Abstract
Smad-interacting protein-1 (SIP1) has been implicated in the development of Mowat-Wilson syndrome whose patients exhibit Hirschsprung disease, an aganglionosis of the large intestine, as well as other phenotypes. We have identified and cloned two sip1 orthologues in zebrafish. Both sip1 orthologues are expressed maternally and have dynamic zygotic expression patterns that are temporally and spatially distinct. We have investigated the function of both orthologues using translation and splice-blocking morpholino antisense oligonucleotides. Knockdown of the orthologues causes axial and neural patterning defects consistent with the previously described function of SIP1 as an inhibitor of BMP signaling. In addition, knockdown of both genes leads to a significant reduction/loss of the post-otic cranial neural crest. This results in a subsequent absence of neural crest precursors in the posterior pharyngeal arches and a loss of enteric precursors in the intestine.
Keywords: Zebrafish, Enteric Nervous System, SIP1, Mowat-Wilson Syndrome, Neural Crest
Introduction
Hirschsprung disease (HSCR) (MIM 142623) is a human condition, affecting 1 in 5000 live births, in which patients have aganglionosis of the distal colon (Amiel and Lyonnet, 2001). The disease phenotype results from defects in the normal development of vagal neural crest derived ENS precursors, that subsequently leads to a lack of submucosal and myenteric ganglion cells in the distal part of the gastrointestinal tract. This lack of ENS results in gut obstruction after birth. HSCR is a complex genetic disorder with multiple loci giving rise to the condition. HSCR is usually non-syndromic, but in some cases HSCR occurs as a characteristic phenotype of a larger syndrome (Amiel and Lyonnet, 2001). Haploinsufficiency in the gene which encodes Smad-interacting protein-1 (SIP1; also designated ZFHX1B or ZEB2 (Postigo, 2003)) has been found to cause Mowat-Wilson syndrome (MWS) (MIM 235730), whose patients exhibit HSCR (64% of the cases) along with mental retardation (100% of the cases), cranio-facial dismorphology (100% of the cases), and heart defects (50 % of the cases) (Cacheux et al., 2001; Dastot-Le Moal et al., 2007). To date over 100 sip1 mutations have been described in patients with clinically typical MWS (Wakamatsu et al., 2001; Dastot-Le Moal et al., 2007).
SIP1 is one of the two members of the vertebrate ZFHX1 family (Postigo and Dean, 2000). This family encodes zinc finger and homeodomain/homeodomain-like containing proteins that act primarily as transcriptional repressors (Verschueren et al., 1999; Comijn et al., 2001; (van Grunsven et al., 2003; Vandewalle et al., 2005) but can also act as transcriptional activators in vivo (Long et al., 2005; Yoshimoto et al., 2005). SIP1 was originally identified in mouse by a yeast two-hybrid screen for proteins that bind Smad1 (Verschueren et al., 1999). It interacts with the Bone Morphogenetic Protein (BMP)/TGFβ superfamily signaling pathway by binding the MH2 domain of certain Smads and represses transcription of downstream target genes (Verschueren et al., 1999). SIP1 binds to the activated forms of Smad1 and 5, which are BMP targets, as well as Smad 2 and 3, which are TGFβ/activin/nodal targets (Remacle et al., 1999; Verschueren et al., 1999; Postigo, 2003). It has also been shown that constructs containing the SIP1 CtBP interaction domain (CID) can recruit the co-repressor CtBP (carboxyl terminal binding protein) to the Smad complex in vitro (Postigo et al., 2003). Furthermore, SIP1 has been shown to be present in a CtBP affinity–purified complex (Shi et al., 2003). Finally, SIP1 can also bind the co-activators P300 and pCAF (p300/CBP associated factor) (van Grunsven et al., 2006).
Numerous studies have shown that SIP1 is involved in neural specification, primarily via transcriptional inhibition of the BMP pathway (Eisaki et al., 2000; Van de Putte et al., 2003; Nitta et al., 2004; Nitta et al., 2007; van Grunsven et al., 2007). This is in accordance with the well established fact that attenuation of BMP signaling in the ectoderm is required for the formation of neural tissue (Vonica and Brivanlou, 2006). Recent studies in xenopus have shown that XSIP1 binds directly to the BMP4 proximal promoter and can modulate its activity (van Grunsven et al., 2007). Furthermore, in vitro structure function studies of XSIP1 have determined that the suppression of BMP signaling is mediate through its N-terminus zinc finger domain (Nitta et al., 2007). In addition, overexpressed SIP1 is a direct transcriptional repressor of brachyury (Verschueren et al., 1999), a regulator of mesodermal formation during gastrulation (Smith, 2004). It has also been proposed that sip1 is a direct target of Churchill, a regulator of FGF signaling that indirectly blocks mesodermal formation (Sheng et al., 2003). Thus, SIP1 may induce neural fate in the neurectoderm by suppressing BMP signaling and repressing mesodermal cell fate.
SIP1 can also modulate the Wnt signaling pathway as demonstrated by a recent study in mouse (Miquelajauregui et al., 2007). This study showed that the Wnt antagonist Sfrp1 was ectopically activated when sip1 was specifically inactivated in mouse cortical precursors. By contrast the activity of the noncanonical Wnt effector, JNK, was down regulated in the developing hippocampus of these mutant mice. The investigators were also able to identify a SIP1 protein/Sfrp1 DNA complex using chromatin immunoprecipiation (Miquelajauregui et al., 2007).
