Abstract
Biotin synthase (BS) is an S-adenosylmethionine (AdoMet)-dependent radical enzyme that catalyzes the addition of sulfur to dethiobiotin. In common with other AdoMet radical enzymes, BS contains a [4Fe-4S] cluster that is coordinated by a highly conserved CxxxCxxC sequence motif and by the methionyl amine and carboxylate of AdoMet. The close association of the [4Fe-4S]+ cluster with AdoMet facilitates reductive cleavage of the sulfonium and the generation of transient 5′-deoxyadenosyl radicals, which are then proposed to sequentially abstract hydrogen atoms from the substrate to produce carbon radicals at C9 and C6 of dethiobiotin. BS also contains a [2Fe-2S]2+ cluster located ∼4-5 Å from dethiobiotin, and we have proposed that a bridging sulfide of this cluster quenches the substrate radicals, leading to formation of the thiophane ring of biotin. In BS from E. coli, the [2Fe-2S]2+ cluster is coordinated by cysteines 97, 128, and 188, and the atypical metal ligand, arginine 260. The evolutionary conservation of an arginine guanidinium as a metal ligand suggests a novel role for this residue in tuning the reactivity or stability of the [2Fe-2S]2+ cluster. In the present work, we explore the effects of mutagenesis of Arg260 to Ala, Cys, His, and Met. Although perturbations in a number of characteristics of the [2Fe-2S]2+ cluster and the proteins are noted, the reconstituted enzymes have in vitro single-turnover activities that are 30-120% of WT. Further, in vivo expression of each mutant enzyme was sufficient to sustain growth of a bioB- mutant strain on dethiobiotin supplemented medium, suggesting the enzymes were active and efficiently reconstituted by the in vivo iron-sulfur cluster (ISC) assembly system. Although we cannot exclude an as-yet unidentified in vivo role in cluster repair or retention, we can conclude that Arg260 is not essential for the catalytic reaction of BS.
Biotin synthase is an iron-sulfur enzyme that catalyzes the formation of the biotin thiophane ring (1-3). The enzyme contains a cuboidal [4Fe-4S]2+ cluster that is coordinated at a unique Fe atom by the methionyl carboxylate and amine of AdoMet1 (4,5). This covalent interaction facilitates the close association of the AdoMet sulfonium with the [4Fe-4S]+ cluster that is thought to be important for the one-electron reductive cleavage of an AdoMet C-S bond, generating methionine and a transient 5′-deoxyadenosyl radical (6). The substrate dethiobiotin lies in van der Waals contact with the AdoMet ribose, and H-atom transfer from C9 of dethiobiotin to the 5′-deoxyadenosyl radical would generate the observed product, 5′-deoxyadenosine, and a substrate-centered carbon radical. Formation of a new C-S bond on the path to biotin presumably involves a reaction of this substrate radical with a sulfur donor; the precise identity of this sulfur donor remains the most significant unanswered mechanistic question.
Biotin synthase belongs to the SAM radical superfamily, which includes ca. 20 known AdoMet-dependent radical enzymes and perhaps more than 600 unique unidentified enzymes (7). These enzymes share a FeS cluster binding motif, CxxxCxxC, that is found within an extended loop region and binds the [4Fe-4S]2+/+ cluster (Cys53, Cys57, and Cys60 in BS from E. coli) (5, 8). These enzymes also share somewhat weaker conservation of residues involved in AdoMet binding, including a hydrophobic residue at position 7 of the cluster motif (Tyr59 in BS) that forms a portion of the adenine binding pocket, and Asp/Glu and Asn/Gln residues (Asp155 and Asn153 in BS) that hydrogen bond to the ribose hydroxyl groups (8). In addition to residues shared with other AdoMet radical enzymes, BS sequences share conserved residues involved in binding DTB that includes the motif N(H/N)N(L/I)(D/E), where Asn151 and Asn153 hydrogen bond to the ureido ring of dethiobiotin. Finally, BS contains an essential [2Fe-2S]2+ cluster that is coordinated by Cys97, Cys128, Cys188, and Arg260; this motif was initially not identified because it is spread throughout the protein, with each residue originating from a different β strand within the (αβ)8 barrel that surrounds the active site (5). Mutagenesis studies have suggested that Cys97, Cys128, and Cys188 are critical for binding to the [2Fe-2S]2+ cluster, since replacement of each residue with Ser resulted in the generation of an inactive mutant apoprotein(9). Although vibrational spectroscopy had indicated O/N ligation at the fourth coordination position of this cluster (10, 11), the identification of Arg260 was made only through solution of the crystal structure of the E. coli enzyme (5), and we postulated that this unusual metal ligand must have a unique catalytic or structural role.
The closest bridging sulfide of the [2Fe-2S]2+ cluster lies 4.7 Å from C9 of dethiobiotin (5). Several experimental results have suggested that a sulfide from this cluster is incorporated as the thiophane sulfur of biotin. When biotin synthase is purified from media supplemented with (35S)-cysteine, the enzyme contains a 35S-labeled [2Fe-2S]2+ cluster and partial turnover results in formation of (35S)-biotin (12). Likewise, when the purified enzyme is stripped of unlabeled FeS clusters and reconstituted with Fe2+ and 34S2-, then the biotin produced incorporates ∼70% of the expected heavy-atom label (13). More recently, partial turnover of BS containing a [2Fe-2Se]2+ cluster, generated by reconstitution with Fe2+ and sodium selenide, produced biotin that contained 45-70% selenium in place of sulfur (14). Transfer of sulfur from the [2Fe-2S]2+ cluster is accompanied by apparent destruction of the cluster, as detected in the optical spectrum by a broad decrease in absorbance centered at ∼450 nm (15), and in the Mössbauer spectrum by a decrease in the narrow quadropole doublet associated with the [2Fe-2S]2+ cluster (δ=0.29 mm/s, ΔEQ=0.51 mm/s) and an appearance of much broader doublets associated with Fe2+ ion (δ=0.73, 0.86 mm/s, ΔEQ=3.36, 3.65 mm/s) (16).
The conservation of the atypical guanidinium metal ligand Arg260, together with the implication of the [2Fe-2S]2+ cluster as the probable sulfur donor, suggested that this residue played an important role in the enzyme. We postulated several possible roles: (i) the potential positive charge of the guanidinium could be important in modulating the redox potential of the FeS cluster, (ii) the bidentate nature of a guanidinium functional group could facilitate liberation of sulfide from the cluster during turnover and would therefore be catalytically important, (iii) the long Arg260 side chain could be important for closing β strand 7 around the barrel-shaped active site and would thus be structurally important (5), or (iv) protonation of the guanidine following turnover could facilitate opening the active site to allow cluster assembly and/or repair by the endogenous iron-sulfur cluster assembly system (ISC) of E. coli. To test these hypotheses, we mutated Arg260 to Ala, Cys, His, and Met, and conducted a series of experiments designed to examine changes in the properties of the [2Fe-2S]2+ clusters, the stability of the folded protein with and without the [2Fe-2S]2+ cluster, and the importance of this residue for in vitro and in vivo activity.
Materials and Methods
Construction of Mutants
All mutants were constructed by a modified Quickchange method (Stratagene) using the WT plasmid pJJ15-4A (based on pET21d (17)) as a template. Forward and reverse PCR primers (Integrated DNA Technologies) that overlap the Arg260 codon were mixed with template, deoxyribonucleotides, pfu Turbo polymerase and buffer, and subjected to 25 rounds of PCR with 5 minute extension times. Single stranded DNA was digested with DpnI and the resulting mutant plasmids transformed into library competent DH5α and screened on LB/agar plates with 50 μg/mL ampicillin. For each mutant, plasmid DNA was reisolated, the sequence confirmed by sequencing the entire BioB gene insert, and then the plasmids were transformed into BL21(DE3)pLysS (Novagen) for protein expression. The Arg260Cys mutant was poorly expressed in this strain, and was instead expressed in the protease-deficient strain BLR(DE3)pLysS (Novagen). Protein expression and purification was performed as previously described (17).
