Abstract
Background
The Duffy-binding protein II of Plasmodium vivax (PvDBPII) has been considered as an attractive target for vaccine-mediated immunity despite a possible highly polymorphic nature. Among seven PvDBP domains, domain II has been shown to exhibit a high rate of nonsynonymous polymorphism, which has been suggested to be a potential immune (antibody binding) evasion mechanism. This study aimed to determine the extent of genetic polymorphisms and positive natural selection at domain II of the PvDBP gene among a sampling of Thai P. vivax isolates.
Methods
The PvDBPII gene was PCR amplified and the patterns of polymorphisms were characterized from 30 Thai P. vivax isolates using DNA cloning and sequencing. Phylogenetic analysis of the sequences and positive selection were done using DnaSP ver 4.0 and MEGA ver 4.0 packages.
Results
This study demonstrated a high rate of nonsynonymous polymorphism. Using Sal I as the reference strain, a total of 30 point-mutations were observed in the PvDBPII gene among the set of Thai P. vivax isolates, of which 25 nonsynonymous and five synonymous were found. The highest frequency of polymorphism was found in five variant amino acids (residues D384G, R390H, L424I, W437R, I503K) with the variant L424I having the highest frequency. The difference between the rates of nonsynonymous and synonymous mutations estimated by the Nei and Gojobori's method suggested that PvDBPII antigen appears to be under selective pressure. Phylogenetic analysis of PvDBPII Thai P. vivax isolates to others found internationally demonstrated six distinct allele groups. Allele groups 4 and 6 were unique to Thailand.
Conclusion
Polymorphisms within PvDBPII indicated that Thai vivax malaria parasites are genetically diverse. Phylogenetic analysis of DNA sequences using the Neighbour-Joining method demonstrated that Thai isolates shared distinct alleles with P. vivax isolates from different geographical areas. The study reported here will be valuable for the development of PvDBPII-based malaria vaccine.
Background
Molecular mechanisms of invasion by Plasmodium vivax merozoites are mediated by the Duffy-binding-like (DBL) family of homologous erythrocyte binding protein (EBPs) located within the micronemes of merozoites that recognize specific receptors on red blood cell [1]. PvDBP is a 140-kDa protein belonging to a family of erythrocyte binding proteins characterized by a functionally conserved cysteine-rich region. The similarity among DBL families of EBPs is the greatest within two adjacent cysteine-rich domains, designated as the amino cysteine-rich domain (N-cys) and carboxyl cysteine-rich domain (C-cys) [2]. The amino cysteine-rich domain has been identified as the principle adhesion region binding to erythrocyte receptors, while the function of the carboxyl cysteine-rich domain remains unclear. This amino-cysteine-rich domain is present within the 330 amino acid region II, which has been shown to contain the binding motifs necessary for adherence to the Duffy antigen receptor for chemokines (DARC), required for erythrocyte invasion [3]. Critical binding motifs in PvDBPII have been mapped to a 170 amino acid spanning region between cysteines 4 and 7 (amino acids 291–460) [4]. Cysteine residues are conserved within the identified binding motif, whereas other amino acids are highly polymorphic, having a high ratio of nonsynonymous to synonymous mutations [5].
The polymorphic nature of the PvDBP, particularly its region II (DBPII) is a major impediment to the development of a broadly protective vaccine against vivax malaria. A study in Papua New Guinea showed 43 and 18 unique nonsynonymous mutations in 10 Madang [6] and 24 Wosera vivax isolates [7], respectively, whereas 19 unique nonsynonymous mutations were described in 10 Colombian vivax isolates [8]. Often the mutations were found in corresponding locations in the protein sequence of isolates from different geographical areas. Amino acids substitutions at D384G, K386(N/Q), N417K, L424I, W437R, and I503K were found in both Papua New Guinea isolates and Colombia isolates [5-8]. Such results demonstrate that many similar alleles are widely distributed among P. vivax from different geographical areas. Polymorphic patterns in the PvDBP cluster within the critical erythrocyte-binding segment imply the existence of a selection pressure on PvDBP [9]. Under the influence of immune pressure, the distribution of existing antigenic polymorphisms affects the population structure in which newly selected mutants spread and may provide insight to the evolution and selection of parasite populations over time [10]. Polymorphisms of PvDBPII require further exploration among vivax isolates from different geographic areas, particularly south-east Asia where a large proportion of vivax infections take place. Therefore, the gene polymorphisms within the region II domain of the P. vivax DBP among Thai P. vivax isolates were investigated.
