Abstract
The correlation between telomerase activity and human tumors has led to the hypothesis that tumor growth requires reactivation of telomerase and that telomerase inhibitors represent a class of chemotherapeutic agents. Herein, we examine the effects of inhibition of telomerase inside human cells. Peptide nucleic acid and 2′-O-MeRNA oligomers inhibit telomerase, leading to progressive telomere shortening and causing immortal human breast epithelial cells to undergo apoptosis with increasing frequency until no cells remain. Telomere shortening is reversible: if inhibitor addition is terminated, telomeres regain their initial lengths. Our results validate telomerase as a target for the discovery of anticancer drugs and supply general insights into the properties that successful agents will require regardless of chemical type. Chemically similar oligonucleotides are in clinical trials and have well characterized pharmacokinetics, making the inhibitors we describe practical lead compounds for testing for an antitelomerase chemotherapeutic strategy.
Human telomerase is a ribonucleoprotein that adds repeated units of TTAGGG to the ends of telomeres (1, 2). Telomerase activity has been found in almost all human tumors but not in adjacent normal cells (3, 4). This correlation has led to the hypotheses that reactivation of telomerase is necessary for sustained cell proliferation in many tumors and that telomerase is an exceptional target for a class of chemotherapeutic agents that act by an unidentified mechanism (5). Supporting these hypotheses is the observation that early-stage neuroblastomas have little or no telomerase activity, and this minimal activity generally correlates with a favorable outcome, whereas the late-stage disease has high telomerase activity and correlates with a poor outcome (6). A similar linkage between telomerase activity and poor clinical outcome has been reported for ordinary meningiomas (7), and other studies have suggested that telomerase activity is correlated with the pathologic stage (8–11) or tumor aggressiveness (11, 12).
Additional evidence for the importance of telomerase activity for sustained cell growth comes from studies of mice that lack the mouse telomerase RNA component (mTR). Depending on their genetic backgrounds, these mice survived for four to six generations with few detectable phenotypic changes (13, 14); however, by the seventh generation, highly proliferative organ systems like the testis, bone marrow, and spleen appeared abnormal, and the mice were no longer able to reproduce (15). In some genetic backgrounds, an increased incidence of neural tube defects limited viability after only four generations (14). Other studies of these mice revealed shortened life spans, an increased incidence of chromosomal abnormalities, and a slight increase in spontaneous malignancies (16). The long lag between loss of telomerase activity and a detrimental phenotype suggests that telomerase will be a difficult target for effective chemotherapy, but the mice used in these experiments possessed much longer telomeres than those found in cancerous human cells, and inhibition of telomerase in human cancer would be expected to produce effects more rapidly. Furthermore, recent reports of the effect of the mTR deletion in different genetic backgrounds (17, 18) suggest that the role of telomerase in cancer development could be highly context dependent (19).
Mutation of the RNA component of telomerase of Tetrahymena (20), Kluyveromyces lactis (21), Saccharomyces cerevisiae (22), and human cells (23) also leads to decreased cell proliferation, whereas expression of antisense RNA complementary to the human telomerase RNA component (hTR) caused decreased proliferation of HeLa cells after 23 to 26 doublings (24). Conversely, transfection of cells with the gene encoding the human telomerase reverse transcriptase component (hTERT) and subsequent expression of active telomerase have been shown to extend the life spans of normal human fibroblasts and epithelial cells (25–27). Most recently, expression of telomerase in conjunction with expression of simian virus 40 large T oncoprotein and an oncogenic allele of H-ras has been shown to promote tumorigenic conversion of normal human cells (28). Thus, the lack of telomerase expression seems to curb growth of rapidly proliferating cells eventually, whereas an increase in telomerase permits indefinite proliferation.
A potential concern for the telomerase–cancer connection is the observation that a few rare tumors (29) and some experimentally immortalized cell lines (30) lack detectable telomerase activity. Thus, although telomerase activity can confer extended life span to cells, other mechanisms may also exist. A pathway termed ALT (alternative lengthening of telomeres) has been proposed to account for this phenomenon. Evidence for the existence of this pathway is found in yeast in which the gene for est1, a protein involved in telomere-length maintenance, has been deleted (31). The cells divide normally until telomeres shorten sufficiently to affect proliferation. At this stage, most cells die, but some rare survivors continue to proliferate through a recombination mechanism to maintain telomere length.