Determination of Extinction Coefficients and Fe and S content
Samples of WT and mutant proteins were diluted to ∼5 mg/mL in 25 mM Tris.HCl, 25 mM NaCl, pH 8.0, and accurate UV/visible absorbance spectra recorded on a Varian Cary 50Bio spectrophotometer. Each sample was then analyzed for protein content using the Bradford assay with a commercial BSA standard (BioRad). Protein concentrations were divided by the correction factor of 1.10, previously determined by quantitative amino acid analysis of the WT protein (18), and extinction coefficients were then calculated from an average and standard deviation of 15 analyses for each protein. The same samples were then subjected to iron and sulfide analysis using previously described methods (18), with ratios reported representing the average and standard deviation for 9 samples, and also incorporate the uncertainty in the protein concentrations as derived from the calculated extinction coefficients.
Activity Assays
Biotin synthase (25 μM in 250 μl final volume, 0.26 mg protein) was added to septum-covered vials containing 50 mM Tris.HCl, 30 mM KCl, 5 mM DTT, pH 7.5 and degassed under argon. The [4Fe-4S]2+ cluster was reconstituted by incubation with 0.4 mM Na2S and 0.4 mM FeCl3 for 5 min. Flavodoxin (10 μM), ferredoxin (flavodoxin):NADP+ oxidoreductase (2 μM), NADPH (2 mM), and AdoMet (0.5 mM) were added while the vials were continuously degassed. Turnover was initiated by addition of DTB (0.25 mM), and the vials were incubated in a 37°C water bath for 5 - 240 min. Potassium acetate, pH 4.5 (25 μl of 4.5 M stock) was added to quench the reaction and precipitate the protein. The vials were incubated on ice to ensure full precipitation, the precipitate removed by centrifugation for 10 min at 18,000 × g, and the supernatant transferred to an HPLC autosampler vial. HPLC analysis for biotin and DTB was performed on a Waters Atlantis dC18 reversed-phase column (3.0 × 150 mm, 5 μm) with a 20 min linear gradient from 5 - 20% acetonitrile in H2O containing 5 mM H3PO4 and UV detection at 205 nm. Fresh external standards containing biotin (10-250 μM) and DTB (10-250 μM) were prepared and analyzed prior to and following each set of samples to ensure accurate identification and quantitation of biotin.
Circular Dichroism Spectroscopy
Apoprotein samples (100 μM) were prepared by adding EDTA (5 mM), sodium dithionite (1 mM), and methyl viologen (5 μM), and incubating 1 hr under nitrogen. As-isolated [2Fe-2S]2+ protein and apoprotein were exchanged into anaerobic 10 mM MOPS, 25 mM NaF, pH 7.5 on a Sephadex G-25 column, and then diluted to 5 μM in the same buffer. Samples were transferred to a sealed cuvette under nitrogen and CD spectra collected with a Jasco J-810 circular dichroism spectrometer with a Peltier temperature controller. Each spectrum is an average of 3 scans from 260-190 nm with 20 nm/min scanning speed, 2 sec response time, 1 nm bandwidth. Temperature melts were recorded from 0-98°C at 222 nm with a 2°C data pitch, 2 sec response time, 1 nm bandwidth. Thermal melts were fit to the Gibbs-Helmholtz equation as previously described (19).
Electrochemical Analysis of the [2Fe-2S]2+ Cluster
The electrochemical potential for reduction of the [2Fe-2S]2+ cluster was determined by titration with sodium dithionite in the presence of methyl viologen (MV) by a modification of the method of Drummond and Matthews (20). BS (50 μM in 50 mM Tris, 200 mM NaCl, pH 8.0) and methyl viologen (50 μM) were placed in an anaerobic cuvette (1.6 mL total volume) under argon, and for each spectrum, dithionite (2 μl of a 6 mM stock) was added with a syringe through a rubber septum, the solution mixed by brief mechanical stirring, allowed to equilibrate for 2 min, and a spectrum collected from 300 to 800 nm. Spectra were separately collected for oxidized and fully reduced protein, and for oxidized and 50% reduced MV. The change in absorbance at 620 nm is largely due to the reduced MV radical cation (ε620 = 12135 M-1 cm-1), and this data was used to calculate the ratio [MVred]/[MVox] and the cell potential, Eh, using a midpoint potential for MV of Em = -440 mV (21). The change in absorbance at 454 nm is largely due to changes in the protein spectrum, with a minor contribution from MVred. This minor MVred component was subtracted from the data, the remaining absorbance change was normalized by dividing by the initial absorbance, and the resulting data fit to the Nernst equation to obtain the apparent potential for reduction of the [2Fe-2S]2+ cluster. For these fits, the extrapolated final protein absorbance was based upon spectra of fully reduced protein obtained in the absence of MVred.
EPR Spectroscopy
WT and R260 mutant proteins (200 μM in 320 μl final volume) were diluted in 50 mM Tris.HCl, pH 8.0 under argon. Sodium dithionite (1 mM) and methyl viologen (1 μM) were added and the samples incubated in a 15 °C water bath. At 5 or 60 min, samples were transferred to EPR tubes, frozen in a hexane/liquid N2 slurry, and stored in a liquid N2 dewar until spectra could be collected. X-band EPR spectra were collected with a Bruker ESP300E spectrometer over the temperature range 4 - 50 K, with 12 K chosen as the optimal temperature for observing the reduced FeS cluster signal. Temperature was regulated with a liquid He transfer system (Oxford ESR Model 900) with a cryostat positioned in a TE102 cavity. The parameters for high-field spectra were P = 16 mW, modulation amplitude = 16 G, time constant = 163 ms, conversion time = 81.98 ms, modulation frequency = 100 kHz, with each spectrum the sum of 3 scans, while the parameters for low-field spectra were identical with the exceptions of P = 50 mW, time constant = 655 ms, with each spectrum the sum of 6 scans.
In Vivo Activity of Mutant Biotin Synthase
Strain 364 is a bioB- derivative of Escherichia coli K12 originally isolated by Max Eisenberg (Columbia University) and kindly provided by Max Gottesmann (Columbia University). Although this strain is unpublished, the preparation method and properties are identical to strain 310 as described by Rolfe and Eisenberg (22), however strain 364 is more resistant to spontaneous reversion (M. Gottesmann, personal communication). This strain was lysogenized with λDE3 to generate bio364(DE3), which was then complemented by transformation with either pJJ15-4A (WT BioB), plasmids containing each R260 mutant gene, or for a negative control, pET21d, by electroporation on a Bio-Rad Gene Pulser II at 200 Ω, 25 μF, 1.8 V with a 0.1 cm cuvette. Cells were initially grown on LB/agar plates containing ampicillin (50 μg/ml). One colony was transferred to a 20 ml culture containing 0.4% glucose, 0.4% vitamin-free casamino acids, M9 salts, 10 μM FeCl3, an essential metal supplement, ampicillin (50 μg/ml), and DTB (5 μM) and grown overnight to partially deplete the endogenous biotin levels. The overnight cultures were diluted to an initial OD600 = 0.05 in 35 ml of the same medium containing IPTG (1 mM) and either no DTB or biotin, DTB (5 μM), or biotin (5 μM). Cultures were grown at 37 °C with gentle agitation and samples were removed at intervals for determination of the OD600. Samples were also removed at OD600 = 0.8 for Western blot analysis of BioB and biotinylated-AccB content (supporting information).