Methods
P. vivax samples and DNA preparation
Plasmodium vivax isolates were obtained from malaria patients with informed consent at the outpatient clinic, Hospital for Tropical Diseases, Faculty of Tropical Medicine, Mahidol University, Bangkok, from May, 2002 to June, 2003 under a protocol approval by the Ethics Committee, Faculty of Tropical Medicine, Mahidol University. The confirmation of P. vivax infection was done by microscopic examination of the parasites in thick and thin blood smears. Approximately 1 ml of venous blood was collected from each individual in 0.5 M ethylene-diamine tetra-acetic acid (EDTA) tubes and kept at -20°C until use. Genomic DNA was extracted from 200 μl of blood using a High Pure PCR Template Preparation Kit (Roche, Mannhein, Germany) according to the manufacturer's protocol.
Genotyping of P. vivax isolates at the PvDBPII locus
P. vivax DBPII gene was amplified by PCR using the oligonucleotide primers, DBPII-F; 5'-CACCACGATCTCTAGTGCTATTATA-3' and DBPII-R; 5'-TGTCACAACTTCCTGAGTATT-3' [11]. PCR cycling conditions were 94°C for 3 min, 55°C for 1 min, and 72°C for 1 min for 35 cycles followed by a 10 min extension at 72°C. PCR products were stained with 0.1 μg/ml ethidium bromide and visualized by digital photography under ultraviolet light. The PCR products were run adjacently to 0.1 μg/lane of a 2-log DNA ladder (NewEngland BioLabs, Inc. MA, USA) as a standard DNA marker.
Purification of PCR products, DNA cloning and DNA sequencing of PvDBPII gene
PCR products were purified by QIAquick PCR purification Kit (Qiagen Inc., Valencia, CA) following the manufacturer's instructions. The PCR products were cloned directly into pCR-Blunt II-TOPO cloning vector using a Zero Blunt TOPO PCR cloning kit (Invitrogen, La Jolla, CA). For DNA sequencing of the species-specific amplicons, excess dNTPs and unincorporated primers were eliminated using the QIAquick PCR purification kit (Qiagen Inc., Valencia, CA). DNA sequencing was performed by fluorescence based methodologies using MegaBACE DNA Analysis Systems (Amersham Biosciences, Piscataway, NJ). The sequences have been submitted to the GenBank under the accession number EF219451, EF368159–368180, EF379127–379135. Due to the possibility of multiple genotypes within parasites taken from one individual, direct sequencing was not performed. Instead, at least five clones were sequenced from each corresponding PCR product and sequences of these had to be identical to meet our quality control standards.
Analysis of PvDBPII gene sequences
The alignment of complete sequences of 30 PvDBPII genes was analysed by Sequencher ver 4.2 software (Gene Code Corporation, Ann Arbor, MI) [12,13]. Sequences were aligned using CLUSTALX Multiple Sequence Alignment Program developed by the National Center for Biotechnology Information (NCBI, Bethesda, MD). The percent similarity was assessed using BLASTN (NCBI, Bethesda, MD) and BioEdit ver 7.0 software (Tom Hall Isis Therapeutics, Isis Pharmaceuticals, CA), DnaSP ver 4.0 [14] and MEGA ver 4.0 Beta program [15].
Analysis of natural selection
Evidence of positive natural selection was determined by comparing the rate of nonsynonymous and synonymous substitutions in region II of the P. vivax DBP gene. The rates of substitutions were estimated using the Nei and Gojobori's method [16] with the Jukes and Cantor correction [17,18] as implemented in the MEGA program. Standard error was determined by 1000 bootstrap replications. Negative selection acting on most coding genes can be identified typically when the rate on nonsynonymous mutations is less than the rate of synonymous mutations (kn <Ks). However, when positive selection is acting on a gene, the rate of nonsynonymous mutations will exceed that of synonymous mutations (kn > Ks). Two other tests of neutrality were performed by Tajima's D test, Fu and Li's D- and F-tests on DnaSP 4 software using P. knowlesi DBP as an outgroup. Tajima's D test compares the estimation of nucleotide diversity calculated in two ways (θ calculated from the number of segregating sites and π calculated from average pairwise nucleotide diversity) in order to test for a departure from neutrality. Fu and Li's D and F test, departures from neutrality are identified as a deviation between estimates of θ (derived from the number of mutations in external branches of the phylogeny and from the total number of mutations giving the index D or from the average pairwise diversity π giving the index F) [19].