Telomerase will be an unusually challenging target for drug development because of the long lag period expected before telomeres would shorten sufficiently to produce detrimental effects on cell growth and because of the possibility of alternate mechanisms for maintenance of telomeres. Because of this uncertainty, discovery of the potential of telomerase as a target for human therapy requires development of potent and selective synthetic inhibitors and their testing inside cells. To confirm action through a telomerase-dependent mechanism, inhibitors must meet the following criteria: (i) inhibitors should reduce telomerase activity but, initially, should not affect cell growth rates; (ii) addition of inhibitors should lead to progressive shortening of telomeres with each cell division; (iii) addition of inhibitors should cause cells to die or undergo growth arrest; (iv) the time necessary to observe decreased proliferation should vary depending on initial telomere length; and (v) chemically related molecules that do not inhibit telomerase activity should not cause decreased cell proliferation or telomere shortening.
Herein, we examine how inhibition of telomerase by exogenously added molecules affects cellular phenotypes. We observe that 2′-O-MeRNA and peptide nucleic acid (PNA) oligomers complementary to the template region of hTR inhibit telomerase and cause telomeres to shorten and that extended treatment with 2′-O-MeRNA promotes cell death through apoptosis. Inhibition of cell proliferation suggests that oligonucleotides are a viable approach to antitelomerase therapy, and our results should encourage and guide further testing of other approaches to telomerase inhibition such as nucleotide analogues (32), G quadruplex interactive agents (33, 34), or other small molecules.
Materials and Methods
Cell Lines.
The human mammary epithelial (HME) cells used for these experiments were spontaneously immortalized from an epithelial culture derived from normal breast tissue from a patient with Li-Fraumeni syndrome (35). Cells were grown in serum-free medium (MCDB from GIBCO) supplemented with 0.4% bovine pituitary extract (Hammond Cell Tech, Alameda, CA), 10 ng/ml epidermal growth factor (GIBCO), 5 μg/ml insulin (Sigma), 0.5 μg/ml hydrocortisone (Sigma), 5 μg/ml transferrin, and 50 μg/ml gentamicin (Sigma). The medium was changed every 2–3 days. Cells were used between population doublings 100 and 150. The prostate-tumor-derived DU145 cells were maintained in DMEM containing 10% (vol/vol) FCS, 500 units/ml penicillin (Sigma), and 0.1 μg/ml streptomycin (Sigma) and incubated at 5% CO2 at 37°C. Detection and quantitation of apoptosis by flow cytometry were performed with the ApoAlert Annexin V Apoptosis kit (CLONTECH).
Oligonucleotides and PNAs.
We purchased 2′-O-MeRNA oligonucleotides from Oligos Etc. and Oligo Therapeutics (Wilsonville, OR). The sequence of the match phosphorothioate modified 2′-O-MeRNA is 5′-CAGUUAGGGUUAG-3′; the mismatch sequence is 5′-CAGUUAGAAUUAG-3′, where the underlined nucleotides possess phosphorothioate linkages. Match and mismatch PNAs (36) of similar sequences and nontemplate directed PNA AGCGGGCCAGCAGCTG were synthesized automatically with a PerSeptive Biosystems (Framingham, MA) Expedite 8909 Synthesizer by using Fmoc protocols and reagents obtained from PE Biosystems. PNAs were purified by HPLC and characterized by matrix-assisted laser desorption time-of-flight mass spectrometry by using a Voyager-DE mass spectrometry workstation (PE Biosystems) as described (37). The sequence of the match PNA is Gly-CAGTTAGGGTTAG-Lys; the sequence of the mismatch is Gly-CAGTTAGAATTAG-Lys. DNA oligonucleotides for transfection of PNA/DNA complexes were obtained from Life Technologies (Gaithersburg, MD). The sequence of the DNA oligonucleotide complexed to the match PNA is (5′–3′) TCTAACCCTAA; the sequence of the DNA oligonucleotide complexed to the mismatch PNA is (5′–3′) TCTAATTCTAA.
Uptake of 2′-O-MeRNA and PNA into Cells.
HME50-5E and HME50-hTERT cells were transfected with 2′-O-MeRNA and mismatch control oligomers after the FuGENE6 Transfection Reagent protocol (Roche Molecular Biochemicals). Briefly, 5 × 104 cells were allowed to adhere overnight in appropriate media. The next day, the oligomers were incubated for 15 min with the previously combined FuGENE6 Transfection Reagent and PBS. After changing to fresh medium, the oligomer mixture was added dropwise to the cells. Cells were harvested after 4 days, counted, and replated, and telomerase activity was determined.
DU145 cells were plated at 25,000 cells per well in a 24-well plate in DMEM supplemented with 10% (vol/vol) FCS, 500 units/ml penicillin, and 0.1 μg/ml streptomycin. For DNA/PNA transfections 100 μM PNA was hybridized with 109 μM appropriate DNA oligonucleotide in 0.5× PBS. After allowing the cells to adhere, they were transfected with 2.0 μl (7 μg/ml) of Lipofectamine (Life Technologies) and 0.5 μM 2′-O-MeRNA oligonucleotide (38) or 1 μM PNA/DNA complex (39) in 200 μl of total Opti-Mem (Life Technologies) according to the manufacturer’s directions. After 6 h at 37°C, the transfecting mixture was removed, and medium without antibiotics and with 20% (vol/vol) serum was added. Cells were then harvested 12–15 h later after three washes with PBS and treatment with trypsin or allowed to grow for 3 days. Cells were harvested after 3 days, counted, replated, and assayed for telomerase activity.