SIP1 has also been shown to have an important role in epithelial-mesenchymal transition (EMT). EMT is required during embryogenesis in a number of different developmental processes including gastrulation and neural crest formation (Le Douarin, 1982; Selleck MA, 1996; Pla et al., 2001). EMT is also a critical step in metastasis of tumors (Thiery, 2002). For EMT to occur, homophilic E-cadherin-mediated cell-cell adhesion contacts must dissociate. SIP1, along with several other transcriptional repressors, such as Snail, E12/E47 and Twist, mediate EMT by repressing the transcription of E-cadherins (Cano et al., 2000; Comijn et al., 2001; Perez-Moreno et al., 2001; Guaita et al., 2002; Yang et al., 2004; Sivertsen et al., 2006). In human gastric carcinomas, a statistically significant association has been found between Slug up-regulation and the expression of SIP1 and Snail. This suggests that Slug may act synergistically with SIP1 and Snail in the down regulation of E-cadherin (Come et al., 2006; Castro Alves et al., 2007).
sip1 orthologues have been cloned in drosophila, xenopus, chick, mouse, and human (Fortini et al., 1991; Verschueren et al., 1999; Eisaki et al., 2000; van Grunsven et al., 2000; Cacheux et al., 2001; Tylzanowski et al., 2003; Liu et al., 2006). The drosophila sip1orthologue, known as zfh-1, shows a complex embryonic expression pattern in the mesoderm and the nervous system (Fortini et al., 1991). Phenotypic analysis of zfh-1 mutant fly embryos has revealed that this gene is required for the proper differentiation of a number of mesoderm-derived tissues including the heart (Lai et al., 1993; Broihier et al., 1998; Su et al., 1999). Recently, it has been shown that expression of mouse sip1 in zfh1 mutant flies can rescue normal heart development, demonstrating some potential functional conservation of SIP1 biological activities across vertebrate and invertebrate species (Liu et al., 2006).
In xenopus, sip1 is strongly expressed in neural tissue and the neural crest (van Grunsven et al., 2000). Knock down of XSIP1 inhibits SOXD expression and ultimately inhibits neural differentiation (Nitta et al., 2004). In mouse, SIP1 has been shown to be required for the specification of vagal neural crest and is required for the normal migratory behavior of cranial neural crest cells. sip1 knockout mice die at embryonic age 9.5 due to cardio-vascular dysfunction. Additionally, the neural tube fails to close in these mice and they have shortened somites (Van de Putte et al., 2003; Maruhashi et al., 2005). More recently a conditional SIP1 knockout mouse has been generated in which zfhx1b function was specifically removed in the neural crest. These mice have specific abnormalities in craniofacial, heart and melanocyte development, as well as defects in the ENS and the sympatho-adrenal lineage, which are reminiscent of Mowat-Wilson syndrome in humans (Van de Putte et al., 2007). In early human embryos, the sip1 homologue (zfhx1b) is expressed in neural crest derived cells, including ENS and facial neurectoderm, as well as in the central nervous system and other tissues (Espinosa-Parrilla et al., 2002).
In this study we report the identification of two zebrafish orthologues of sip1. We have performed expression analysis and morpholino knockdown in order to characterize the role of SIP1 in development of the zebrafish nervous system and cranial neural crest derived structures.
Results and Discussion
Two zebrafish orthologues were identified from a TBLAST X search of the zebrafish genome. We used RT-PCR to isolate fragments of these predicted genes followed by 3′ and 5′ RACE to identify the complete ORF. The two predicted zebrafish SIP1 proteins share 64.5% of homology. Comparison of their predicted amino acid sequence with that of human sip1 (zfhx1b), using Clustal W method (DNA star; MegAlign) revealed that sip1b is more similar to ZFHX1B than sip1a (sip1b has a 72.8% homology, while sip1a has a 61.9% homology). Further analysis of the functional domains in the zebrafish sip1 orthologues show that both the amino and the carboxy terminal zinc finger domains are highly conserved (respectively 90% and 98.75% sequence homology for sip1a and 95% and 100% for sip1b). Both zebrafish orthologues contain the previously identified homeodomain-like domain, but there is a higher homology in sip1b (94.2%) than in sip1a (61.5%). Similarly, a CtBP binding domain is found in both orthologues but the sequence conservation is weaker. sip1a has a 42.3% homology in this domain, while sip1b has a 71.1% homology. Finally, the proposed Smad-binding domain is also present in both sip1a and sip1b. There is a 79.8% homology in this domain for sip1a and a 86.1% homology for sip1b. It remains to be tested whether these amino acid differences in the functional domains result in different functional activities for the zebrafish sip1 orthologues.
RT-PCR analysis of sip1a and sip1b expression shows that both genes are expressed maternally and zygotically through 96hpf, the latest embryonic age we examined in this study. RT-PCR using primers flaking exon9 revealed that there are two alternate transcripts for both sip1 genes (Fig. 1A). This alternative splicing of exon9 (Amino acid 1032 to 1057, Supplemental Fig. 1, orange box) adds or removes a zing finger in the carboxy terminal zinc finger domain and is likely to have functional significance.