Phylogenetic analysis
Phylogenetic analysis was used to investigate the associations of PvDBPII gene with sequences elucidated from different geographical regions. The gene tree was constructed using regions common to all available PvDBPII sequences. Forty five PvDBPII genes sequences found in GenBank were compiled and compared to Thai isolates including the sequences from a reference strain, Sal I, one individual sequence from Vietnam, Indonesia, Brazil and India as well as 2, 3, 3 and 31 sequences from Bangladesh, Colombia (COL), South Korea (SK) and Papua New Guinea (PNG), respectively (Table 1). A phylogenetic tree was derived from the aligned nucleotide sequences using the Neighbour-Joining (NJ) method with 1000 bootstrap replicates, the Tamura's three-parameter distance model as implemented in the MEGA ver 4.0 Beta program [20].
Table 1.
PvDBPII sequences deposited in GenBank used in the study
| species | locality | GenBank accession number |
| P. vivax | Papua New Guinea | AF289480–289653, AF469522–469602, AY970848–970925, AF291096, AF695565, DQ156519 |
| P. vivax | Colombia | AY341907, DQ156513, AY341899 |
| P. vivax | Korea | AF215737, AF215738, DQ156523 |
| P. vivax | India | DQ156514 |
| P. vivax | Brazil | DQ153520 |
| P. vivax | Vietnam | DQ156518 |
| P. vivax | Indonesia | DQ156521 |
| P. vivax | Sal I | M37514, DQ156512 |
Results
PCR products and genetic polymorphisms of PvDBPII gene among Thai isolates
The PCR amplification of the P. vivax DBPII gene from 30 Thai isolates yielded DNA fragments of approximately 990 base pairs in length (Figure 1). A 900 bp was selected after excluding primer regions and adjacent bases which did not give uniformly reliable sequences. DNA analysis in comparison to the PvDBP gene of Sal I reference strain revealed amino acid polymorphisms across the entire domain II of PvDBP protein (300 amino acids) among Thai isolates. Among the 30 polymorphic sites, 29 showed monomorphic mutation (changed into one amino acid-type) and only a position 386(N/Q) showed dimorphic mutation (changed into two amino acid-types) (Table 2). Among 300 amino acids within PvDBPII, 25 nonsynonymous (representing 83% of all isolates) and five synonymous mutations were identified (Table 2). High frequencies of variant amino acids (>50%) were found and included L424I (86%), D384G (76%), W437R (63%), R390H (56%), and I503K (56%) residue polymorphisms.
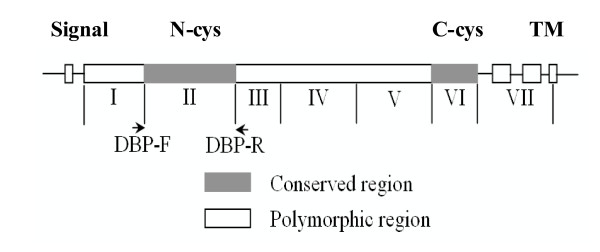
Figure 1.
Schematic diagram of PvDBPII gene showing the location of the primers.
Table 2.
Amino acid changes found in PvDBPII gene among 30 Thai isolates, comparing to DBP Sal I sequence.