Measurement of Telomerase Activity and Telomere Length.
Telomerase activity was measured by telomere repeat amplification protocol by using the TRAPeze telomerase detection kit (Intergen, Purchase NY; ref. 40). After the extension of the substrate by telomerase, the products were amplified by PCR in the presence of an 32P-end-labeled TS primer, resolved on 10% polyacrylamide gels, and revealed by exposure to a PhosphorImager cassette (Molecular Dynamics). Telomerase activity was calculated as the ratio of the intensity of telomerase ladders to the intensity of the 36-bp internal standard. Percentage of inhibition was calculated by comparing telomerase activity of oligomer-treated cells with telomerase activity of cells treated with lipid alone. The levels of telomerase activity were within the linear range of the TRAP assay (40). Mean telomere length was evaluated by using telomere restriction fragment analysis, a variation of standard Southern analysis, and was quantitated as described (41). Digested samples were resolved on a 0.7% agarose gel and hybridized to a telomeric probe [[32P](TTAGGG)4 oligonucleotide].
Results
Effect of 2′-O-MeRNA on Cell Proliferation.
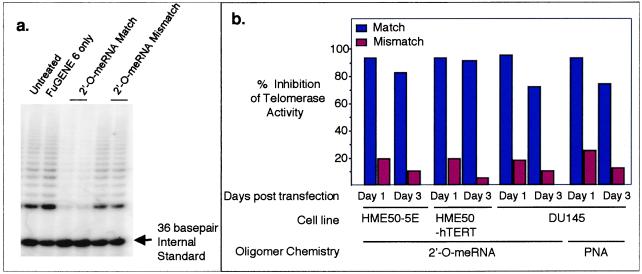
We used cationic lipids to introduce 2′-O-MeRNA complementary to the template region of hTR into the immortalized human cell lines HME50-5E and DU145. We also introduced 2′-O-MeRNA into HME50-hTERT cells, a cell line generated by infecting preimmortal HME50 cells with the gene encoding hTERT. A 2′-O-MeRNA containing mismatched bases was employed as a control for the sequence-specificity of inhibitor action. Analysis of telomerase inhibition by TRAP indicated that the complementary 2′-O-MeRNA oligomer blocked more than 95% of telomerase activity 1 day after transfection, and more than 70% of activity was inhibited 3 days after transfection (Fig. 1). The 2′-O-MeRNA oligomers used in these studies are stable to degradation inside cells (38); thus, it is likely that the reduced inhibition is due to dilution of inhibitor as the cell population increases.
Figure 1.
(a) Inhibition of telomerase activity by 2′-O-MeRNA oligomers delivered into HME50-5E cells by using FuGENE6 lipid, measured 3 days after transfection. Similar results were observed on introduction of 2′-O-MeRNA oligomers into HME50-hTERT cells and on introduction of 2′-O-MeRNA or PNA oligomers into DU145 cells. (b) Inhibition of telomerase activity in HME50-5E and DU145 cells by 2′-O-MeRNA and PNA oligomers as detected by the TRAP assay. HME50-5E and DU145 cells were collected 1 and 3 days after transfection with 2′-O-MeRNA or PNA oligomers. Telomerase activity was quantitated as described (38).