Results
Choice of Mutants
The role of Arg260 as a metal ligand to the [2Fe-2S]2+ cluster in BS is unique and unconventional. More common coordination motifs for [2Fe-2S] clusters include all cysteine coordination, as observed in the [2Fe-2S] ferredoxins, and two histidine and two cysteine coordination, as observed in Rieske iron-sulfur proteins. Thus we chose to explore whether substitution with these more common FeS cluster ligands would affect the role of the [2Fe-2S]2+ cluster in catalysis by characterizing the mutants Arg260Cys and Arg260His. However, an examination of the structure of BS suggested that the Arg260Cys thiolate would probably not extend to coordinate the [2Fe-2S]2+ cluster without a major structural rearrangement, and so we also generated an Arg260Met mutant, where although the thioether of Met is not a strong metal ligand, the length of the amino acid might potentially allow for a more isosteric substitution for Arg and for metal coordination. Finally, we eliminated the protein-derived metal ligand altogether through generation of an Arg260Ala mutant, most likely leaving a pocket in the protein that could be filled with one or more water molecules that might substitute as metal ligands.
All Arg260 Mutant Proteins Contain a [2Fe-2S]2+ Cluster
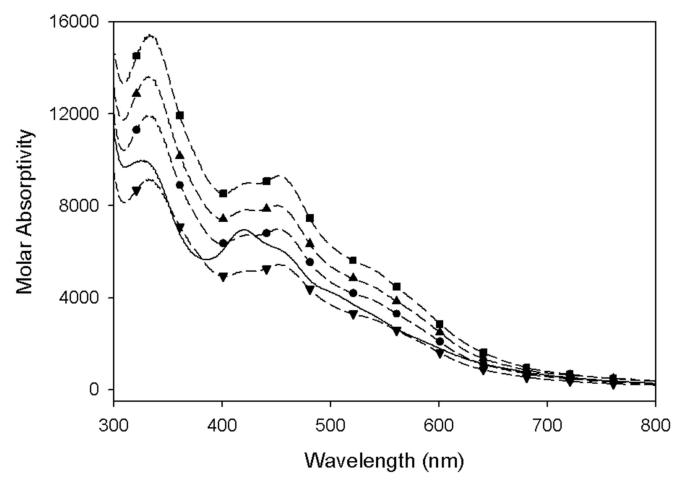
The presence of an air-stable [2Fe-2S]2+ cluster is a distinctive feature of active WT BS. This cluster has been observed in vivo by Mössbauer spectroscopy (23) and is the dominant spectroscopic feature of the aerobically purified enzyme (24). Further, mutagenesis of the [2Fe-2S]2+ cluster ligands Cys97, Cys128, or Cys188 to Ser or Ala has been reported to result in aerobically purified protein that does not contain this cluster and is inactive (9, 25). To ascertain the cluster content of the Arg260 mutant proteins, we relied on UV/visible spectra and Fe and S content, as compared to the WT enzyme. UV/visible spectra of the WT and each mutant (Figure 1) display characteristic bands at 322, 415, and 452 nm, with the WT, Arg260Ala, and Arg260His exhibiting nearly indistinguishable spectra and comparable extinction coefficients (Table 1). Fe and S analysis also confirm comparable levels of [2Fe-2S]2+ cluster for these mutants as compared to WT (Table 1). The Arg260Cys mutant exhibits a UV/visible spectrum similar to WT, but with an extinction coefficient that is only ∼75% of that for the WT protein, and the Fe analysis also suggests a decreased total cluster content (∼70% Fe content as compared to WT). Since we did not observe further degradation of the cluster in the purified protein, we conclude that the [2Fe-2S]2+ cluster in the Arg260Cys mutant is not inherently unstable, but rather the protein is not obtaining a full complement of [2Fe-2S]2+ cluster in vivo during induced protein expression. Since protein overexpression levels are similar for all mutants, this effect is more likely due to a problem with recognition by or cluster transfer from the in vivo FeS cluster assembly system. The Arg260Met mutant also does not obtain a full complement of FeS cluster, as indicated by somewhat low Fe content and extinction coefficient. In addition, the UV/visible spectrum of the Arg260Met mutant exhibits an increased absorbance at 415 nm relative to the 452 nm band, such that the ratio of these bands goes from ε415/ε452 = 0.93 in the WT protein to ε415/ε452 = 1.09 in the Arg260Met mutant. In this respect, the spectrum becomes more like the tetrathiolate coordinated [2Fe-2S]2+ ferredoxins, with ε410/ε460 ≈ 1.14 (26), which we interpret as indicating that the thioether of the Arg260Met mutant is contributing to the ligand-to-metal charge-transfer bands of the [2Fe-2S]2+ cluster and is therefore a covalent ligand to the cluster. In contrast, the spectrum of the Arg260Cys mutant is shaped similarly to WT, and we therefore conclude that there is no covalent interaction between the newly introduced Cys thiolate and the [2Fe-2S]2+ cluster.
Figure 1.
UV/visible spectra of WT and Arg260 mutants of biotin synthase. Samples from top to bottom: R260H (-■-), R260A (-▲-), WT (-●-),R260M (—), R260C (-▼-). Spectra have been scaled according to extinction coefficients determined by the Bradford protein assay.
Table 1.
Properties of WT and Arg260 Mutants of Biotin Synthase
| Protein | Fe/monomer | S/monomer | Ext. Coeff. (M-1 cm-1) | Tm (°C) | Em (mV) | Initial Activity (nmol min-1 mg-1) | Yield (nmol per nmol BS) | Doubling Time (min) | |
|---|---|---|---|---|---|---|---|---|---|
| [2Fe-2S] | Apoprotein | ||||||||
| WT | 1.6 ± 0.2 | 2.9 ± 0.6 | 7000 ± 400 | 58 ± 2 | 46 ± 2 | -502 ± 30 | 0.18 | 0.50 | 83 |
| R260A | 1.8 ± 0.2 | 3.3 ± 0.6 | 8000 ± 500 | 63 | 49 | -504 ± 10 | 0.15 | 0.40 | 83 |
| R260H | 2.0 ± 0.2 | 3.8 ± 0.6 | 9300 ± 500 | 61 | 49 | -502 ± 30 | 0.22 | 0.54 | 86 |
| R260M | 1.3 ± 0.1 | 2.6 ± 0.5 | 6100 ± 300 | 51 | 53 | -460 ± 15 | 0.17 | 0.27 | 89 |
| R260C | 1.1 ± 0.1 | 2.5 ± 0.5 | 5400 ± 400 | 50 | 47 | -509 ± 30 | 0.062 | 0.25 | 88 |
Electrochemical Analysis of the Reduction of the [2Fe-2S]2+ Cluster
In WT BS, the [2Fe-2S]2+ cluster exhibits unusual electrochemical properties. The cluster is sequestered away from solvent within the (αβ)8 barrel of the protein, and in the presence of the strong chemical reductant sodium dithionite, is slowly reduced over several minutes with a previously reported reduction potential of -430 mV (18). However, the reduced [2Fe-2S]+ cluster is not stable and rapidly dissociates from the protein. Since the oxidized [2Fe-2S]2+ cluster is proposed to be a necessary starting point for catalysis, the low reduction potential may be an important property that prevents non-catalytic reduction of the cluster and subsequent inactivation of the protein.