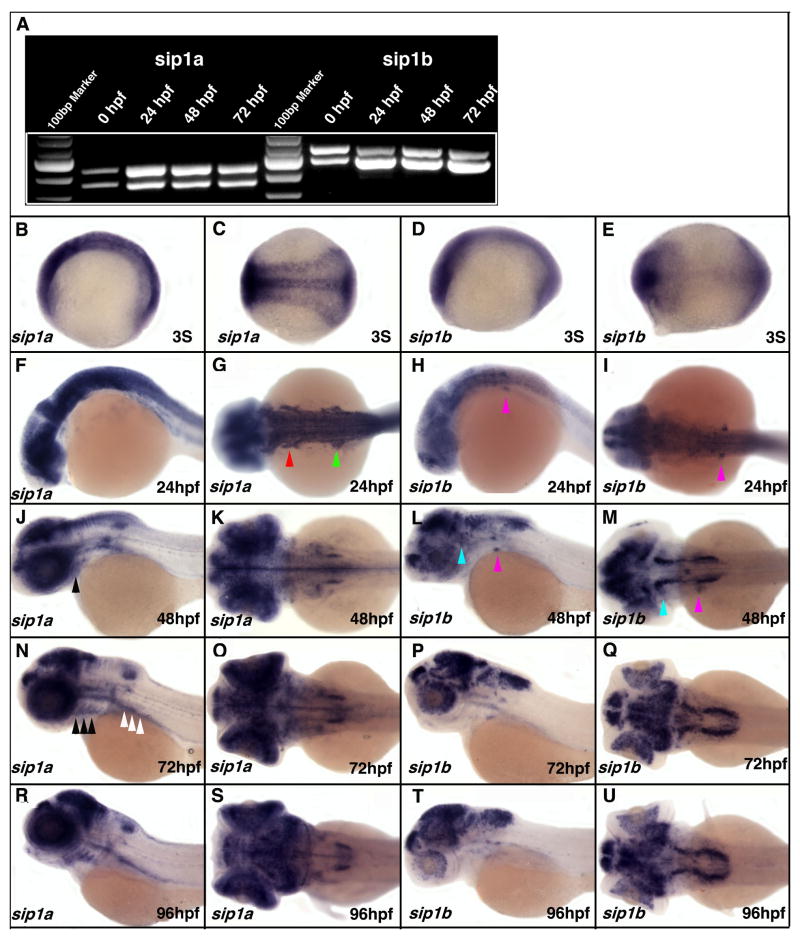
Figure 1. RT-PCR and in situ analysis of the spatial and temporal expression pattern of zebrafish sip1a and sip1b.
(A) RT-PCR of zebrafish sip1a and sip1b with primers flaking Exon9 using mRNA isolated from wild type embryos at 0, 24, 48 and 72hpf. Each gene displayed two bands corresponding to two alternatively spliced mRNAs. (B–U) Wholemount in situ hybridized embryos hybridized with either a sip1a (B, C, F, G, J, K, N, O, R, S) or sip1b (D, E, H, I, L, M, P, Q, T, U) antisense probes at the indicated developmental stages. The first and third columns are lateral views; the second and fourth columns are dorsal views. Anterior is to the left. Red arrowhead indicate pre-otic neural crest (G), green arrowhead indicate post-otic neural crest (G), pink arrowheads indicate VII/VIII cranial ganglia (H, I, L, M), light blue arrowheads indicate V cranial ganglia (L, M), black arrowheads indicate pharyngeal arches (J, N), white arrow heads indicate gut mesendoderm (N).
To determine the spatial expression pattern of sip1a and sip1b we undertook wholemount in situ hybridization study. At the 3 somite stage, the earliest stage examined in this study, the strongest expression of sip1a is in the anterior- and posterior-most parts of the embryo, as well as in the midline and lateral edges of the neuroectoderm (Fig. 1B, C). At this stage sip1b is predominantly expressed in the anterior- and posterior-most parts of the embryo (Fig. 1D, E). By 20–24hpf, expression of both genes is mainly restricted to the CNS, with higher expression in the anterior part (Fig. 1F–G). The high level of CNS expression of both genes continues through out all stages examined, however, from 36hpf sip1b becomes more specifically localized to discrete fore, mid and hindbrain regions, while sip1a becomes more discretely localized in the CNS around 48–72hpf. While the two genes have overlapping expression in some CNS regions, much of their expression pattern in other tissues and in the PNS is distinct. sip1a is strongly expressed in the pre- and post-otic neural crest at 24hpf, whereas sip1b is only present in the post-otic stream (Fig. 1 F–I; arrowheads). sip1b is, however, expressed in the VII/VIII cranial ganglia from 24hpf and the V ganglia by 48hpf, while sip1a is never specifically expressed in the cranial ganglia (Fig. 1F-M). A significant difference in expression is also seen in the pharyngeal arches where sip1a is expressed from 48hpf, while sip1b is not expressed at any stage (Fig. 1J, L, N, P, R, T). Similarly sip1a is expressed in the gut mesendoderm from 48hpf, while sip1b is not (Fig. 1J, L, N, P, R, T). Given the patterns of expression of sip1a and sip1b during early embryogenesis, it is possible that they both may be involved in axis formation and embryonic patterning by regulating BMP/TGFβ signaling. Later in embryogenesis, their differing expression patterns suggest that sip1a and sip1b have distinct roles in subsequent aspects of zebrafish development.
To determine the in vivo function of the zebrafish orthologues, we generated and injected both translation blocking morpholinos (TBMO) and splice-blocking morpholinos (SBMO). Similar phenotypes were obtained for both types of morpholinos but given the predominant zygotic expression of both orthologues, based on RT-PCR expression results (Fig. 1A ), and the ability to monitor the splice-blocking activity by RT-PCR, we focused on the SBMO morphant phenotypes.