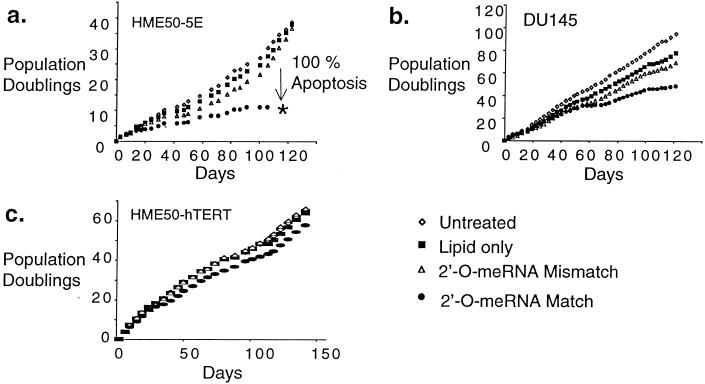
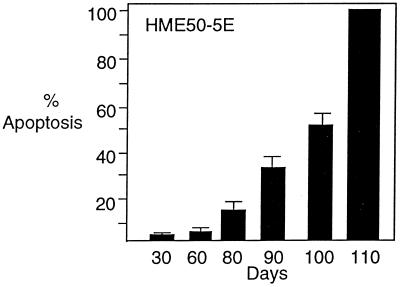
To determine whether inhibition of telomerase would ultimately limit proliferation, we transfected HME50-5E, DU145, and HME50-hTERT cells with 2′-O-MeRNA oligomers at 3- to 4-day intervals for 120 days. Because of the extreme length of these experiments, stringent precautions were taken to avoid contamination of cultured cells, and all experiments were completed successfully without interruption. After 15–25 days, proliferation of HME50-5E cells treated with the complementary oligomer began to slow, and after 110 days, no treated cells remained (Fig. 2a). The growth of cells treated with the control oligomer containing mismatch bases was not affected. During the first 60 days after initial transfection, less than 5% of cells treated with the fully complementary 2′-O-MeRNA underwent apoptosis. By 80 days, however, 14% of cells treated with the match oligomer were observed to undergo apoptosis, and this percentage increased to 56% after 100 days with cell viability lost after 110 days (Fig. 3). By contrast, cells treated with lipid alone or the 2′-O-MeRNA oligomer containing mismatched bases underwent apoptosis at a rate of 2–3% throughout the entire experiment. To confirm that the effects of inhibitor addition were reproducible, treatment of HME50-5E cells with 2′-O-MeRNA was repeated, and a similar decrease in proliferation was observed.
Figure 2.
Effects on cell growth after long-term transfection with match or mismatch 2′-O-MeRNA. (a) HME50-5E cells transfected with 2′-O-MeRNA oligomers. (b) DU145 cells transfected with 2′-O-MeRNA oligomers. (c) HME50-hTERT cells transfected with 2′-O-MeRNA oligomers.
Figure 3.
Increase of apoptosis of HME50-5E cells after repeated transfection with fully complementary 2′-O-MeRNA. Levels of apoptosis were measured by staining with 4′6-diamidino-2-phenylindole followed by microscopy. Background apoptosis of cells that were untreated or that were treated with 2′-O-MeRNA containing mismatched bases was 2–3%. Levels of apoptosis were confirmed by flow cytometry with the ApoAlert Annexin V Apoptosis kit (CLONTECH).
Addition of complementary 2′-O-MeRNA to DU145 cells caused proliferation of cells to begin to slow after 60 days (Fig. 2b). By the end of the experiment (after 120 days), these cells had undergone 20 fewer population doublings than had cells treated with mismatch-containing oligomer. HME50-5E cells have a mean terminal restriction fragment (TRF) length of 2,000 bp, whereas DU145 cells possess a mean length of 3,600 bp. TRF length corresponds qualitatively to telomere length, and the finding that proliferation of treated HME50-5E cells decreases more dramatically than does cell proliferation of treated DU145 cells may be due to HME50-5E cells possessing shorter telomeres. Consistent with this hypothesis, HME50-hTERT cells have a much longer mean TRF length (7.6 kilobases) than either HME50-5E or DU145 cells, and we observed no significant reduction in their growth rates during the 120-day treatment period (Fig. 2c).
Telomere Shortening On Addition of 2′-O-MeRNA.
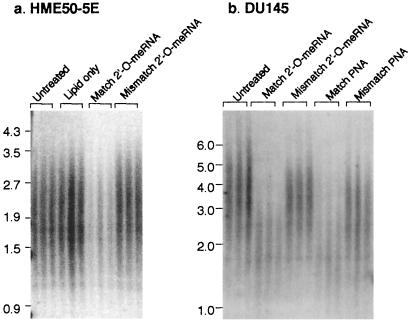
To support the hypothesis that the decrease in cell proliferation is caused by the 2′-O-MeRNA oligomer through a telomerase-dependent mechanism rather than being caused by long-term toxicity that is independent of telomerase inhibition, we monitored the effect of inhibition on telomere length. Within 60 days of treatment, the mean telomere length of HME50-5E cells treated with 2′-O-MeRNA complementary to hTR was reduced from 2,000 to 1,700 bp (Fig. 4a). This decrease in measured TRF length may be an underestimate of the total loss of chromosomal DNA, because the TRF assay does not monitor erosion of subtelomeric regions and because relatively little telomeric DNA remained to hybridize with radiolabeled probe. The TRF length of cells treated with 2-O-MeRNA containing mismatches relative to hTR remained at 2,000 bp, and the signal intensity of probe hybridization was undiminished.
Figure 4.
