–255 [DOI] [PubMed] [Google Scholar]
- Jelitto T, Sonnewald U, Willmitzer L, Hajirezeai M, Stitt M (1992) Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing Escherichia coli pyrophosphatase in their cytosol. Planta 188 238–244 [DOI] [PubMed] [Google Scholar]
- Kalckar HM (1944) Adenylpyrophosphatase and myokinase. J Biol Chem 153 355–367 [Google Scholar]
- Kalsi G, Etzler ME (2000) Localization of a Nod factor-binding protein in legume roots and factors influencing its distribution and expression. Plant Physiol 124 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettlun AM, Uribe L, Calvo V, Silva S, Rivera J, Mancilla M, Valenzuela MA, Traversocori A (1982) Properties of 2 apyrases from Solanum tuberosum. Phytochemistry 21 551–558 [Google Scholar]
- Kim SY, Sivaguru M, Stacey G (2006) Extracellular ATP in plants: visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol 142 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P (2005) Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 102 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XJ, Prat S, Willmitzer L, Frommer WB (1990) Cis regulatory elements directing tuber-specific and sucrose-inducible expression of a chimeric class-I patatin promoter GUS-gene fusion. Mol Gen Genet 223 401–406 [DOI] [PubMed] [Google Scholar]
- Loef I, Stitt M, Geigenberger P (1999) Orotate leads to a specific increase in uridine nucleotide levels and a stimulation of sucrose degradation and starch synthesis in discs from growing potato tubers. Planta 209 314–323 [DOI] [PubMed] [Google Scholar]
- Marcus AJ, Broekman MJ, Drosopoulos JH, Islam N, Alyonycheva TN, Safier LB, Hajjar KA, Posnett DN, Schoenborn MA, Schooley KA, et al (1997) The endothelial cell ecto-ADPase responsible for inhibition of platelet function is CD39. J Clin Invest 99 1351–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattanovich D, Ruker F, Machado AD, Laimer M, Regner F, Steinkellner H, Himmler G, Katinger H (1989) Efficient transformation of Agrobacterium spp by electroporation. Nucleic Acids Res 17 6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlvin CB, Stacey G (2005) Transgenic expression of the soybean apyrase in Lotus japonicus enhances nodulation. Plant Physiol 137 1456–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkouropoulos G, Shirsat AH (2003) The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217 356–366 [DOI] [PubMed] [Google Scholar]
- Molnar J, Lorand L (1961) Studies on apyrases. Arch Biochem Biophys 93 353–363 [DOI] [PubMed] [Google Scholar]
- Neckelmann G, Orellana A (1998) Metabolism of uridine 5′-diphosphate-glucose in Golgi vesicles from pea stems. Plant Physiol 117 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Tiessen A, Fernie AR, Geigenberger P (2008) Decreased expression of plastidial adenylate kinase in potato tubers results in an enhanced rate of respiration and a stimulation of starch synthesis that is attributable to post-translational redox-activation of ADP-glucose pyrophosphorylase. J Exp Bot 59 315–325 [DOI] [PubMed] [Google Scholar]
- Orellana A, Neckelmann G, Norambuena L (1997) Topography and function of Golgi uridine-5′-diphosphatase from pea stems. Plant Physiol 114 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesner L (1995) Ecto-ATPases: identities and functions. Int Rev Cytol 158 141–214 [DOI] [PubMed] [Google Scholar]
- Riewe D, Grosman L, Zauber H, Wucke C, Fernie AR, Geigenberger P (2008) Metabolic and developmental adaptations of growing potato tubers in response to specific manipulations of the adenylate energy status. Plant Physiol 146 1579–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K, Shirsat AH (2006) Increased extensin levels in Arabidopsis affect inflorescence stem thickening and height. J Exp Bot 57 537–545 [DOI] [PubMed] [Google Scholar]