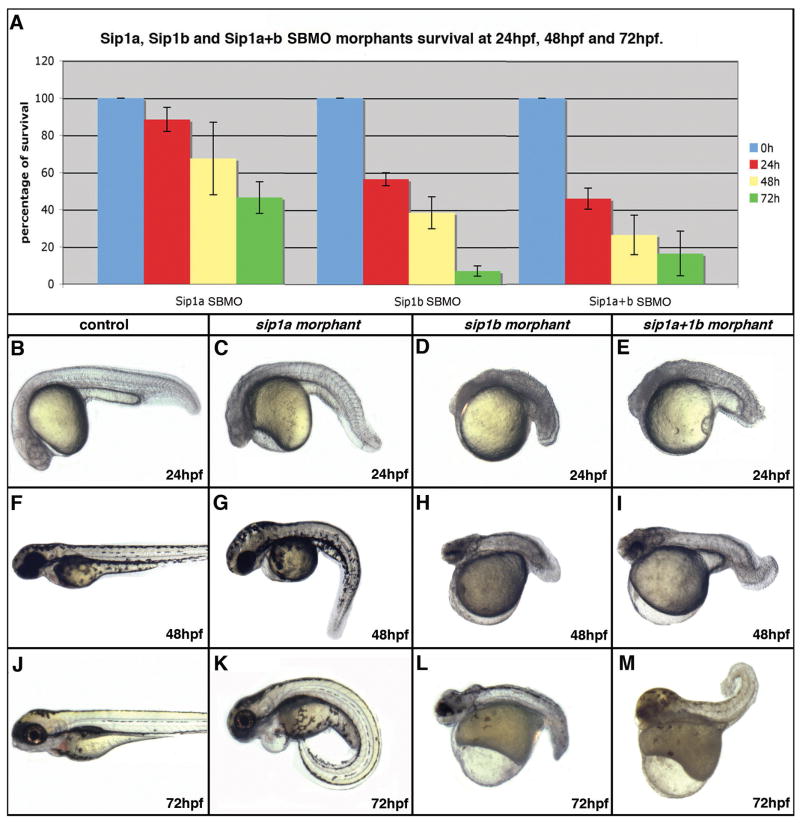
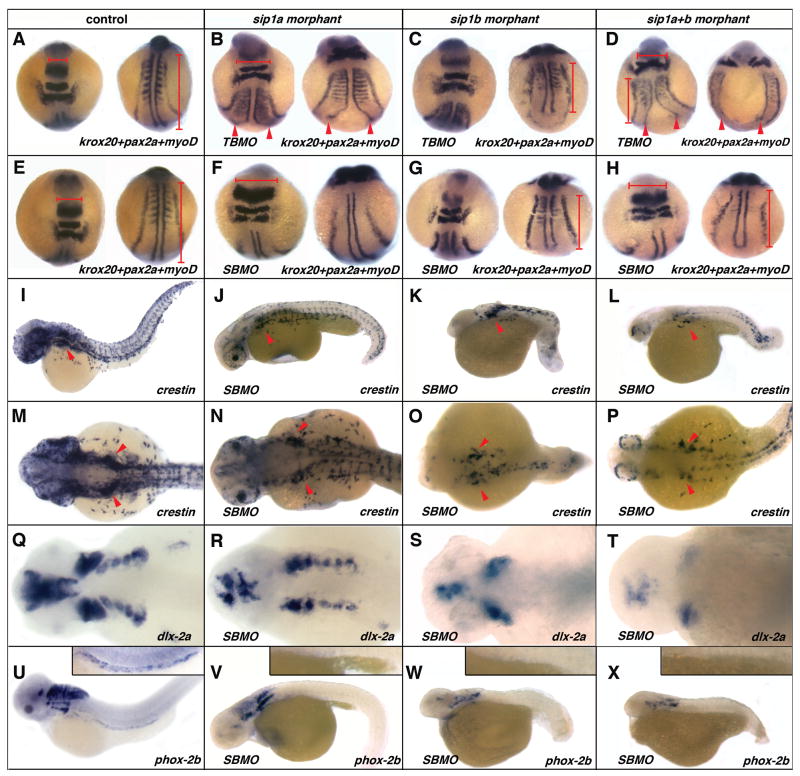
sip1a and sip1b morphants have early embryonic patterning defects that result in high mortality at an early stage of development. 50% of sip1a morphants, 90% of sip1b morphants and 85% of sip1a plus sip1b morphants are dead by 72hpf (Fig. 2A). sip1 morphants have defects in anterior CNS and tailbud formation (Fig. 2B–M). Given these axis patterning defects, both genes are likely to be involved in the patterning of the early embryo. To further investigate the axial patterning defects in sip1 morphants, we performed in situ hybridization on 8 somite stage embryos using a mix of krox20-pax2a-myod riboprobes. sip1a TBMO morphants have defects in the midline and are consistently flattened when compared with controls, suggesting a role for sip1a in convergence during early embryogenesis (Fig. 3A,B). This result is also consistent with the pattern of sip1a expression at the 3 somite stage (Fig. 1B, C). sip1b TBMO morphants have comparatively normal midline convergence but are consistently shorter as compared to controls, suggesting that sip1b is required for normal extension. Moreover, myod in situ revealed that sip1b TBMO morphants also have misshaped somites, suggesting a role for sip1b in early myogenesis (Fig. 3A,C). At this stage, double TBMO morphants have a combination of both sip1a and sip1b phenotypes, with convergence defects, in particular in the midline, as well as a shortened axis and somites defects (Fig. 3 D). These results are in accordance with previous studies that have shown that (i) convergence-extension in zebrafish is regulated by a gradient of BMP activity (Myers et al., 2002) and that (ii) Sip1 is a inhibitor of BMP signaling (Eisaki et al., 2000; Van de Putte et al., 2003; Nitta et al., 2004; Nitta et al., 2007; van Grunsven et al., 2007).
Figure 2. Effect of sip1a and sip1b splice blocking antisense morpholino oligonucleotide injection on survival and morphological development at 24–72hpf.
(A) A bar graph showing the percent of surviving sip1a SBMO morphants, sip1b SBMO morphants and sip1a sip1b double morphants at 24, 48 and 72 hpf. The numbers represent the percent of injected embryos surviving at each specific time point ± s.e.m. based on 3 independent experiments. (B–M) Lateral views of control (B, F, J) sip1a morphant (C, G, K) sip1b morphant (D, H, L) and sip1a sip1b double morphant (E, I, M) embryos at 24hpf (B–E), 48hpf (F–I) and 72 (J–M) hpf. Anterior is to the left.
Figure 3. Effect of sip1a and sip1b translation and splice blocking morpholino antisense oligonucleotide injection on axial patterning, and neural crest specification and migration.