Measurement of telomere restriction fragment length (TRF) in HME50-5E and DU145 cells treated with 2′-O-MeRNA and PNA oligomers. (a) HME50-5E cells that had been treated with 2′-O-MeRNA oligomers for 60 days, with the results of independent experiments shown in triplicate. (b) DU145 cells treated with 2′-O-MeRNA and PNA oligomers for 76 days, with the results of independent experiments shown in triplicate. In parts a and b, the signal intensity in the lanes showing the outcome of treatment with fully complementary oligomer is weak, because telomeres have eroded and few telomeric repeats remain to hybridize with radiolabeled probe. Equivalent amounts of chromosomal DNA were loaded in each lane. TRF lengths are expressed as kilobase pairs.
We also examined the mean TRF length of DU145 cells that had been treated with 2′-O-MeRNA oligomers for 76 days. Because DU145 cells possess longer telomeres, we were able to observe a dramatic erosion of telomere length, with the mean TRF length of cells treated with the 2′-O-MeRNA complementary to the template region of hTR decreasing from 3,600 bp to 2,200 bp (Fig. 4b). As observed with HME50-5E cells, the signal intensity of probe hybridization was greatly reduced because of the reduction in telomeric repeats. We also examined the effect of inhibitor treatment on HME50-hTERT cells and found that TRF length decreased from 7,600 to 6,800 bp after 75 days (results not shown).
The 2′-O-MeRNA inhibitor contained four phosphorothioate linkages, a chemical moiety noted for its propensity to produce misleading cellular effects that seem sequence-specific but are actually unrelated to binding to the intended target (42). Demonstrating that decreased cell proliferation derives from a telomerase-dependent mechanism involving binding of inhibitor to the RNA template is a critical prerequisite for embarking on lengthy studies in animals and humans. Therefore, to provide further evidence that the reduction of telomere length was due to Watson–Crick recognition of the template region of hTR, we tested the effects of addition of match and mismatch PNAs (36).
PNAs are ideal agents for confirming the specificity of oligonucleotides, because they possess a neutral amide backbone and have a much lower propensity for making unwanted interactions with macromolecules that bind the repetitive anionic linkages of DNA and RNA. PNAs cannot be introduced into cells by direct complexation with cationic lipid, because PNA backbone linkages are uncharged; however, PNAs can be hybridized to DNA oligonucleotides and introduced into cells as cargo on complexation of DNA with lipid (39). Addition of a PNA that was complementary to the hTR template to DU145 cells inhibited over 90% of telomerase activity after 1 day (Fig. 1b) and caused telomere shortening similar to that caused by addition of complementary 2′-O-MeRNA (Fig. 4b). A PNA directed to a nontemplate region of hTR inhibited only 50% of telomerase activity after 1 day and did not cause telomeres to shorten (results not shown).
Effects of Terminating Inhibitor Addition.
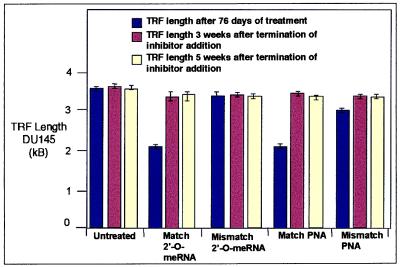
An inherent advantage of using synthetic inhibitors to reduce gene expression rather than genetic knockouts is that the phenotypic effects of regaining function can be evaluated by terminating inhibitor addition. After 76 days, we stopped adding inhibitor to DU145 cells that had been treated with fully complementary or mismatch-containing 2′-O-MeRNA or PNA oligomers. After 3 weeks, we noted that the eroded telomeres of cells treated with fully complementary oligomers had returned to approximately their initial lengths (Fig. 5). Telomeres remained near their initial lengths when measured a second time at 5 weeks. We also evaluated the effect of inhibitor withdrawal on proliferation of HME50-5E cells that had been treated with the fully complementary 2′-O-MeRNA until population growth was static. We observed that, within 3 weeks of terminating inhibitor treatment, previously static HME50-5E cells regained the ability to grow at the same rate as cells treated with mismatch-containing 2′-O-MeRNA.
Figure 5.
The TRF length of DU145 as a function of inhibitor addition. TRF lengths were measured after 76 days of addition of fully complementary or mismatch-containing 2′-O-MeRNA or PNA oligomers. TRF lengths were then measured again 3 or 5 weeks after terminating oligomer addition. kB, kilobase.
Discussion
Telomerase as a Target for Chemotherapy.
Cancer remains a major cause of morbidity and mortality in spite of substantial progress toward understanding the molecular basis of the disease. The discovery of new drugs is urgent, and telomerase inhibitors have the potential to provide an additional option for chemotherapy. Telomerase inhibitors might not only limit growth of human tumors directly but might also act in a synergistic fashion with existing inhibitors and amplify their efficacy. For example, after initial chemotherapy or surgery, telomerase inhibitors might be used in an adjuvant setting to limit the recovery of residual cancer cells, making them more susceptible to attack by the immune system or killing by existing chemotherapeutic agents. The use of telomerase inhibitors is particularly attractive for situations of ongoing cell turnover, as occurs with tumor-static antiangiogenesis inhibitors. Systemic administration of telomerase inhibitors will also inhibit telomerase in normal stem cells. This inhibition may result in few side effects, because the relatively short telomeres of tumors may erode to a critical length before irreversible harm is done to other cells.