- Roberts NJ, Brigham J, Wu B, Murphy JB, Volpin H, Phillips DA, Etzler ME (1999) A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes. Mol Gen Genet 262 261–267 [DOI] [PubMed] [Google Scholar]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L (1989) Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J 8 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter A, Usadel B, Baebler S, Stitt M, Gruden K (2007) Adaptation of the MapMan ontology to biotic stress responses: application in solanaceous species. Plant Methods 3 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumeau D, Maher EA, Kelman A, Showalter AM (1990) Extensin and phenylalanine ammonia-lyase gene expression altered in potato tubers in response to wounding, hypoxia, and Erwinia carotovora infection. Plant Physiol 93 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato AJ, Perez-Ruiz JM, Spinola MC, Cejudo FJ (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279 43821–43827 [DOI] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21 2067–2075 [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Wood CC, Roeb GW, Udvardi MK (2002) Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol 130 1788–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CJ, Steinebrunner I, Wang X, Stout SC, Roux SJ (2006) Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol 140 1222–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebrunner I, Jeter C, Song C, Roux SJ (2000) Molecular and biochemical comparison of two different apyrases from Arabidopsis thaliana. Plant Physiol Biochem 38 913–922 [Google Scholar]
- Steinebrunner I, Wu J, Sun Y, Corbett A, Roux SJ (2003) Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol 131 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Muller-Rober B, Willmitzer L, Hill SA (1999) The contribution of adenosine 5′-diphosphoglucose pyrophosphorylase to the control of starch synthesis in potato tubers. Planta 209 330–337 [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Thomas C, Rajagopal A, Windsor B, Dudler R, Lloyd A, Roux SJ (2000) A role for ectophosphatase in xenobiotic resistance. Plant Cell 12 519–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Sun Y, Naus K, Lloyd A, Roux S (1999) Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol 119 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farre EM, Geigenberger P (2002) Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden J, Mohlmann T, Kampfenkel K, Henrichs G, Neuhaus HE (1998) Altered plastidic ATP/ADP-transporter activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. Plant J 16 531–540 [Google Scholar]
- Todorov LD, Mihaylova-Todorova S, Westfall TD, Sneddon P, Kennedy C, Bjur RA, Westfall DP (1997) Neuronal release of soluble nucleotidases and their role in neurotransmitter inactivation. Nature 387 76–79 [DOI] [PubMed] [Google Scholar]
- Updegraff DM (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32 420–424 [DOI] [PubMed] [Google Scholar]
- Voelker C, Schmidt D, Mueller-Roeber B, Czempinski K (2006) Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J 48 296–306 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–590 [DOI] [PubMed] [Google Scholar]
- Windsor B, Roux SJ, Lloyd A (2003) Multiherbicide tolerance conferred by AtPgp1 and apyrase overexpression in Arabidopsis thaliana. Nat Biotechnol 21 428–433 [DOI] [PubMed] [Google Scholar]
- Wolf C, Hennig M, Romanovicz D, Steinebrunner I (2007) Developmental defects and seedling lethality in apyrase AtAPY1 and AtAPY2 double knockout mutants. Plant Mol Biol 64 657–672 [DOI] [PubMed] [Google Scholar]
- Wu H, de Graaf B, Mariani C, Cheung AY (2001) Hydroxyproline-rich glycoproteins in plant reproductive tissues: structure, functions and regulation. Cell Mol Life Sci 58 1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Steinebrunner I, Sun Y, Butterfield T, Torres J, Arnold D, Gonzalez A, Jacob F, Reichler S, Roux SJ (2007) Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiol 144 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Greiten C, Schmitz I, Haferkamp I, Neuhaus HE, Hausler RE, Flugge UI, Ludewig F (2006) Impact of glucose 6-phosphate import into amyloplasts on potato tuber starch content. In DP Bourque, ed, 8th International Congress of Plant Molecular Biology, Adelaide, Australia. International Society for Plant Molecular Biology, Athens, GA, p 93
- Zrenner R, Willmitzer L, Sonnewald U (1993) Analysis of the expression of potato uridinediphosphate-glucose pyrophosphorylase and its inhibition by antisense RNA. Planta 190 247–252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.