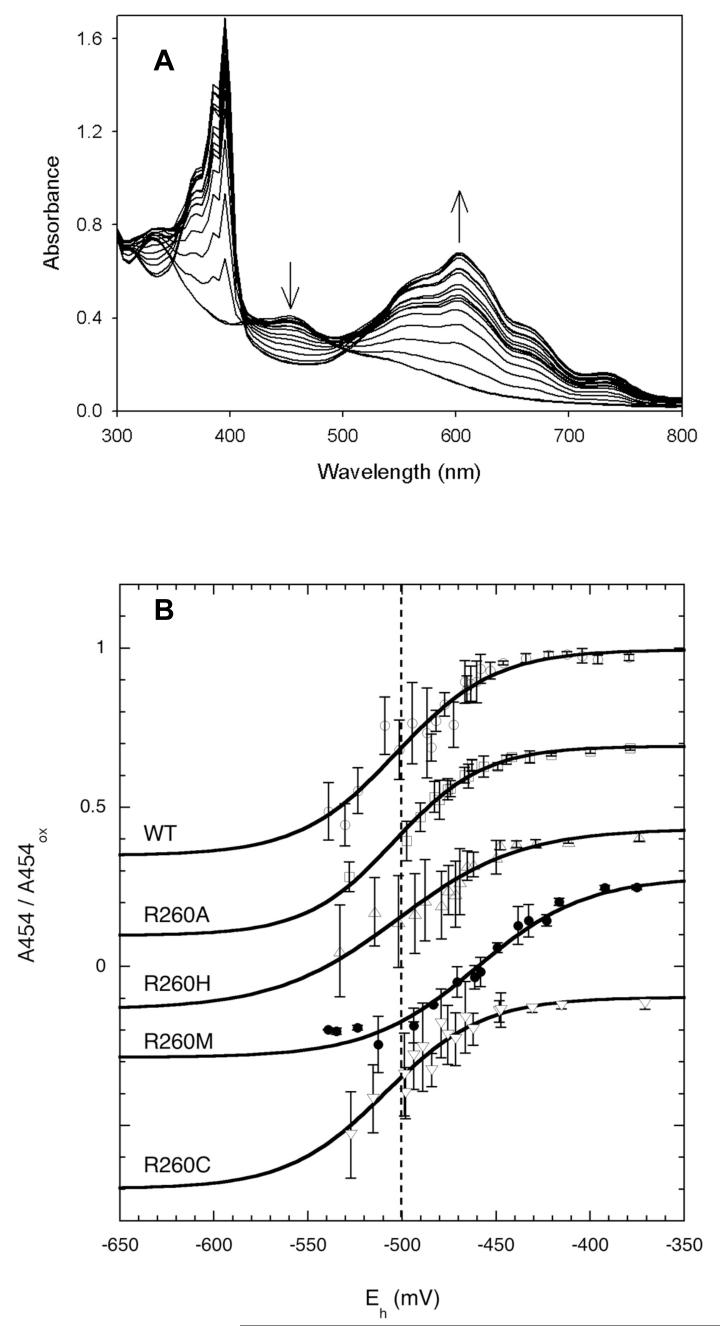
In the present study, we employed an electrode-independent method for determining electrochemical midpoint potentials in which methyl viologen (MV) is used as an internal redox indicator dye. This method allows calculation of the cell potential, Eh, based on the concentration of reduced MV observed in the UV/visible spectrum as a sharp peak at 400 nm and a broad peak centered at ∼600 nm (Figure 2A), and using the Nernst equation and a midpoint potential for MV of -440 mV (21). MVred has a spectral window at 430-480 nm that allows observation of changes in the spectrum of the [2Fe-2S]2+ cluster at 454 nm. Minor corrections are necessary to account for dilution of the sample and for a slight increase in absorbance at 454 nm due to MVred, and we chose protein and MV concentrations (50 μM each) that ensured that this correction was <20% of the total spectral change. Electrochemical titration is accomplished by the addition of substoichiometric amounts of sodium dithionite, which reduces methyl viologen rapidly, and then allowing the methyl viologen to reduce the [2Fe-2S]2+ cluster for ∼2 min after each addition. In principle, this method allows a more rapid determination of the midpoint potential than standard electrode based methods, where equilibration with the electrode limits the speed of the experiment.
Figure 2.
Electrochemical reduction potentials of the [2Fe-2S]2+ cluster. A. Spectra were recorded during titration of WT (50 μM) and methyl viologen (50 μM) in 50 mM Tris.HCl, 200 mM NaCl, pH 8.0 with 6 mM sodium dithionite (2 μl per addition). B. Reduction of the [2Fe-2S]2+ cluster in WT and Arg260 mutants as detected by the decrease in absorbance at 454 nm. For each sample, the increase in absorbance at 620 nm was used to calculate the concentration of reduced methyl viologen radical and the cell potential (Eh) as described in the Materials and Methods. Each data point shows an average and standard deviation for 3 titrations, and data were normalized by dividing by the initial absorbance of the oxidized protein. The vertical line shows the WT Em = -502 mV for comparison with other curves; mutant data is arbitrarily offset along the vertical axis to allow comparison with WT.
Data derived from titration of WT BS and each Arg260 mutant are shown in Figure 2. During titration of WT [2Fe-2S]2+ cluster, we observed formation of significant amounts of MVred (600 nm) prior to any reduction of the FeS cluster (454 nm), suggesting the protein reduction potential was lower than that for MV. Using an extinction coefficient for the reduced protein of ε454=2800 (based on separate reduction of the protein in the absence of MV), we calculate a reduction potential of -502 ± 30 mV (based on 3 titrations). This value is significantly lower than we had previously reported based on Ag/AgCl electrode methods, and we can only speculate that in those prior experiments, protein reduction was much more rapid than electrode equilibration, leading to erroneously high values for the reduction potentials. Titrations of Arg260Ala, Arg260His, and Arg260Cys gave reduction potentials that are identical to WT, within the error of the experiment. In contrast, mutation of Arg260 to Met resulted in an increase in the reduction potential to -460 ± 15 mV. As noted in the previous section, the spectral properties of the Arg260Cys mutant indicate that the thiol does not coordinate the FeS cluster, and we speculate that both the Arg260Cys and Arg260Ala mutants retain a bound hydroxide or water in the coordination position normally occupied by Arg. Thus, our interpretation is that all of the mutant proteins with O/N ligands retain approximately the same low midpoint potential. On the other hand, Met is a neutral sulfur ligand that likely fills the space of Arg and excludes water, and in this mutant we observe a destabilization of the oxidized [2Fe-2S]2+ state by ∼45 mV.
Electron Paramagnetic Resonance Analysis of the Reduced Proteins
Reduction of the [2Fe-2S]2+ cluster does not lead to the formation of a stable reduced cluster in WT BS. Instead, we have previously observed Fe2+/3+ in the buffer immediately following reduction, and we observe slow formation of a substoichiometric amount of reduced [4Fe-4S]+ cluster over 1-2 hr (17). This [4Fe-4S]+ cluster is not coordinated to the same site as the original [2Fe-2S]2+ cluster, but rather to the conserved CxxxCxxC motif that coordinates a [4Fe-4S] cluster in the active enzyme (27). We thought it might be possible that the loss of the cluster upon reduction is due to weak coordination of the Arg ligand, and therefore we might expect that one or more Arg260 mutants would retain the reduced [2Fe-2S]+ cluster.