Oligonucleotides as Telomerase Inhibitors.
Telomerase is an ideal target for oligonucleotides, because its RNA template is essential for activity and is intrinsically accessible to binding by nucleic acids. Oligonucleotides are being tested in 13 ongoing clinical trials (43), and recently, one oligonucleotide, Fomivirsen, has been approved for the treatment of cytomegalovirus retinitis (44). In addition, 2′-O-MeRNA and other 2′-O-alkyl-derivatized oligomers bind more tightly to complementary RNA sequences than do analogous DNA oligomers and have improved resistance to degradation by nucleases, reducing the need for phosphorothioate linkages and improving the selectivity of antisense effects (45, 46); 2′-O-alkyl RNA is currently being used in two phase II clinical trials including one trial directed against the R1-α subunit of protein kinase A, an application similar to the use of telomerase inhibitors to prevent tumor regrowth. Experience with 2′-O-alkyl oligomers in clinical trials, combined with our observation of both decreased cell proliferation and increased apoptosis, will encourage the testing of antitelomerase oligonucleotides in animals and humans.
Phenotypic Effects Are Mediated by a Sequence-Specific, Telomerase-Dependent Mechanism.
Use of oligonucleotides to affect cellular phenotypes is often met with skepticism, because non-sequence-specific effects and misleading phenotypes have plagued their use. Tu et al. (47) have performed a comprehensive survey of the literature, leading Stein (48) to estimate that non-sequence-selective effects may be at least partially implicated in results reported in 94% of the 2,026 studies published through 1997. Indeed, we have previously observed that oligonucleotides that contain uniform phosphorothioate oligonucleotides inhibit telomerase in a non-sequence-specific fashion, presumably through binding to hTERT (38).
We offer multiple independent lines of evidence in the present study that the effects we observe are due to Watson–Crick recognition of telomerase. Treatment with match 2′-O-MeRNA and PNA oligomers cause much greater decreases in telomere length and cell proliferation than does treatment with oligomers that contain mismatched bases or treatment with a less efficient PNA inhibitor directed to a nontemplate region of hTR. The fact that inhibition of telomerase has a more rapid and pronounced effect on proliferation of HME50-5E cells, which have shorter telomeres than DU145 cells, supports a mechanism of decreased proliferation involving telomere shortening rather than nonselective toxicity. Similarly, even though the genetic background of HME50-hTERT cells is similar to that of HME50-5E cells, proliferation of treated HME50-hTERT cells did not decrease, presumably because their initial telomere length of HME50-hTERT cells is approximately 4,000 bp longer. PNA and 2′-O-MeRNA oligomers cause similar reductions in telomere length, suggesting that the effects are caused by Watson–Crick recognition rather than by non-base-pair-mediated contacts with backbone phosphodiester or phosphorothioate linkages. The use of PNAs to confirm the mechanism of oligonucleotide action has important general implications for the use of oligonucleotides within cells, because validating the mechanism of oligonucleotide action is a key problem for antisense research. If neutral and anionic oligomers produce the same effects, they likely derive from specific Watson–Crick base pairing rather than misleading non-sequence-specific interactions. Recent work with PNAs introduced into cells by electroporation has shown decreased cell viability, supporting our findings (49).
General Implications for Development of Telomerase Inhibitors.
Typically, when a putative antiproliferative agent is applied to cells, an effect is observed within hours or days. Telomerase is an unusually challenging target for drug discovery, because a cellular response that depends on telomere shortening will require weeks to become apparent. Although this outcome should not have serious consequences for administration of proven drugs to patients accustomed to chronic treatment of residual disease, it greatly complicates the assays needed to develop and test such drugs. Our primary finding is that telomerase is a viable target for synthetic agents aimed at controlling proliferation of immortal human cells. No evidence for adoption of an ALT (alternative lengthening of telomeres) pathway for telomere maintenance was observed, suggesting that this pathway may not be readily adopted by some immortal cell types. The decreased proliferation we observe should encourage further development and testing of other promising classes of compounds such as nucleoside derivatives directed to hTERT (32) or small molecules designed to disrupt telomere synthesis by promoting G quadruplex formation (33, 34).