(A, E, I, M, Q, U) wild-type control, (B) sip1a TBMO morphant, (F, J, N, R, V) sip1a SBMO morphant, (C) sip1b TBMO morphant, (G, K, O, S) sip1b SBMO morphant embryos and (D, H, L, P, T) sip1a sip1b double morphant embryos. (A–H) dorsal views of 3 somite stage embryos that have been hybridized with a krox20, pax2a and myoD riboprobes mix. (I–L) Lateral views of 36hpf embryos that have been hybridized with riboprobes for crestin. (M–P) Lateral and (Q–T) dorsal views of 48hpf embryos that have been hybridized with riboprobes for dlx-2a. (U–X) Lateral views of 55hpf embryos that have been hybridized with riboprobes for phox2b. Insert in (U–X) are close ups of the intestine of these embryos. Red arrowheads (B–D) point to convergence defects in the midline. Red horizontal bars (A, B, D, E, F, H) highlight flattening of the morphant embryos in the hindbrain region, as compared to the control. Red vertical bars (A, C, D, E, G, H) highlight shorter axis in morphant embryos, as compared to control. Red arrowheads (I–P) indicate the vagal/post-otic neural crest. Anterior is to the left.
sip1a SBMO morphants have a much milder phenotype than TBMO morphants. sip1a SBMO morphants have an abnormally curved axis, abnormal CNS development, as well as heart defects at later stages of development (Fig. 2C, G, K; Fig. 3J, V). In situ analysis of 8 somites stage morphants has revealed that there is no midline convergence defect in sipa1a SMBO injected embryos as compared to TBMO injected embryos, suggesting that sip1a maternal transcripts are critically required for normal midline convergence. sip1a SBMO are, however, still flattened compared to the controls, which suggests that both maternal and zygotic sip1a are involved in regulation of convergence movements (Fig. 3F–H). Finally, sip1a morphants also display somites defects, as seen by myod in situ (Fig. 3F–H).
sip1b morphants (TBMO and SBMO) are consistently shorter compared to sip1a morphants or controls, suggesting a specific role for sip1b in axis extension (Fig. 2D, H, L; Fig. 3K, O, W). Furthermore, sip1b morphants have clear dorso-ventral patterning defects similar to embryos where BMP signaling is perturbed (Schier and Talbot, 2005). Co-injection of the sip1a and sip1b morpholinos produces a phenotype that is a combination of both indiviudal morpholino phenotypes (Fig. 2E, I, M; Fig. 3D, H, L, P, T, X). These results suggest that both zebrafish orthologues have acquired independent biological activities after they arose through the proposed teleost genome duplication event (Ohno, 1993; Postlethwait et al., 2000).
To determine if the sip1 genes have a role in neural crest formation and patterning in zebrafish, we examined crestin expression in morphant embryos. At 36hpf, crestin expression is perturbed in sip1 morphants. The disruption is greatest in the trunk region in sip1a morphants, while neural crest specification is severely perturbed at all axial levels in the sip1b morphants and sip1a sip1b double morphants (Fig. 3A–H). Further analysis of the specification and migration of the neural crest at later stages of development show that the initial migration and patterning of the neural crest in the pharyngeal arches is only slightly reduced in sip1a morphants, as determined by dlx-2a expression in 36hpf morphant embryos. By contrast, in sip1b morphants and sip1a and sip1b double morphants there is a complete loss of dlx-2a expression in the posterior pharyngeal arches consistent with the loss of vagal/post otic neural crest (Fig. 3Q–T).
To determine if the development of the ENS is abnormal in morphant embryos, the pattern of phox2b expression was examined in 55hpf embryos. Both sip1a and sip1b morphants as well as double morphants completely lack any phox2b expressing cells in the intestine and have also reduced or lack of phox2b expression in cranial ganglia (Fig 3 U–X). We were unable to determine if the later differentiation of the ENS precursors at 96hpf was similarly affected in the sip1 morphants due to the high level of embryonic death in the morphant embryos (Fig. 2A). Taken together these results suggest that sip1 genes are important for normal vagal/post-otic neural crest derivatives formation. This is consistent with the sip1 knockout mouse phenotype in which the vagal/post-otic neural crest is absent (Van de Putte et al., 2003; Van de Putte et al., 2007). The neural crest cells defects observed the SIP1 knockout mouse and zebrafish morphants may have arisen due to interference with BMP4 and −7 signaling. Signaling from both of these BMP ligands is required for the normal induction of neural crest precursors on the neural ridge (LaBonne and Bronner-Fraser, 1999; Van de Putte et al., 2007). Alternatively, the neural crest defects may also have derived from a failure of EMT in the neural tissue that gives rise to neural crest, a process that potentially requires SIP1 activity (Le Douarin, 1982; Selleck MA, 1996; Pla et al., 2001). Both hypotheses are not mutually exclusive.
In conclusion, we have identified and sequenced two orthologues of sip1 in zebrafish. These two orthologues most likely arose due to the proposed genome duplication event that occurred in the ray-finned fishes lineage (Ohno, 1993; Postlethwait et al., 2000). Amino acid sequence comparisons show that both orthologues contain all of the previously identified SIP1 functional domains (Verschueren et al., 1999). While the partially overlapping pattern of expression of both sip1 orthologues suggests that these genes have some common biological activities and may be partially functionally redundant, we have found that sip1a and sip1b play disctinct roles in axis formation at early embryonic stages. sip1a is required in convergence and midline formation, whereas sip1b is required for axis extension. In addition both morphants have somites defects. Interestingly, shortened somites have been reported in Sip1−/− mouse knockout (Maruhashi et al., 2005). We also found that both sip1 genes play a critical role in neural patterning and neural crest formation in zebrafish. These results are consistent with the expression pattern of sip1a and sip1b in the early nervous system and with SIP1,s biological function as a BMP signaling pathway inhibitor (Postigo et al., 2003). These results are also consistent with SIP1,s functions in neural development previously described in xenopus and mouse (Van de Putte et al., 2003; Nitta et al., 2004; Nitta et al., 2007; van Grunsven et al., 2007). Finally, the phenotypes observed in zebrafish sip1 morphants are consistent with those observed in Mowat-Wilson syndrome patients (Yamada et al., 2001). Future studies will permit a more thorough investigation of SIP1,s role in zebrafish embryogenesis, specifically with regard to its function in neural/neural crest development. These studies will potentially lead to a better understanding of the mechanisms that underlie the Mowat-Wilson syndrome clinical phenotypes.