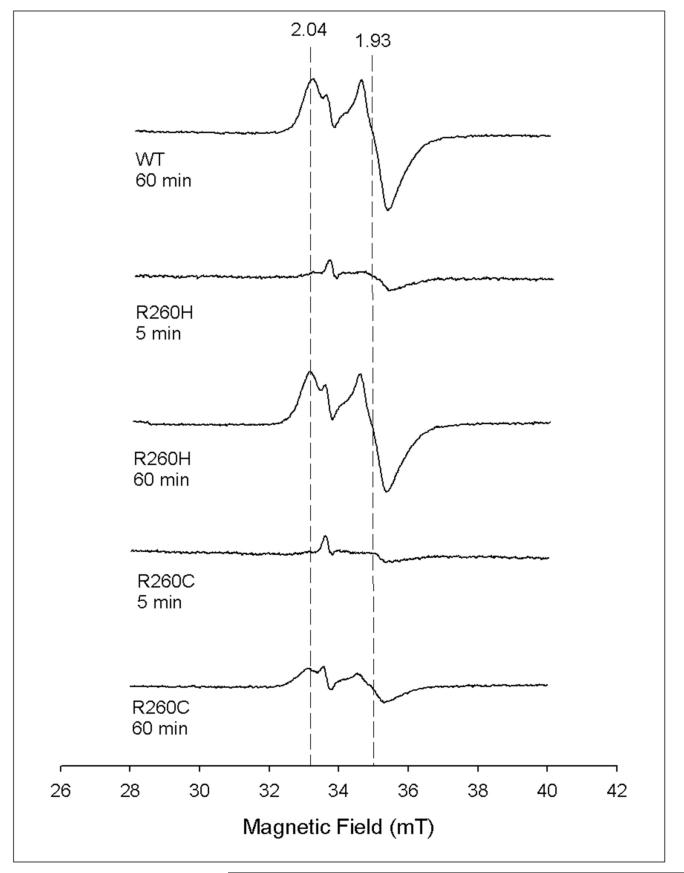
We reduced each protein with dithionite, in the presence of a catalytic amount of MV to speed electron delivery to the buried cluster, and observed bleaching of the distinctive brown color of the protein over 2-5 min, suggesting reduction of the [2Fe-2S]2+ cluster. Samples were frozen at 5 and 60 min and analyzed by EPR spectroscopy (Figure 3). The WT protein develops a signal after 60 min that is identical to previously reported spectra, exhibiting an axial spectrum with g values of 2.04 and 1.93 and relaxation properties that indicate a fast-relaxing [4Fe-4S]+ cluster (Tmax = 12 K). All of the mutant proteins gave identical spectra after 60 min incubation (Arg260His and Arg260Cys are shown for comparison), with g values and relaxation properties that were indistinguishable from the WT. The only significant differences were in the yield of [4Fe-4S]+ cluster, which can be rationalized on the basis of the Fe content of the original oxidized protein (Table 1). Interestingly, this axial signal is in low abundance after 5 min despite the fact that the brown color of the [2Fe-2S]2+ cluster has been lost. Temperature and power saturation studies indicate that no observable paramagnetic species are present, and we would suggest that after 5 min most of the Fe is present as high-spin Fe2+ (S=2) that is not detected by perpendicular-mode EPR spectroscopy as used in this study. We conclude that neither the WT nor any of the Arg260 mutants retains a tightly-bound and stable [2Fe-2S]+ cluster.
Figure 3.
EPR spectra of reduced WT, R260H, and R260C indicate formation of a [4Fe-4S]+ cluster. Each protein (200 μM in 320 μl) was mixed with methyl viologen (1 μM) in anaerobic 50 mM Tris.HCl, pH 8.0 and reduced with sodium dithionite (1 mM) for the indicated times at 15 °C, and then flash-frozen in an EPR tube. The small, sharp signal at g = 2 in all samples is due to reduced methyl viologen. Spectra of oxidized proteins showed no EPR signals.
Arg260Met and Arg260Cys Mutations Decrease the Thermal Stability
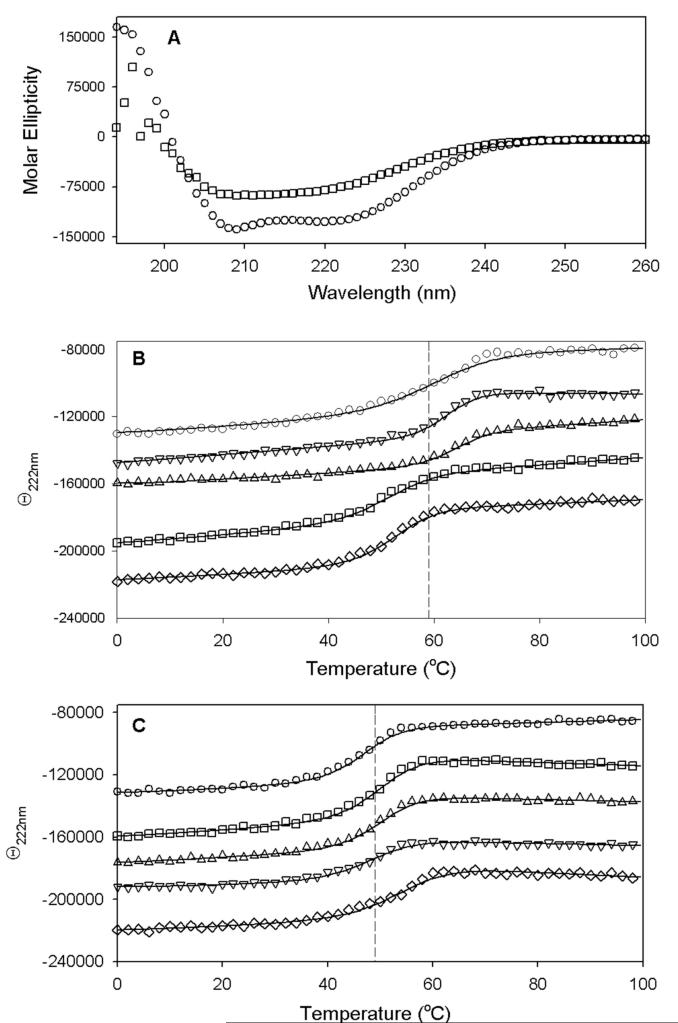
One proposed role for a long Arg metal ligand is to facilitate binding a small [2Fe-2S]2+ cluster within a geometrically larger (αβ)8 barrel, potentially stabilizing both the bound cluster and the folded protein. If this were the case, one might expect that mutation of Arg260 would result in either weak binding of the cluster, which was not observed, or destabilization of the protein fold. We examined the stability of the folded protein using CD spectroscopy, both in the presence and the absence of the [2Fe-2S]2+ cluster. The WT and mutant proteins exhibit a CD spectrum with negative bands at 208 and 222 nm (Figure 4a), typical of proteins with a high α-helical content. When the temperature is increased, each protein undergoes a cooperative transition to an unfolded state (Figures 4b and 4c) with a positive band at 195 nm and some negative intensity at ∼210-220 nm, suggesting predominantly random coil but with retention of some minor folded structures. The WT protein containing a [2Fe-2S]2+ cluster unfolds at 58 °C, while the apoprotein unfolds at 46 °C, suggesting that the [2Fe-2S]2+ cluster assists in binding together and stabilizing the protein structure. The Arg260Ala and Arg260His mutants behave in a similar manner, although these mutations appear to stabilize both forms of the protein by ∼3-5 °C. In contrast, the Arg260Met and Arg260Cys mutants destabilize the protein containing a [2Fe-2S]2+ cluster such that it has a stability approximately equivalent to apoprotein. In the case of the Arg260Met, this could be due to the hydrophobic nature of the side-chain, which is unable to hydrogen bond to other hydrophilic groups within the β8 barrel (Arg95, Ser43, Ser218, and Ser283). The case for Arg260Cys is less obvious, since it behaves in other respects like the Arg260Ala mutant. One possibility is that at higher temperatures, the propensity for coordination of the Arg260Cys thiolate to Fe might actually trigger unfolding of the protein since this coordination probably cannot be satisfied within the native fold.
Figure 4.
Protein stability determined by thermal denaturation in a circular dichroism spectrophotometer. Each protein (5 μM in anaerobic 10 mM MOPS, 25 mM NaF, pH 7.5) either contained the original complement of [2Fe-2S]2+ cluster or was reductively stripped of this cluster as described in the Materials and Methods. (A) Initial CD spectrum of WT [2Fe-2S]2+ BS recorded at 20 °C (○) and final melted spectrum recorded at 98 °C (□). (B) WT and R260 mutant proteins containing a [2Fe-2S]2+ cluster were subjected to a thermal melt from 0 to 98 °C monitored at 222 nm: WT (○), Arg260Ala (▽), Arg260His (△), Arg260Met (□), Arg260Cys (◇). (C) WT and R260 mutant apoproteins were subjected to a thermal melt from 0 to 98 °C monitored at 222 nm: WT (○), Arg260Ala (▽), Arg260His (△), Arg260Met (□), Arg260Cys (◇). Melt data are fit to a two-state cooperative unfolding model to determine the Tm values listed in Table 1.