| Position of the amino acid | ▼ | ▽ | ▼ | ||||||||||||
| 268 | 276 | 281 | 306 | 308 | 333 | 351 | 367 | 371 | 375 | 378 | 384 | 385 | 386 | 390 | |
| SalI | CGT | AAC | GTT | TTT | AGG | CTT | AGT | ATC | AAA | AAT | CGC | GAT | GAA | AAG | CGT |
| Thai | AGT | AAT | GTA | TTG | AGT | TTT | TGT | ACC | GAA | GAT | CGT | GGT | AAA | AAT/CAG | CAT |
| SalI | R | N | V | F | R | L | S | I | K | N | R | D | E | K | R |
| Thai | S | N | V | L | S | F | C | T | E | D | R | G | K | N/Q | H |
| a | (1/30) | (1/30) | (1/30) | (2/30) | (8/30) | (14/30) | (1/30) | (2/30) | (6/30) | (7/30) | (30/30) | (23/30) | (14/30) | (13/30) | (17/30) |
| b | 3 | 3 | 3 | 6 | 26 | 46 | 3 | 6 | 20 | 23 | 100 | 76 | 46 | 43 | 56 |
| Position of the amino acid | ▼ | ▼ | ▼ | ||||||||||||
| 398 | 404 | 417 | 419 | 424 | 433 | 436 | 437 | 447 | 464 | 475 | 486 | 503 | 507 | 513 | |
| Sal I | TCT | ACA | AAT | ATA | TTA | CAG | AGA | TGG | TCA | ATC | CCA | CAA | ATA | AAC | ACG |
| Thai | ACT | AGA | AAA | ATG | ATA | AAG | ACA | CGG | TCC | ATA | GCA | GAA | AAA | CAC | AAG |
| Sal I | S | T | N | I | L | Q | R | W | S | I | P | Q | I | N | T |
| Thai | T | R | K | M | I | K | T | R | S | I | A | E | K | H | K |
| a | (3/30) | (3/30) | (11/30) | (1/30) | (26/30) | (1/30) | (1/30) | (19/30) | (1/30) | (1/30) | (3/30) | (2/30) | (17/30) | (1/30) | (1/30) |
| b | 10 | 10 | 36 | 3 | 86 | 3 | 3 | 63 | 3 | 3 | 10 | 6 | 56 | 3 | 3 |
a Showing the detection frequency of each amino acids variant on PvDBPII protein among 30 Thai isolates.
b Percentage of detection frequency calculated from value in panel "a".
▼ High prevalence (>50%) of variant amino acids.
▽ Dimorphic mutation (changed into two amino acid-types)
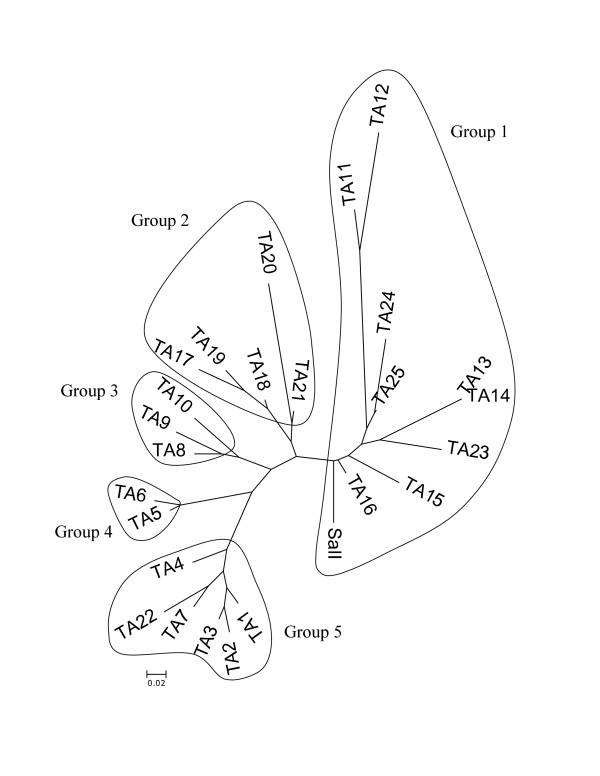
Based on the variation of nonsynonymous mutations and in comparison to Sal I the Thai haplotyes were designated as TA (TA1-TA25). TA1, TA5 and TA11 haplotypes were identified in two isolates while TA17 was identified in three isolates. The other 21 haplotypes were identified in single isolates (Table 3). The cluster analysis of PvDBPII amino acid sequences among 25 haplotypes based on Neighbour-Joining method was organized into five main groups (Figure 2).
Table 3.
PvDBPII haplotypes observed among 30 Thai isolates.