Our results have broad implications for how telomerase inhibitors must act within cells to be effective and on how protocols for animal and clinical testing must be designed. We observe telomere shortening and decreased cell proliferation, even though between 5% and 30% of telomerase activity remained after treatment with inhibitor. However, addition of a PNA directed to a nontemplate region of hTR, which is a less potent inhibitor, does not lead to telomere erosion, and telomeres rapidly regain their lengths once inhibitor addition is terminated. Taken together, these results suggest that, although obtaining a striking phenotypic effect does not require full inhibition of telomerase, potency remains an important design consideration. Inhibitors will be most effective against tumor cells with short telomeres, and inhibition of telomerase must be maintained at a sustained high level over time to prevent the rapid regrowth of telomeres. Regrowth to initial telomere length on cessation of inhibitor addition suggests that there is a “set point” for telomere length in human cells and that the telomeres of stem cells that erode during therapy may recover once treatment is terminated. HME50-5E cells were derived from HME50 preimmortal cells that had one mutant p53 allele and lost the second allele during spontaneous immortalization, whereas DU145 cells have two mutant alleles, indicating that functional p53 is not necessary for the reduced proliferation of these cell lines on treatment with telomerase inhibitors.
Conclusions.
The potential for telomerase to be a target for anticancer chemotherapy has engendered a debate that evokes both great enthusiasm and great skepticism. Our findings indicate that telomerase is a viable target for chemotherapeutic drugs and are important for two reasons. The first is that our data will encourage investment in the demanding long-term studies needed to discover and test other classes of inhibitors and guide the design of protocols that optimize the likelihood of obtaining definitive results in animals. The second is that oligonucleotides seem to be excellent candidates for antitelomerase drugs in their own right. The exposed RNA template of hTR makes telomerase an ideal target for oligonucleotides, and the presence of similar oligonucleotides in clinical trials suggests that it should be possible to test potent antitelomerase oligonucleotides in vivo in the near future.
Acknowledgments
This work was supported by Robert A. Welch Foundation Grant I-1244 (to D.R.C.), by National Institutes of Health Grant CA74908 (to D.R.C.), by National Cancer Institute Grant CN-85143 (to J.W.S.), and by a grant from Geron Corporation (to J.W.S.). A.E.P. was supported by National Institutes of Health Training Grant T32GM07062-23. D.R.C. is an assistant investigator with the Howard Hughes Medical Institute. J.W.S. is an Ellison Medical Foundation Senior Scholar. B.-S.H. was supported by a postdoctoral fellowship award from the Susan B. Komen Foundation.
Abbreviations
- mTR and hTR
mouse and human telomerase RNA component
- hTERT
human telomerase reverse transcriptase component
- PNA
peptide nucleic acid
- HME
human mammary epithelial
- TRF
terminal restriction fragment
References
- 1.Morin G B. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 2.Bryan T M, Cech T R. Curr OpIN Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- 3.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L C, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 4.Counter C M, Hirte H W, Bachetti S, Harley C B. Proc Natl Acad Sci USA. 1994;91:2900–2904. doi: 10.1073/pnas.91.8.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Autexier C, Greider C W. Trends Biol Sci. 1996;21:387–391. [PubMed] [Google Scholar]
- 6.Hiyama E, Hiyama K, Yokoyama T, Matsuura Y, Piatyszek M A, Shay J W. Nat Med. 1995;1:249–255. doi: 10.1038/nm0395-249. [DOI] [PubMed] [Google Scholar]
- 7.Langford L A, Piatyszek M A, Xu R, Schold S C, Wright W E, Shay J W. Hum Pathol. 1997;28:416–420. doi: 10.1016/s0046-8177(97)90029-0. [DOI] [PubMed] [Google Scholar]
- 8.Albanell J, Lonardo F, Rusch V, Engelhardt M, Langenfeld J, Han W, Klimstra D, Venkatraman E, Moore M A S, Dmitrovsky E. J Natl Cancer Inst. 1997;89:1609–1615. doi: 10.1093/jnci/89.21.1609. [DOI] [PubMed] [Google Scholar]
- 9.Tahara H, Kuniyasu H, Yokozaki H, Yasui W, Shay J W, Ide T, Tahara E. Clin Cancer Res. 1995;1:1245–1251. [PubMed] [Google Scholar]
- 10.Engelhardt M, Drullinsky P, Guillem J, Moore M A. Clin Cancer Res. 1997;3:1931–1941. [PubMed] [Google Scholar]
- 11.Ohyashiki J H, Ohyashiki K, Iwama I, Hayashi S, Toyama K, Shay J W. Clin Cancer Res. 1997;3:619–625. [PubMed] [Google Scholar]