All of the Arg260 Mutants are Active for In Vitro Biotin Synthesis
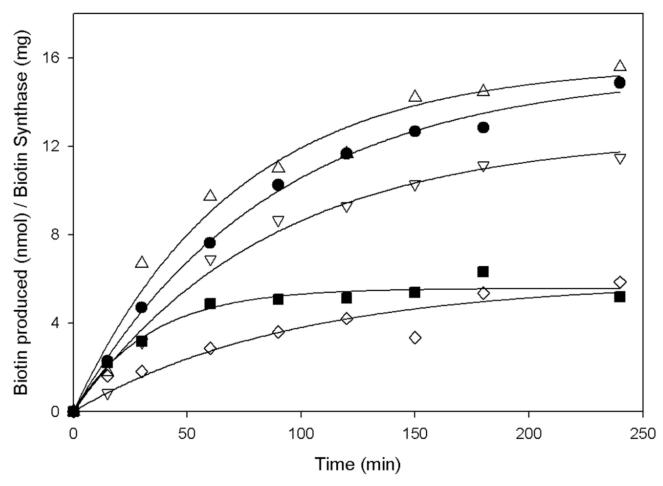
Although we had observed various changes in the properties of the [2Fe-2S]2+ cluster as a result of each Arg260 mutation, evolutionary conservation is concerned only with the end result: can this particular mutant protein make sufficient biotin for survival of the organism? We assayed WT and mutant proteins using our in vitro assay system that involves in situ reconstitution of the [4Fe-4S]2+ cluster under argon and reduction of the system using the native E. coli redox proteins, followed by addition of AdoMet and DTB to trigger turnover. We monitored the time-dependence of biotin formation (Figure 5), fit the data to a single exponential function, and report the “initial rate” of this single turnover derived from the initial slope (0 - 30 min), and the yield of biotin after 180 min (Table 1). We observe that all of the mutants can make biotin, and that the yield of biotin is approximately correlated with the fractional [2Fe-2S]2+ cluster content of the original protein. The initial rate of biotin synthesis is, within the error of the data, identical for the WT and all of the mutants (∼0.2 ± 0.05 nmol/min/mg of BS), except for the Arg260Cys mutant, which turns over ∼3-fold slower than WT. However, none of these mutations is significantly defective for in vitro biotin synthesis.
Figure 5.
Time-dependent activity of WT and R260 mutants: WT (●), Arg260Ala (▽), Arg260His (△), Arg260Met (■), Arg260Cys (◇). Each protein (25 μM) was incubated with DTB, AdoMet, flavodoxin, ferredoxin (flavodoxin):NADP+ oxidoreductase, and NADPH as described in the Materials and Methods. At intervals, aliquots were removed, quenched with sodium acetate (pH 4.5), and analyzed for biotin content by HPLC. Curves are fit to a single exponential function assuming ≤1 turnover per monomer. Activities reported in Table 1 are based on the initial rate of the early linear portion of each curve.
All of the Arg260 Mutants Can Supply Biotin In Vivo
Since Arg260 is apparently conserved, and yet it was not required for biotin formation in vitro, we initially suspected that our assay might not sufficiently replicate in vivo conditions, such that the Arg260 mutants could possibly not be active enough to complement biotin biosynthesis in vivo. We had previously obtained a strain defective in the biotin synthase catalyzed step of the biotin biosynthetic pathway (E. coli K12 strain 364 from Max Eisenberg, Columbia University). Although this strain has not been fully characterized, we do not observe BS by Western blot analysis and suspect the strain carries a mutation that affects expression or stability of the enzyme. Phenotypically, this strain grows extremely slowly on glucose/M9 media, the growth is not stimulated by adding DTB, but regains WT growth rates in the presence of biotin.
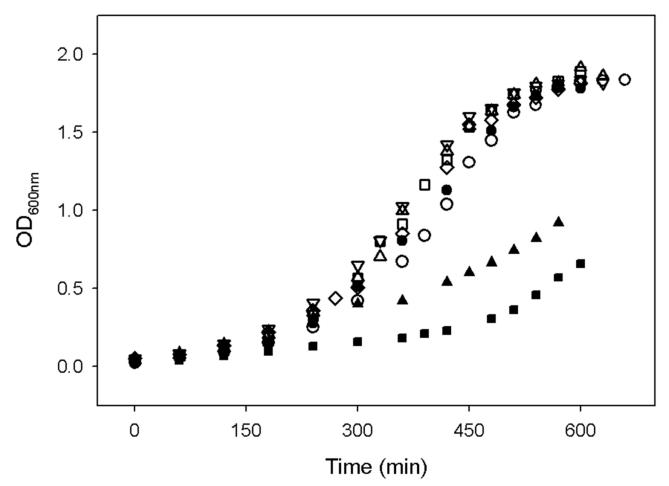
We transformed our WT and mutant plasmids (derivatives of pET21d) into strain 364 that had been lysogenized with λDE3 (Novagen). These strains, along with a control that contained the empty pET21d plasmid, were grown under 3 conditions: glucose/casamino acids/M9 media only, glucose/casamino acids/M9 + DTB, and glucose/casamino acids/M9 + biotin. All media also contained ampicillin, IPTG to induce protein expression, and FeCl3 to facilitate cluster assembly. The control strain that does not contain a plasmid-borne copy of BioB grows very slowly in the absence of exogenous biotin (Figure 6), doubling time (tD) = 240 min), suggesting that the defective chromosomal copy of BioB is capable of extremely limited biotin production. When plasmids expressing either WT or Arg260 mutants are introduced, the strains grow slightly faster in the absence of added DTB or biotin (tD = 175 min), suggesting that the chromosomal bio operon is producing small amounts of DTB that is then converted to biotin by the overexpressed proteins. When DTB is added to the medium, we see growth of WT and mutant BioB-overexpressing strains at rates that are experimentally indistinguishable from each other or from controls grown in the presence of biotin (tD=85 min). These data indicate that the overexpressed Arg260 mutants can carry out sufficient biotin production in vivo to sustain maximal growth rates in glucose/M9 media. It should be noted, however, that expression of protein from pET plasmids produces artificially high enzyme levels (∼100-fold higher than endogenous biotin synthase) that may swamp out minor differences in the rate of biotin synthesis. Therefore we can only interpret these results qualitatively: mutation of Arg260 does not abolish catalysis in vivo.
Figure 6.
Growth of bio364(DE3) mutant strain (bioB-) in the absence of biotin, with and without complementation with a pET21d-derived plasmid expressing WT or R260 mutant BS. Growth is at 37 °C in glucose (0.4%), casamino acids (0.4%), M9 salts, microbiological metal supplement, ampicillin (50 μg/ml), and IPTG (1 mM) supplemented with either biotin (5 μM) or DTB (5 μM). Data displayed: -bioB -DTB -biotin (■), -bioB +biotin (●), WT +DTB (□), R260A +DTB (△), R260H +DTB (▽), R260M +DTB (◇), R260C +DTB (○), R260A -DTB (◆).
Discussion
Although the presence of a [2Fe-2S]2+ cluster was one of the first features noted in recombinant biotin synthase (24), the chemical properties and catalytic role of this cluster has been a subject of considerable debate. Most AdoMet radical enzymes contain an air-sensitive [4Fe-4S]2+/+ cluster in their active forms, and it was initially believed that the observed [2Fe-2S]2+ cluster in BS was a stable oxidative degradation product of the essential [4Fe-4S] cluster. Consistent with this proposal, reduction of the [2Fe-2S]2+ cluster results in formation of a [4Fe-4S]2+/+ cluster (11), and it was presumed that this chemistry occurred within the conserved CxxxCxxC sequence motif. However, subsequent mutagenesis (9), electrochemical analysis (18), selective isotopic substitution (10, 27), and x-ray crystallography (5) indicated that the initial [2Fe-2S]2+ cluster and resulting [4Fe-4S]2+ cluster reside in distinct sites. Resonance Raman spectroscopy had provided evidence that the [2Fe-2S]2+ cluster was coordinated by 3 cysteine thiolate ligands and one unidentified O/N ligand (10, 11); the ligands to the [2Fe-2S]2+ cluster were definitively assigned as Cys97, Cys128, Cys188, and Arg260 upon solution of the crystal structure of E. coli BS (5).