| Haplotype name | Amino acid haplotype* | No. of isolate observed (%) |
| TA 1 | RFRFSIKDGKNHSTKIIQRRPQKNT | 2(6.6) |
| TA 2 | RFSFSIKDGKNHSTKIIQRRPQKNT | 1(3.3) |
| TA3 | RFSFSIKDGKKHSTKIIQRRPQKNT | 1(3.3) |
| TA 4 | RFRLSIKDGKNHSTKIIQRRPQKNT | 1(3.3) |
| TA 5 | RFRFSIENGKNRSTNIIQRRPQINT | 2(6.6) |
| TA6 | RFRFSIENGKNRSTKIIQRRPQINT | 1(3.3) |
| TA 7 | RFSFSIKDGKNHSTNIIQRRPQINT | 1(3.3) |
| TA 8 | RFRLSIENGEKHSTKIIQRRPQKNT | 1(3.3) |
| TA 9 | RFRLSIENGEQRSTKIIQRRPQKNT | 1(3.3) |
| TA 10 | RFRLSIENGEKRSTKMIQRRPQINT | 1(3.3) |
| TA 11 | RLSLLSIKNGKNHTRNIIQRWPQINT | 2(6.6) |
| TA 12 | SFSLSIKNGKNHTRNIQRWPEINT | 1(3.3) |
| TA 13 | RFRLSTKNGEKHSTNIIQRWPQKHT | 1(3.3) |
| TA 14 | RFRLSTKNGEKHSTNIIQRWPQKHT | 1(3.3) |
| TA 15 | RFSLSIKNGEKRSTNIIQRWPQKNT | 1(3.3) |
| TA 16 | RFRLSIKNGEKRSTNIIQRWPQINT | 1(3.3) |
| TA 17 | RFRFSIKNDEKRSTNIIQRRAQKNT | 3(10) |
| TA 18 | RFRLSIKNDEKRSTNIIQRRPQKNT | 1(3.3) |
| TA19 | RFRFSIKNDKKRSTNIIQRRPQKNT | 1(3.3) |
| TA 20 | RFRLSIKNDKKRSTKIIKTRPQINT | 1(3.3) |
| TA 21 | RFRLSIKNDKKRSTNIIQRWPQINT | 1(3.3) |
| TA 22 | RFSFSIKDGKNHSTNILQRRPQKNT | 1(3.3) |
| TA23 | RFRLSIKNGEKHSTNILQRWPQKNK | 1(3.3) |
| TA 24 | RFRLCIKNGEKHSTNILQRWPEINT | 1(3.3) |
| TA 25 | RFRLSIKNGEKHSTNILQRWPQINT | 1(3.3) |
* Amino acid residues included in the haplotypes are as follows: 268, 306, 308, 333, 351, 367, 371, 375, 384, 385, 386, 390, 398, 404, 417, 419, 424, 433, 436, 437, 475, 486, 503, 507, 513.
Figure 2.
Neighbour-Joining tree of PvDBPII amino acid sequence of 25 variants observed among 30 Thai isolates. The PvDBPII haplotype for each variant could be seen in Table 3.
Evidence of natural selection
To investigate whether natural selection contributed to generation of this diversity, the ratio of nonsynonymous (Kn) to synonymous substitutions (Ks) was compared between Thai and Sal I isolates. The number of nonsynonymous substitutions per nonsynonymous sites (Kn) (0.00956) exceeded the number of synonymous substitutions per synonymous sites (Ks) (0.00754). The Kn/Ks ratio was 1.270 predicting that positive selection may be occurring in region II of DBP favouring the fixation of amino acid replacement in certain areas of the protein. The statistic tests detecting departure from neutrality showed evidence that the observed polymorphism might be trending natural selection. Tajima's D value was -0.084549 with P > 0.10. The Fu and Li, D and F values were -0.92031 with P > 0.10 and -0.7654 with P > 0.10, respectively.
Phylogenetic tree of PvDBPII
Phylogenetic tree analysis of DNA sequences derived from region II of the P. vivax DBP constructs based on the Neighbour-Joining method using Tamura's three-parameter distance is shown in Figure 3. Variation of DNA sequence was categorized into nine distinct alleles groups. Thai isolates were widely distributed amongst groups 1, 3, 5 and 7 also containing isolates from different geographical regions. PvDBPII DNA sequence of Thai isolates was related to isolates sets from Papua New Guinea and India (group 1), Korea (group 3), Colombia (group 5) and a second Papua New Guinea set (group 7). The Sal I reference strain used in this study was included in group 2 composed of isolates from Colombia, Korea, Bangladesh, Indonesia, and Vietnam, but bootstrap analysis demonstrated that the Thai isolates were distinct from this group. Most isolates from Papua New Guinea formed a distinctive group comprised of 8 and 9.
Figure 3.
Phylogenetic tree of PvDBPII obtaining from Thai P. vivax isolates and other isolates from different geographical malaria endemic areas, i.e. PNG (Papua New Guinea), India, COL (Colombia), Korea, Bangladesh, Brazil, Indonesia, Vietnam, and Sal I. Numbers at nodes indicate percentage support of 1000 bootstrap replicates (bootstrap support values below 10 are not presented).