Methods
Animal maintenance
Zebrafish are kept and bred under standard conditions at 28.5°C (Westerfield, 1993). Embryos were staged and fixed at specific hours post fertilization (hpf). To better visualize in situ hybridization results, embryos were grown in 0.2 mM 1-phenyl-2-thiourea (Sigma) to inhibit pigment formation (Westerfield, 1993).
Isolation of zebrafish SIP1a and SIP1b orthologues
A TBLAST X search was performed on the Zebrafish Genome Browser on the World Wide Web to identify zebrafish SIP1 orthologues using published mouse and human sequences. To clone the complete ORFs of the zebrafish orthologues, multiple RT-PCR primers were designed to amplify up 5′ and 3′ overlapping segments of the open reading frame based on the predicted sequences. The cDNA segments were subcloned and sequenced. Sequencher DNA sequence analysis software was used to assemble the resulting sequences. RACE (rapid amplification of cDNA ends) was used to amplify the 5′ and 3′ ends of the open reading frame. RACE cDNA was isolated from 72hpf embryos using a Smart RACE cDNA Amplification Kit (Clonetech). The resulting PCR products were subcloned and sequenced to complete the open reading frame sequence for the orthologues. The continuity of full-length sequence assembled from the sequences was confirmed by RT-PCR on single stranded cDNA isolated from 48 hpf embryos.
Sequence data has been submitted to GenBank (Sip1a EU379558 and Sip1b EU379559). Homology studies were completed using publicly accessible programs from SDSC Biology Workbench. ClustalW was used to align the amino acid sequences of the zebrafish, Xenopus, chick, mouse, and human orthologues. Boxshade was used to identify any homology between the sequences. Drawtree and Drawgram were used to conduct a phylogenetic analysis (supplemental data, Fig. 2).
Expression analysis
To determine the temporal expression of sip1a and sip1b, RT-PCR was performed at various time points with primers used to amplify up a segment of the open reading frame flanking Exon9 (supplemental data, Fig. 3). The following primers were used:
sip1a forward, 5′-GGCTTATACTTACGCAGCAGGA-3′
sip1a reverse, 5′-TGCAGTAGGAATATCGGTGGTT-3′
sip1b forward, 5′-CACACATGGCCTACACGTACGCGG-3′
sip1b reverse, 5′-GGTACGCCCGGTTCAGCAGCATCT-3′
The predicted fragments size were as follow: for sip1a 440bp with exon9 and 362bp without; sip1b 540bp with exon9 and 462bp without.
To determine the spatial expression patterns of sip1a and sip1b, antisense Digoxigenin-labeled probes for both genes were generated and whole-mount in situ hybridization was performed as described by Thisse et al. (1993).
sip1a and sip1b antisense oligonucleotide injections
Translation blocking morpholinos and splice-blocking morpholinos were generated and injected to determine the effects of knocking down SIP1 protein levels. (Nasevicius and Ekker, 2000). Morpholino antisense oligonucleotides (Gene Tools) with the following sequences were designed to correspond to the translational start sites:
sip1a morpholino 5′ GTCCGCATGGGCTCCGCTCCTTCAT 3′
sip1b morpholino 5′ GATCAGCTCCCGCATTGATAAACGT 3′
Splice-blocking morpholino antisense oligonucleotides (Gene Tools) were designed to the following sequences corresponding to the splice donor site at the predicted exon4/exon5 junction for sip1a and exon3/exon4 for sip1b:
sip1a splice blocking morpholino: 5′ ACAGTTGATTGCCTACCGTTTTCAT 3′
sip1b splice blocking morpholino: 5′ CTCAGACATTCTCACCGTTTTCCTC 3′
The morpholinos were diluted in sterile filtered water over a range of concentrations from 1μg/μl to 10μg/μl. Approximately 1nL of diluted morpholino was injected at the one to two-cell stage using a gas-driven microinjection apparatus to determine the effects of knocking down sip1a, sip1b or both. We determined the dilution of the morpholinos at which we saw a consistent knockdown of sip1a and sip1b was as follows: sip1a TBMO: 4μg/μl SBMO: 5μg/μl; sip1b TBMO: 5μg/μl SBMO: 7μg/μl. The same final concentrations of each morpholino were used in double morphant injections. The standard control morpholino from Genetools as well as two 5 base pair mismatch morpholinos (one for sip1a one for sip1b) were injected as a negative controls for the morpholino experiments.
5 bp mismatch morpholino antisense oligonucleotides (Gene Tools) were as follow:
sip1a 5bp MM morpholino: 5′ GTgCGgATGGGaTCCGaTCgTTCA 3′
sip1b 5bp MM morpholino: 5′ GATgAGgTCCaGCATTcATAAAgGT 3′
The following primers were designed for RT-PCR to verify the effectiveness of the splice-blocking morpholino:
sip1a forward, 5′-CCCGACATCTCTCTTCATGG-3′
sip1a reverse, 5′-GCCCCTTCATTTAGCAGTTG-3′
sip1b forward, 5′-TGGGGACCGATGTATCTCTG-3′
sip1b reverse, 5′-CCAGAACCCTGGTTGAGAAT-3′.
Whole-mount in situ hybridization
To further characterize the affect of the sip1a and sip1b morpholinos, whole-mount in situ hybridization was performed with makers for various tissues including forebrain, hindbrain, myogenesis, neural crest and ENS precursors. Digoxigenin-labeled riboprobes used in this study were synthesized from templates, linearized and transcribed with the appropriate polymerase. References for the markers used are as follow: crestin (Rubinstein et al., 2000), phox2b (Shepherd et al., 2004); dlx2a (Akimenko et al., 1994), krox20 (Oxtoby and Jowett, 1993) myoD (Weinberg et al., 1996), pax2a (Krauss et al., 1991). Whole-mount in situ hybridization was performed as described by Thisse et al. (1993).