The finding that the O/N ligand to the [2Fe-2S]2+ cluster was an arginine guanidinium was unprecedented in the biochemical literature. Guanidine species have been employed as ligands in transition metal complexes, and have been observed to ligate various metals in aqueous solution, but have not been observed in proteins. One might expect that the high pKa of arginine (pKa = 12.5) would present a formidable energetic barrier to guanidinium deprotonation and metal coordination, although this could be partially compensated through formation of the ligand-to-metal bond and through steric encapsulation within the protein fold. In light of these potential energetic problems, the apparent evolutionary conservation of Arg260 in BS suggested that this residue played an important structural or catalytic role. From a structural viewpoint, arginine has a long side-chain that can extend across the β8 barrel and allow the [2Fe-2S]2+ cluster to be bound in an asymmetric location within the enzyme active site. In addition, the guanidinium could potentially tune the physicochemical properties of the cluster through hydrogen bonds or electrostatic effects. Alternatively, from a catalytic viewpoint, the bidentate nature of guanidine could allow Arg260 to play a catalytic role, perhaps in facilitating release of sulfur from the FeS cluster for subsequent incorporation into biotin. In light of these proposals, we chose to explore whether the substitution of arginine with other potential metal ligands would have a significant effect on the properties of the cluster or the enzyme.
We noted only one mutation that affected the properties of the [2Fe-2S]2+ cluster: Arg260Met. Methionine is approximately isosteric with arginine, and the optical spectrum shows a perturbation in electronic transitions that suggests coordination of the FeS cluster by the methionine thioether. This coordination appears to destabilize the oxidized cluster, resulting in an increase the apparent reduction potential of the [2Fe-2S]2+ cluster by ∼45 mV. We also noted two mutations that adversely affect the stability of the [2Fe-2S]2+ cluster-containing protein: Arg260Met and Arg260Cys. While the WT protein is ∼12 °C more stable with the [2Fe-2S]2+ cluster bound as compared to apoprotein, both of these mutant proteins show no stabilization upon binding the [2Fe-2S]2+ cluster. These same mutant proteins have a low FeS cluster content of ∼60-70% when purified under conditions comparable to WT protein. Together these observations indicate that binding of the [2Fe-2S]2+ cluster to the apoprotein is less favorable for the Arg260Met and Arg260Cys mutants as compared to WT. In contrast, the Arg260Ala and Arg260His mutants show no dysfunction. In fact, the Arg260His mutant appears to have a higher cluster content, higher thermal stability, and marginally higher activity than WT enzyme.
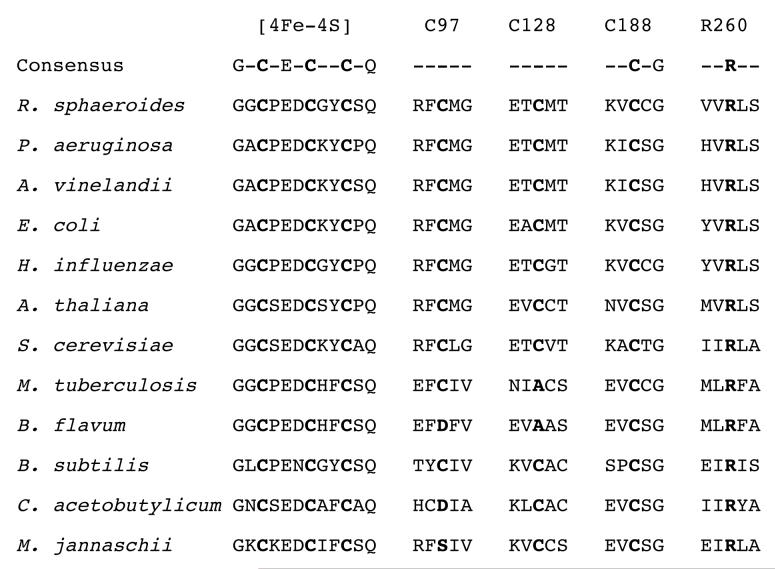
Although at the time this work was initiated, the available microbial sequence databases indicated that the ligands to the [2Fe-2S]2+ cluster were absolutely conserved, sequences from more diverse species have since become available that suggest some tolerance in these residues. A sequence alignment of 186 annotated BS sequences (excluding only those missing the DTB binding motif that we therefore assume are misannotated) is available online as supporting information and is abbreviated inFigure 7. Some variation appears to be tolerated at Cys97 and Cys128. At Cys97, serine or aspartate appears as the probable cluster ligand in 5% of the sequences, while at Cys128, alanine appears in 4% of the sequences. Only one of the sequences has non-cysteinyl ligands simultaneously at both positions. However, Arg260 is absolutely conserved in all organisms, from plants to yeast to anaerobic thermophilic bacteria. Given our finding that the Arg260His mutant appears to offer an optimal combination of stability and activity, one might question why histidine does not appear naturally at this position. Assuming that the primordial residue was arginine, then 2 of the 6 Arg codons (CGU and CGC) could be converted to His (CAU and CAC) by a single base transversion of G→A. However, virtually all known mutagenic outcomes for guanine damage generate a G→T transversion, resulting in an Arg→Leu mutation (28) Clearly, Leu would not be capable of functioning as a metal ligand, probably resulting in either misfolded protein or poor binding of the FeS cluster, and therefore we would expect that a G→T transversion would not be tolerated at the Arg260 codon. In the absence of this direct mutagenic path, the evolutionary conversion of arginine to histidine through single-base mutations would necessitate a less probable process with several intervening mutations.
Figure 7.
Alignment of the FeS cluster ligands from selected BS protein sequences. The residues numbered at the top refer to the E. coli protein. The consensus sequence shows the residues absolutely conserved among all 186 annotated BS sequences (see supporting information for a full alignment).
The finding that the ligand set that coordinates the [2Fe-2S]2+ cluster has little affect on activity is puzzling if one thinks of this FeS cluster as a catalytic cofactor. The primary and secondary coordination shell of an FeS cluster often play a significant role in tuning the properties of an FeS cluster, and in particular the electrochemical behavior (29), and one might assume that evolutionary pressure has resulted in an optimal ligand set that tunes the cluster for the desired reaction. In the case of biotin synthase, we have observed that all of the Arg260 mutants contain a [2Fe-2S]2+ cluster that is assembled in vivo, and that the observed in vitro activity primarily correlates with the amount of cluster present. Likewise, in contrast with results from other labs (9, 25), we have observed that mutations at Cys97, Cys128, and Cys188 that preserve a stable [2Fe-2S]2+ cluster are also active, while double and triple mutants that lack this cluster are inactive (J. Jarrett, unpublished results). Perhaps it would be better to think of these protein residues as forming the binding pocket for the third “substrate,” a [2Fe-2S]2+ cluster. When one mutates a residue in the substrate binding pocket of an enzyme, one expects to affect the Km and/or Kd for the substrate, but not necessarily the kcat for catalysis. In this case, the [2Fe-2S]2+ cluster binds with such high affinity to the WT and mutant enzymes that we are unable to detect significant differences in binding or activity with an in vitro assay. In vivo, BS must compete with all other FeS proteins for binding the [2Fe-2S]2+ cluster “substrate.” Since BS is present at much lower concentrations in vivo, and the [2Fe-2S]2+ cluster is likely delivered through the actions of the ISC system, one possibility is that the ligand set may be tuned to facilitate this sensitive cluster handoff in the complex and competitive cellular environment.
Supplemental Material
Acknowledgments
We thank Dr. Jason T. Wan and Jennifer Dashnau for early work in constructing and characterizing select mutants, and Dr. Devrim Eren for advice concerning the in vivo activity studies.
Footnotes
This research has been supported by the NIH (R01 GM59175 to J.T.J.) and a fellowship from the David and Lucille Packard Foundation (J.T.J.).