Discussion
The process invasion into erythrocyte is essential for survival of P. vivax parasite and requires DBP to bind the Duffy blood group antigen that act as erythrocyte receptor, Duffy antigen receptor for chemokines (DARC). Plasmodium vivax utilizes genetic diversity of individual functional proteins to evade the host immune system and resist many anti-malarial drugs [21]. In this study, polymorphisms and natural selection was assessed and suggested positive natural selection in PvDBPII of Thai isolates. Although, non significant could be observed trending positive selection. As this is only a glimpse at the overall population, a wide range study including more subjects from different areas of Thailand would give a more detailed picture.
Analysis of 300 amino acids of PvDBPII protein relative to the Sal I reference strain, resulted in 25 nonsynonymous mutations being found. In addition, 142 nonsynonymous mutations were reported in 76 Papua New Guinea isolates [22]. The nonsynonymous mutations in PvDBPII might have an effect on parasite binding to erythrocyte receptors. Nonsynonymous mutations often times result in amino acid substitutions that alter charge and therefore host immune/antibody recognition [6]. Thus, excessive stable polymorphism generation within the PvDBPII ligand domain from high rate of nonsynonymous polymorphisms may promote the parasite escape of host immune response [9].
The high frequency (>50%) of L424I (86%), D384G (76%), W437R (63%), R390H (56%), and I503K (56%) residues were found in Thai isolates relative to Sal I PvDBP sequence. The finding is in contrast to previous studies showing R308S (67%), D384G (66%), and S447K (59%) in Papua New Guinea, D384G (59%) in Colombia and D384G (85%) in Brazil isolates [23]. Of the following variants N417K, W437R and I503K, all having been shown to be involved in evasion to antibody neutralization, variant N417K was found in low frequency (36%) while W437R and I503K were high (63% and 56% respectively) in Thai isolates. These results were different from what has been currently reported in that the low frequencies of variants N417K (27.5%) and W437R (27.5%) and the high frequency of variant I503K (55%) found in Brazil and low frequencies of all three variants (N417K, W437R, I503K) found in either Colombia (47%, 18%, 12%, respectively) and/or Papua New Guinea (23%, 26%, 29%, respectively) (Table 4) [23]. N417K, W437R and I503K have recently been shown to not be directly involved in erythrocyte binding, but possibly in inhibition of antibody binding to erythrocyte receptor [24]. Moreover, the variant amino acid T404R was identified in PvDBPII domains of three Thai isolates (Table 2) which is similar to the recent data in Papua New Guinea [22]. Other Thai variants, namely R308S, K371E, D384G, E385K, N386(N/Q), R390H, T404R, N417K, L424I, W437R, S447S and I503K, were also found in isolates of other regions, demonstrating commonality with a progenitor clone (Table 4). The other 18 polymorphic residues of PvDBPII ligand domain were recognized only among Thai isolates, indicative of divergence from a progenitor clone.
Table 4.
Frequencies of the most common variant amino acids in PvDBPII, comparing to Sal I sequence (accession number M37519)
| # | R308S | D384G | K386N | K386Q | N417K | L424I | W437R | S447K | I503K |
| Thai | 26 | 76 | 40 | 3 | 36 | 86 | 63 | 0 | 56 |
| COL* | 0 | 59 | 23 | 0 | 47 | 47 | 18 | 0 | 12 |
| PNG* | 67 | 66 | 8 | 11 | 23 | 34 | 26 | 59 | 29 |
| Brazil* | 12.5 | 85 | 12.5 | 0 | 27.5 | 32.5 | 27.5 | 0 | 55 |
* The frequencies of the common variant amino acid data were referable according to Sousa et al. 2006.
# The first letter represented the amino acid in that position in Sal I sequences, and the other letter represents the substituted amino acid.
COL: Colombia, PNG: Papua New Guinea
The study showed that 25 PvDBPII haplotypes clustered into five main groups (dominant haplotypes) (Table 3) on the basis of nonsynonymous mutations is similar, but not entirely the same, with data from Papua New Guinea isolates whose 27 different PvDBPII haplotypes clustering into three dominant PvDBPII haplotypes. Potentially dominant PvDBPII haplotypes observed in these populations could be less immunogenic, thereby may help promoting parasite success and escaping the host immune response [22].