Supplementary Material
Acknowledgments
We like to thank Andreas Fritz and Robert Esterberg for helpful discussion and comments on the manuscript. This work has been supported by the NIH under Grant No. 1R01 DK067285-01A1 awarded to Dr. Iain Shepherd, by the Howard Hughes Medical Institute under Grant No. 52003727 awarded to Meaghann Guyote through the SURE program, and by the SIRE Grant awarded by Emory University to Meaghann Guyote.
References
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel J, Lyonnet S. Hirschsprung disease, associated syndromes, and genetics: a review. J Med Genet. 2001;38:729–739. doi: 10.1136/jmg.38.11.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broihier HT, Moore LA, Van Doren M, Newman S, Lehmann R. zfh-1 is required for germ cell migration and gonadal mesoderm development in Drosophila. Development. 1998;125:655–666. doi: 10.1242/dev.125.4.655. [DOI] [PubMed] [Google Scholar]
- Cacheux V, Dastot-Le Moal F, Kaariainen H, Bondurand N, Rintala R, Boissier B, Wilson M, Mowat D, Goossens M. Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum Mol Genet. 2001;10:1503–1510. doi: 10.1093/hmg/10.14.1503. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Castro Alves C, Rosivatz E, Schott C, Hollweck R, Becker I, Sarbia M, Carneiro F, Becker KF. Slug is overexpressed in gastric carcinomas and may act synergistically with SIP1 and Snail in the down-regulation of E-cadherin. J Pathol. 2007;211:507–515. doi: 10.1002/path.2138. [DOI] [PubMed] [Google Scholar]
- Come C, Magnino F, Bibeau F, De Santa Barbara P, Becker KF, Theillet C, Savagner P. Snail and slug play distinct roles during breast carcinoma progression. Clin Cancer Res. 2006;12:5395–5402. doi: 10.1158/1078-0432.CCR-06-0478. [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Dastot-Le Moal F, Wilson M, Mowat D, Collot N, Niel F, Goossens M. ZFHX1B mutations in patients with Mowat-Wilson syndrome. Hum Mutat. 2007;28:313–321. doi: 10.1002/humu.20452. [DOI] [PubMed] [Google Scholar]
- Eisaki A, Kuroda H, Fukui A, Asashima M. XSIP1, a member of two-handed zinc finger proteins, induced anterior neural markers in Xenopus laevis animal cap. Biochem Biophys Res Commun. 2000;271:151–157. doi: 10.1006/bbrc.2000.2545. [DOI] [PubMed] [Google Scholar]
- Espinosa-Parrilla Y, Amiel J, Auge J, Encha-Razavi F, Munnich A, Lyonnet S, Vekemans M, Attie-Bitach T. Expression of the SMADIP1 gene during early human development. Mech Dev. 2002;114:187–191. doi: 10.1016/s0925-4773(02)00062-x. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Lai ZC, Rubin GM. The Drosophila zfh-1 and zfh-2 genes encode novel proteins containing both zinc-finger and homeodomain motifs. Mech Dev. 1991;34:113–122. doi: 10.1016/0925-4773(91)90048-b. [DOI] [PubMed] [Google Scholar]
- Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Moens U, Ericson JU, Fjose A. Zebrafish pax[zf-a]: a paired box-containing gene expressed in the neural tube. Embo J. 1991;10:3609–3619. doi: 10.1002/j.1460-2075.1991.tb04927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Molecular mechanisms of neural crest formation. Annu Rev Cell Dev Biol. 1999;15:81–112. doi: 10.1146/annurev.cellbio.15.1.81. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Rushton E, Bate M, Rubin GM. Loss of function of the Drosophila zfh-1 gene results in abnormal development of mesodermally derived tissues. Proc Natl Acad Sci U S A. 1993;90:4122–4126. doi: 10.1073/pnas.90.9.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N. The Neural Crest. Cambridge: Cambridge University Press; 1982. [Google Scholar]
- Liu M, Su M, Lyons GE, Bodmer R. Functional conservation of zinc-finger homeodomain gene zfh1/SIP1 in Drosophila heart development. Dev Genes Evol. 2006;216:683–693. doi: 10.1007/s00427-006-0096-1. [DOI] [PubMed] [Google Scholar]
- Long J, Zuo D, Park M. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J Biol Chem. 2005;280:35477–35489. doi: 10.1074/jbc.M504477200. [DOI] [PubMed] [Google Scholar]
- Maruhashi M, Van De Putte T, Huylebroeck D, Kondoh H, Higashi Y. Involvement of SIP1 in positioning of somite boundaries in the mouse embryo. Dev Dyn. 2005;234:332–338. doi: 10.1002/dvdy.20546. [DOI] [PubMed] [Google Scholar]
- Miquelajauregui A, Van de Putte T, Polyakov A, Nityanandam A, Boppana S, Seuntjens E, Karabinos A, Higashi Y, Huylebroeck D, Tarabykin V. Smad-interacting protein-1 (Zfhx1b) acts upstream of Wnt signaling in the mouse hippocampus and controls its formation. Proc Natl Acad Sci U S A. 2007;104:12919–12924. doi: 10.1073/pnas.0609863104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers DC, Sepich DS, Solnica-Krezel L. Bmp activity gradient regulates convergent extension during zebrafish gastrulation. Dev Biol. 2002;243:81–98. doi: 10.1006/dbio.2001.0523. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nitta KR, Takahashi S, Haramoto Y, Fukuda M, Tanegashima K, Onuma Y, Asashima M. The N-terminus zinc finger domain of Xenopus SIP1 is important for neural induction, but not for suppression of Xbra expression. Int J Dev Biol. 2007;51:321–325. doi: 10.1387/ijdb.062252kn. [DOI] [PubMed] [Google Scholar]
- Nitta KR, Tanegashima K, Takahashi S, Asashima M. XSIP1 is essential for early neural gene expression and neural differentiation by suppression of BMP signaling. Dev Biol. 2004;275:258–267. doi: 10.1016/j.ydbio.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Ohno S. Patterns in genome evolution. Curr Opin Genet Dev. 1993;3:911–914. doi: 10.1016/0959-437x(93)90013-f. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- Pla P, Moore R, Morali OG, Grille S, Martinozzi S, Delmas V, Larue L. Cadherins in neural crest cell development and transformation. J Cell Physiol. 2001;189:121–132. doi: 10.1002/jcp.10008. [DOI] [PubMed] [Google Scholar]
- Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. Embo J. 2003;22:2443–2452. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Dean DC. Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc Natl Acad Sci U S A. 2000;97:6391–6396. doi: 10.1073/pnas.97.12.6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. Embo J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, Smith JC, Huylebroeck D. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. Embo J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AL, Lee D, Luo R, Henion PD, Halpern ME. Genes dependent on zebrafish cyclops function identified by AFLP differential gene expression screen. Genesis. 2000;26:86–97. doi: 10.1002/(sici)1526-968x(200001)26:1<86::aid-gene11>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Selleck MAB-FM. The genesis of avian neural crest cells: a classic embryonic induction. Proc Natl Acad Sci U S A. 1996;93:9352–9357. doi: 10.1073/pnas.93.18.9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, dos Reis M, Stern CD. Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell. 2003;115:603–613. doi: 10.1016/s0092-8674(03)00927-9. [DOI] [PubMed] [Google Scholar]
- Shepherd IT, Pietsch J, Elworthy S, Kelsh RN, Raible DW. Roles for GFRalpha1 receptors in zebrafish enteric nervous system development. Development. 2004;131:241–249. doi: 10.1242/dev.00912. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Sivertsen S, Hadar R, Elloul S, Vintman L, Bedrossian C, Reich R, Davidson B. Expression of Snail, Slug and Sip1 in malignant mesothelioma effusions is associated with matrix metalloproteinase, but not with cadherin expression. Lung Cancer. 2006;54:309–317. doi: 10.1016/j.lungcan.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Smith JC. Role of T-box gnes during gastrulation. In: Stern CD, editor. Gastrulation: From Cells to Embryo. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2004. pp. 571–580. [Google Scholar]
- Su MT, Fujioka M, Goto T, Bodmer R. The Drosophila homeobox genes zfh-1 and even-skipped are required for cardiac-specific differentiation of a numb-dependent lineage decision. Development. 1999;126:3241–3251. doi: 10.1242/dev.126.14.3241. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Van de Putte T, Francis A, Nelles L, van Grunsven LA, Huylebroeck D. Neural crest-specific removal of Zfhx1b in mouse leads to a wide range of neurocristopathies reminiscent of Mowat-Wilson syndrome. Hum Mol Genet. 2007;16:1423–1436. doi: 10.1093/hmg/ddm093. [DOI] [PubMed] [Google Scholar]
- Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am J Hum Genet. 2003;72:465–470. doi: 10.1086/346092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grunsven LA, Michiels C, Van de Putte T, Nelles L, Wuytens G, Verschueren K, Huylebroeck D. Interaction between Smad-interacting protein-1 and the corepressor C-terminal binding protein is dispensable for transcriptional repression of E-cadherin. J Biol Chem. 2003;278:26135–26145. doi: 10.1074/jbc.M300597200. [DOI] [PubMed] [Google Scholar]
- van Grunsven LA, Papin C, Avalosse B, Opdecamp K, Huylebroeck D, Smith JC, Bellefroid EJ. XSIP1, a Xenopus zinc finger/homeodomain encoding gene highly expressed during early neural development. Mech Dev. 2000;94:189–193. doi: 10.1016/s0925-4773(00)00318-x. [DOI] [PubMed] [Google Scholar]
- van Grunsven LA, Taelman V, Michiels C, Opdecamp K, Huylebroeck D, Bellefroid EJ. deltaEF1 and SIP1 are differentially expressed and have overlapping activities during Xenopus embryogenesis. Dev Dyn. 2006;235:1491–1500. doi: 10.1002/dvdy.20727. [DOI] [PubMed] [Google Scholar]
- van Grunsven LA, Taelman V, Michiels C, Verstappen G, Souopgui J, Nichane M, Moens E, Opdecamp K, Vanhomwegen J, Kricha S, Huylebroeck D, Bellefroid EJ. XSip1 neuralizing activity involves the co-repressor CtBP and occurs through BMP dependent and independent mechanisms. Dev Biol. 2007;306:34–49. doi: 10.1016/j.ydbio.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, Smith JC, Huylebroeck D. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH. An obligatory caravanserai stop on the silk road to neural induction: inhibition of BMP/GDF signaling. Semin Cell Dev Biol. 2006;17:117–132. doi: 10.1016/j.semcdb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, Taniguchi H, Kitoh H, Mutoh N, Yamanaka T, Mushiake K, Kato K, Sonta S, Nagaya M. Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat Genet. 2001;27:369–370. doi: 10.1038/86860. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Eugene, OR: University of Oregon Press; 1993. [Google Scholar]
- Yamada K, Yamada Y, Nomura N, Miura K, Wakako R, Hayakawa C, Matsumoto A, Kumagai T, Yoshimura I, Miyazaki S, Kato K, Sonta S, Ono H, Yamanaka T, Nagaya M, Wakamatsu N. Nonsense and frameshift mutations in ZFHX1B, encoding Smad-interacting protein 1, cause a complex developmental disorder with a great variety of clinical features. Am J Hum Genet. 2001;69:1178–1185. doi: 10.1086/324343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A, Saigou Y, Higashi Y, Kondoh H. Regulation of ocular lens development by Smad-interacting protein 1 involving Foxe3 activation. Development. 2005;132:4437–4448. doi: 10.1242/dev.02022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.