Supporting Information Available
Western blots demonstrating high-level expression of all Arg260 mutants and equivalent levels of biotinylated AccB (acetyl CoA carboxylase β subunit or BCCP). Sequence alignments of 186 annotated biotin synthase proteins listed in the NCBI database. This material is available free of charge via the Internet at http://pubs.acs.org.
- AdoMet
- S-adenosyl-L-methionine
- BS
- biotin synthase (bioB gene product)
- DTB
- dethiobiotin
- DTT
- dithiothreitol
- EDTA
- ethylenediaminetetraacetic acid
- ISC
- E. coli iron-sulfur cluster assembly proteins
- MOPS
- 4-morpholinepropanesulfonic acid
- MV
- methyl viologen
- Tris
- tris(hydroxymethyl)aminomethane.
References
- (1).Jarrett JT. The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase. Arch Biochem Biophys. 2005;433:312–21. doi: 10.1016/j.abb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- (2).Marquet A. Enzymology of carbon-sulfur bond formation. Curr. Opin. Chem. Biol. 2001;5:541–9. doi: 10.1016/s1367-5931(00)00249-0. [DOI] [PubMed] [Google Scholar]
- (3).Marquet A, Tse Sum Bui B, Florentin D. Biosynthesis of biotin and lipoic acid. Vitam. Horm. 2001;61:51–101. doi: 10.1016/s0083-6729(01)61002-1. [DOI] [PubMed] [Google Scholar]
- (4).Cosper MM, Jameson GN, Davydov R, Eidsness MK, Hoffman BM, Huynh BH, Johnson MK. The [4Fe-4S]2+ cluster in reconstituted biotin synthase binds S-adenosyl-L-methionine. J Am Chem Soc. 2002;124:14006–7. doi: 10.1021/ja0283044. [DOI] [PubMed] [Google Scholar]
- (5).Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–9. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jarrett JT. The generation of 5′-deoxyadenosyl radicals by adenosylmethionine-dependent radical enzymes. Curr Opin Chem Biol. 2003;7:174–82. doi: 10.1016/s1367-5931(03)00022-x. [DOI] [PubMed] [Google Scholar]
- (7).Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Nicolet Y, Drennan CL. AdoMet radical proteins--from structure to evolution--alignment of divergent protein sequences reveals strong secondary structure element conservation. Nucleic Acids Res. 2004;32:4015–25. doi: 10.1093/nar/gkh728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hewitson KS, Baldwin JE, Shaw NM, Roach PL. Mutagenesis of the proposed iron-sulfur cluster binding ligands in Escherichia coli biotin synthase. FEBS Lett. 2000;466:372–6. doi: 10.1016/s0014-5793(00)01101-7. [DOI] [PubMed] [Google Scholar]
- (10).Cosper MM, Jameson GN, Hernandez HL, Krebs C, Huynh BH, Johnson MK. Characterization of the cofactor composition of Escherichia coli biotin synthase. Biochemistry. 2004;43:2007–21. doi: 10.1021/bi0356653. [DOI] [PubMed] [Google Scholar]
- (11).Duin EC, Lafferty ME, Crouse BR, Allen RM, Sanyal I, Flint DH, Johnson MK. [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry. 1997;36:11811–20. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- (12).Gibson KJ, Pelletier DA, Turner IM., Sr. Transfer of sulfur to biotin from biotin synthase (BioB protein) Biochem Biophys Res Commun. 1999;254:632–5. doi: 10.1006/bbrc.1998.9991. [DOI] [PubMed] [Google Scholar]
- (13).Tse Sum Bui B, Florentin D, Fournier F, Ploux O, Mejean A, Marquet A. Biotin synthase mechanism: on the origin of sulphur. FEBS Lett. 1998;440:226–230. doi: 10.1016/s0014-5793(98)01464-1. [DOI] [PubMed] [Google Scholar]
- (14).Tse Sum Bui B, Mattioli TA, Florentin D, Bolbach G, Marquet A. Escherichia coli biotin synthase produces selenobiotin. Further evidence of the involvement of the [2Fe-2S]2+ cluster in the sulfur insertion step. Biochemistry. 2006;45:3824–34. doi: 10.1021/bi052388m. [DOI] [PubMed] [Google Scholar]
- (15).Ugulava NB, Sacanell CJ, Jarrett JT. Spectroscopic changes during a single turnover of biotin synthase: Destruction of a [2Fe-2S] cluster accompanies sulfur insertion. Biochemistry. 2001;40:8352–8358. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Jameson GN, Cosper MM, Hernandez HL, Johnson MK, Huynh BH. Role of the [2Fe-2S] cluster in recombinant Escherichia coli biotin synthase. Biochemistry. 2004;43:2022–31. doi: 10.1021/bi035666v. [DOI] [PubMed] [Google Scholar]
- (17).Ugulava NB, Gibney BR, Jarrett JT. Iron-sulfur cluster interconversions in biotin synthase: dissociation and reassociation of iron during conversion of [2Fe-2S] to [4Fe-4S] clusters. Biochemistry. 2000;39:5206–14. doi: 10.1021/bi9926227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ugulava NB, Gibney BR, Jarrett JT. Biotin synthase contains two distinct iron-sulfur cluster binding sites: Chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry. 2001;40:8343–8351. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Boice JA, Dieckmann GR, DeGrado WF, Fairman R. Thermodynamic analysis of a designed three-stranded coiled coil. Biochemistry. 1996;35:14480–5. doi: 10.1021/bi961831d. [DOI] [PubMed] [Google Scholar]
- (20).Drummond JT, Matthews RG. Nitrous oxide degradation by cobalamin-dependent methionine synthase: characterization of the reactants and products in the inactivation reaction. Biochemistry. 1994;33:3732–41. doi: 10.1021/bi00178a033. [DOI] [PubMed] [Google Scholar]
- (21).Mayhew SG. The redox potential of dithionite and SO2- from equilibrium reactions with flavodoxins, methyl viologen and hydrogen plus hydrogenase. Eur J Biochem. 1978;85:535–47. doi: 10.1111/j.1432-1033.1978.tb12269.x. [DOI] [PubMed] [Google Scholar]
- (22).Rolfe B, Eisenberg MA. Genetic and Biochemical Analysis of the Biotin Loci of Escherichia coli K12. J. Bacteriol. 1968;96:515–524. doi: 10.1128/jb.96.2.515-524.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Benda R, Tse Sum Bui B, Schunemann V, Florentin D, Marquet A, Trautwein AX. Iron-sulfur clusters of biotin synthase in vivo: a Mössbauer study. Biochemistry. 2002;41:15000–6. doi: 10.1021/bi026590q. [DOI] [PubMed] [Google Scholar]
- (24).Sanyal I, Cohen G, Flint DH. Biotin synthase: purification, characterization as a [2Fe-2S] cluster protein, and in vitro activity of the Escherichia coli bioB gene product. Biochemistry. 1994;33:3625–31. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- (25).Hewitson KS, Ollagnier-de Choudens S, Sanakis Y, Shaw NM, Baldwin JE, Munck E, Roach PL, Fontecave M. The iron-sulfur center of biotin synthase: site-directed mutants. J. Biol. Inorg. Chem. 2002;7:83–93. doi: 10.1007/s007750100268. [DOI] [PubMed] [Google Scholar]
- (26).Ta DT, Vickery LE. Cloning, sequencing, and overexpression of a [2Fe-2S] ferredoxin gene from Escherichia coli. J Biol Chem. 1992;267:11120–5. [PubMed] [Google Scholar]
- (27).Ugulava NB, Surerus KK, Jarrett JT. Evidence from Mössbauer spectroscopy for distinct [2Fe-2S]2+ and [4Fe-4S]2+ cluster binding sites in biotin synthase from Escherichia coli. J. Am. Chem. Soc. 2002;124:9050–1. doi: 10.1021/ja027004j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- (29).Torres RA, Lovell T, Noodleman L, Case DA. Density functional and reduction potential calculations of Fe4S4 clusters. J Am Chem Soc. 2003;125:1923–36. doi: 10.1021/ja0211104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.