Phylogenetic analysis of PvDBPII suggested that Thai isolates fell into 6 different alleles groups. Among these, group 1 formed a group with a subset of Papua New Guinea isolates. Groups 4 and 6 were unique among Thai P. vivax isolates. However, the rest of the allele groups that Thai isolates fell into were more related to the isolates from Korea, India, and Colombia than from Papua New Guinea (Figure 3). The observations derived from the PvDBPII phylogeny suggested that single Indian isolate found in group 1 appeared to share a common ancestor close to a subset of Thai and Papua New Guinea isolates, demonstrating common ancestoral origins. Thai and Papua New Guinea isolate subsets in group 7 also may have shared common ancestor heritage. Recently, it was reported that travellers' malaria among foreigners at the Hospital for Tropical Diseases, Bangkok, Thailand including India and Papua New Guinea patients were though to have acquired their infections in their countries. Malaria importations from country to country can occur by either immigration or travel, and changing malaria attack rates in the countries of exposure are likely to influence the incidence of imported disease [25]. Additionally, these results demonstrated that there is some difference in the nucleotide or amino acid variation in PvDBPII among isolates from Thai, Korea, India, Papua New Guinea and Colombia, but most importantly, unique variants from Thailand do exist and should be considered in future vaccine development of PvDBPII. It should be noted that the present highly diverse phylogenetic tree was constructed relying on polymorphisms within a single gene, demonstrating the high degree of topological variation that can be found using this technique, but also highlighting the problems that can be associated with gene sampling in phylogenetic studies [26]. Use of a combination of several genes with different rates of evolutions has been suggested as an efficient way to overcome this difficulty to prove the existence of a unique tree relating these sequences [27]. Finally, it must also be pointed out that in many situations a single-gene phylogeny may be interesting in itself. Awareness of the problems of orthology (genes in different species that derive from a common ancestor) assignment and tree reconstruction artifacts should be considered.
Conclusion
The results indicated that PvDBPII gene among Thai isolates is genetically diverse. Nonsynonymous polymorphism in PvDBPII was the predominant result of which variant L424I showed the highest frequency (86%). Five dominant PvDBPII haplotypes could be clustered among Thai isolates. The high frequency of polymorphisms and the presence of distinct alleles within PvDBPII gene among P. vivax from different geographical areas could provide complication in malaria vaccine development, but should be considered important parts of the development process.
List of abbreviations used
PvDBPII: Domain II of Plasmodium vivax DBP gene; DARC: Duffy antigen receptor for chemokines
Authors' contributions
SRM and SK designed the study, SK, MMF, SRM, KES, DEL and TK, were responsible for the supervision of the work, KES, TK, DEL, and PG were responsible for the sequences and phylogenetic analysis, SK, MMF, SRM, KES, DEL, TK, and PG drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank the patients for their kind participation in this study; staff of the Hospital for Tropical Diseases, Faculty of Tropical Medicine, Mahidol University for their assistance in blood collection and COL Douglas S. Walsh, US Army Medical Research Unit-Kenya for his valuable guidance and comments. This study was supported by the United States Army Medical Research and Materiel Command, Fort Detrick, MD and Faculty of Tropical Medicine, Mahidol University.
Disclaimer
The views of the authors are solely there own, and do not reflect official opinion of the United States Army Medical Research and Materiel Command or the Department of Defense.
Contributor Information
Panita Gosi, Email: panitag@afrims.org.
Srisin Khusmith, Email: tmskm@mahidol.ac.th.
Thareerat Khalambaheti, Email: tmtkl@mahidol.ac.th.
David E Lanar, Email: david.lanar@us.army.mil.
Kurt E Schaecher, Email: Kurt.Schaecher@afrims.org.
Mark M Fukuda, Email: Mark.Fukuda@afrims.org.
Scott R Miller, Email: robert.s.miller@amedd.army.mil.