- 12.Hoos A, Hepp H H, Kaul S, Ahlert T, Bastert G, Wallwiener D. Int J Cancer. 1998;79:8–12. doi: 10.1002/(sici)1097-0215(19980220)79:1<8::aid-ijc2>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Blasco M A, Lee H-W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 14.Herrera E, Samper E, Martin-Caballero J, Flores J M, Lee H W, Blasco M A. EMBO J. 1999;18:1172–1181. doi: 10.1093/emboj/18.5.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H-W, Blasco M A, Gottlieb G J, Horner J W, Greider C W, DePinho R A. Nature (London) 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 16.Rudolph K L, Chang S, Lee H-W, Blasco M, Gottlieb G J, Greider C W, DePinho R A. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg R, Chin L, Femino A, Lee K, Gottlieb G J, Greider C W, DePinho R A. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 18.Chin L, Artandi S E, Shen Q, Tam A, Lee S L, Gottlieb G J, Greider C W, DePinho R A. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 19.de Lange T, Jacks T. Cell. 1999;98:273–275. doi: 10.1016/s0092-8674(00)81955-8. [DOI] [PubMed] [Google Scholar]
- 20.Yu G-L, Bradley J D, Attardi L D, Blackburn E H. Nature (London) 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 21.McEachern M J, Blackburn E H. Nature (London) 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 22.Singer M S, Gottschling D E. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 23.Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Mol Cell Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J, Funk W D, Wang S-S, Weinrich S L, Avilion A A, Chiu C-P, Adams R R, Chang E, Allsopp R C, Yu J, et al. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 25.Bodnar A G, Oullette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 26.Vaziri H, Benchimol S. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 27.Counter C M, Hahn W C, Wei W, Caddle S D, Beijersbergen R L, Landsdorp P M, Sedivy J M, Weinberg R A. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Nature (London) 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 29.Gupta J, Han L P, Wang P, Bacchetti S. J Natl Cancer Inst. 1996;88:1152–1157. doi: 10.1093/jnci/88.16.1152. [DOI] [PubMed] [Google Scholar]
- 30.Bryan T M, Englezou A, Dalla-Pozza L, Dunham M A, Reddel R R. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 31.Lunblad V, Blackburn E H. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 32.Strahl C, Blackburn E H. Mol Cell Biol. 1996;16:53–65. doi: 10.1128/mcb.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D, Thompson B, Cathers B E, Salazar M, Kerwin S M, Trent J O, Jenkins T C, Neidle S, Hurley L H. J Med Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 34.Han F X, Wheelhouse R T, Hurley L H. J Am Chem Soc. 1999;121:3561–3570. [Google Scholar]
- 35.Shay J W, Tomlinson G, Piatyszek M A, Gollahon L S. Mol Cell Biol. 1995;15:425–432. doi: 10.1128/mcb.15.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 37.Mayfield L D, Corey D R. Anal Biochem. 1999;268:401–404. doi: 10.1006/abio.1998.3052. [DOI] [PubMed] [Google Scholar]
- 38.Pitts A E, Corey D R. Proc Natl Acad Sci USA. 1998;95:11549–11554. doi: 10.1073/pnas.95.20.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamilton S E, Simmons C G, Kathriya I, Corey D R. Chem Biol. 1999;6:343–351. doi: 10.1016/S1074-5521(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 40.Holt S E, Norton J C, Wright W E, Shay J W. Methods Cell Sci. 1996;18:237–248. [Google Scholar]
- 41.Shay J W, Brasiskyte D, Ouellette M, Piatyszek M A, Werbin H, Ying Y, Wright W E. Methods Mol Genet. 1994;5:263–268. [Google Scholar]
- 42.Stein C A. Antisense Res Dev. 1997;7:207–209. [Google Scholar]
- 43.Hogrefe R I. Antisense Nucleic Acid Drug Dev. 1999;9:351–357. doi: 10.1089/oli.1.1999.9.351. [DOI] [PubMed] [Google Scholar]
- 44.Marwick C. J Am Med Assoc. 1998;280:871. [Google Scholar]
- 45.Agrawal S, Jiang Z, Zhao Q, Shaw D, Cai Q, Roskey A, Channavajjala L, Saxinger C, Zhang R. Proc Natl Acad Sci USA. 1997;94:2620–2625. doi: 10.1073/pnas.94.6.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou W, Agrawal S. Bioorg Med Chem Lett. 1998;8:3269–3274. doi: 10.1016/s0960-894x(98)00591-5. [DOI] [PubMed] [Google Scholar]
- 47.Tu G C, Cao Q, Zhou F, Israel Y. J Biol Chem. 1998;273:25125–25131. doi: 10.1074/jbc.273.39.25125. [DOI] [PubMed] [Google Scholar]
- 48.Stein C A. Nat Biotechnol. 1999;17:209. doi: 10.1038/6909. [DOI] [PubMed] [Google Scholar]
- 49.Shammas M A, Simmons C S, Corey D R, Shmookler-Reis R J. Oncogene. 1999;18:6191–6200. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]