References
- Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci USA. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang XD, Kaslow DC, Adams JH, Miller LH. Cloning of the Plasmodium vivax duffy receptor. Mol Biochem Parasitol. 1991;44:125–132. doi: 10.1016/0166-6851(91)90228-X. [DOI] [PubMed] [Google Scholar]
- Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. The domain on the duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med. 1996;184:1531–1536. doi: 10.1084/jem.184.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Chitnis CE. Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc Natl Acad Sci USA. 1999;96:14067–14072. doi: 10.1073/pnas.96.24.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kappe SH, al-Yaman F, Prickett MD, Alpers M, Adams JH. Natural variation within the principal adhesion domain of the Plasmodium vivax duffy binding protein. Infect Immun. 1994;62:5581–5586. doi: 10.1128/iai.62.12.5581-5586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole-Tobian JL, Cortes A, Baisor M, Kastens W, Xainli J, Bockarie M, Adams JH, King CL. Age-acquired immunity to a Plasmodium vivax invasion ligand, the duffy binding protein. J Infect Dis. 2002;186:531–539. doi: 10.1086/341776. [DOI] [PubMed] [Google Scholar]
- Xainli J, Adams JH, King CL. The erythrocyte binding motif of Plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol Biochem Parasitol. 2000;111:253–260. doi: 10.1016/S0166-6851(00)00315-7. [DOI] [PubMed] [Google Scholar]
- Ampudia E, Patarroyo MA, Patarroyo ME, Murillo LA. Genetic polymorphism of the duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol Biochem Parasitol. 1996;78:269–272. doi: 10.1016/S0166-6851(96)02611-4. [DOI] [PubMed] [Google Scholar]
- Cole-tobian JL, King CL. Diversity and natural selection in Plasmodium vivax duffy binding protein gene. Mol Biochem Parasitol. 2003;127:121–132. doi: 10.1016/S0166-6851(02)00327-4. [DOI] [PubMed] [Google Scholar]
- Rich SM, Hudson RR, Ayala FJ. Plasmodium falciparum antigenic diversity: evidence of clonal population structure. Proc Natl Acad Sci USA. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Daugherty JR, Ware LA, Lanar DE, Ockenhouse CF. Expression, purification and characterization of a functional region of the Plasmodium vivax duffy binding protein. Mol Biochem Parasitol. 2000;109:179–184. doi: 10.1016/S0166-6851(00)00244-9. [DOI] [PubMed] [Google Scholar]
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Jukes TH. The neutral theory of molecular evolution. Genetics. 2000;154:956–958. doi: 10.1093/genetics/154.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor CR, Fields CA. Genome sequencing conference. III: Evolution and progress. Genomics. 1992;12:419–420. doi: 10.1016/0888-7543(92)90397-B. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM. Phylogenetic analysis. Curr Biol. 1997;7:R129–131. doi: 10.1016/S0960-9822(97)70070-8. [DOI] [PubMed] [Google Scholar]
- VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, King CL, Adams JH. Antigenic drift in the ligand domain of Plasmodium vivax duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis. 2004;190:1556–1562. doi: 10.1086/424852. [DOI] [PubMed] [Google Scholar]
- Cole-Tobian JL, Michon P, Dobod E, Mueller I, King CL. Dynamics of asymptomatic Plasmodium vivax infections and duffy binding protein polymorphisms in relation to parasitemia levels in Papua New Guinean children. Am J Trop Med Hyg. 2007;77:955–962. [PubMed] [Google Scholar]
- Sousa TN, Ceravolo IP, Fontes CJF, Couto A, Carvalho LH, Brito CFA. The pattern of major polymorphisms in the duffy binding protein ligand domain among Plasmodium vivax isolates from the Brazilian Amazon area. Mol Biochem Parasitol. 2006;146:251–254. doi: 10.1016/j.molbiopara.2005.11.006. [DOI] [PubMed] [Google Scholar]
- VanBuskirk KM, Sevova E, Adams JH. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc Natl Acad Sci USA. 2004;101:15754–15759. doi: 10.1073/pnas.0405421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyaphanee W, Krudsood S, Silachamroon U, Pornpininworakij K, Danwiwatdecha P, Chamnachanan S, Wilairatana P, Looareesuwan S. Travelers' malaria among foreigners at the Hospital for Tropical Diseases, Bangkok, Thailand – a 6-year review (2000–2005) Korean J Parasitol. 2006;44:229–232. doi: 10.3347/kjp.2006.44.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Topological variation in single-gene phylogenetic trees. Genome Biol. 2007;8:216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hypsa V. Parasite histories and novel phylogenetic tools: alternative approaches to inferring parasite evolution from molecular markers. Int J Parasitol. 2006;36:141–155. doi: 10.1016/j.ijpara.2005.10.010. [DOI] [PubMed] [Google Scholar]