Abstract
The rice genus, Oryza, which comprises 23 species and 9 recognized genome types, represents an enormous gene pool for genetic improvement of rice cultivars. Clarification of phylogenetic relationships of rice genomes is critical for effective utilization of the wild rice germ plasm. By generating and comparing two nuclear gene (Adh1 and Adh2) trees and a chloroplast gene (matK) tree of all rice species, phylogenetic relationships among the rice genomes were inferred. Origins of the allotetraploid species, which constitute more than one-third of rice species diversity, were reconstructed based on the Adh gene phylogenies. Genome types of the maternal parents of allotetraploid species were determined based on the matK gene tree. The phylogenetic reconstruction largely supports the previous recognition of rice genomes. It further revealed that the EE genome species is most closely related to the DD genome progenitor that gave rise to the CCDD genome. Three species of the CCDD genome may have originated through a single hybridization event, and their maternal parent had the CC genome. The BBCC genome species had different origins, and their maternal parents had either a BB or CC genome. An additional genome type, HHKK, was recognized for Oryza schlechteri and Porteresia coarctata, suggesting that P. coarctata is an Oryza species. The AA genome lineage, which contains cultivated rice, is a recently diverged and rapidly radiated lineage within the rice genus.
Rice is the world’s most important food crop. Future increases in rice production required to feed the growing population will rely primarily on genetic improvement of rice cultivars (1). Recent advances in molecular breeding methods hold tremendous potential for genetic improvement of rice cultivars with beneficial genes from wild rice species (2–4). A clear understanding of the evolutionary relationships of rice species will be essential in directing our effort to search for beneficial genes (3). Furthermore, cultivated rice, Oryza sativa, will soon become the first crop plant with its entire genome sequenced (5). Clarifying evolutionary relationships among genome types of rice species will provide a foundation for future studies of rice genome evolution (6, 7).
The rice genus, Oryza, comprises approximately 23 species distributed in Asia, Africa, Australia, and Central and South America (8, 9). Over the past half century, continued efforts have been devoted to understanding genomic composition and relationships of the rice species. Based on interspecific crossing and subsequent cytogenetic analyses (10, 11) and genomic DNA hybridization (12), 9 types of diploid (2n = 24) genomes and various combinations among them at the tetraploid level (2n = 48) have been recognized for 22 Oryza species (Table 1) (9). Approximately one-third of the rice species were considered to be allotetraploids whose origins through hybridization could confound the reconstruction of phylogenetic relationships within the rice genus. Phylogenetic studies of Oryza, however, have been less extensive than those of other major crop plants, such as maize (13), soybean (14), and cotton (15, 16), and have not been examined with phylogenetic analyses of DNA sequences. Evolutionary relationships among the rice genomes and species were previously estimated by phenetic analysis of morphology, isozyme, and nuclear and chloroplast DNA restriction fragment-length polymorphisms (17–21). However, limitations in the nature of the data and/or methods of data analysis in these studies have hampered an accurate reconstruction of the rice phylogeny, particularly the origins of the allotetraploid species.
Table 1.
Accessions of the Oryza Species
| Genome | Section/species | Accession no. | Locality |
|---|---|---|---|
| Section Oryza | |||
| AA | O. sativa | Au73030 | China |
| O. glaberrima | 104042 | Chad | |
| O. barthii | 104140 | Cameroon | |
| O. glumaepatula | B308,* 100968 | Brazil, Suriname | |
| O. longistaminata | 104977 | Kenya | |
| O. meridionalis | 103317, 101147 | Australia | |
| O. nivara | 106148 | Laos | |
| O. rufipogon | 0413,* 105942 | China, Thailand | |
| BB | O. punctata | 104071 | Cameroon |
| CC | O. officinalis | 105085 | Philippines |
| O. rhizomatis | 105448 | Sri Lanka | |
| BBCC | O. minuta | 101082 | Philippines |
| O. eichingeri | 105160 | Uganda | |
| CCDD | O. alta | 105143 | Guyana |
| O. grandiglumis | 105669 | Brazil | |
| O. latifolia | 105141 | Costa Rica | |
| EE | O. australiensis | 105263 | Australia |
| Section Ridleyanae | |||
| FF | O. brachyantha | 105151 | Sierra Leone |
| HHJJ | O. longiglumis | 105148 | Indonesia |
| O. ridleyi | 100877 | Malaysia | |
| Unknown | O. schlechteri | 82047 | Papua New Guinea |
| Section Granulata | |||
| GG | O. granulata | 2422,* 106469 | China, Vietnam |
| O. meyeriana | 104987 | Malaysia |
Accessions of Oryza species studied, with their accession numbers, collection localities, genome types, and classifications indicated (8, 9). Outgroups and their accession numbers are as follows: Leersia perrierri (105164), Porteresia coarctata (104502), Rhynchoryza subulata (100913), and Zizaniopsis villanenses (85425).
Accessions collected by the authors; the remaining accessions were obtained from the International Rice Research Institute.
Allopolyploidy is a widely documented mechanism of speciation in flowering plants (22, 23). An allotetraploid combines two or more distinct diploid genomes and originates through hybridization of diverged diploid species coupled with an increase in chromosome numbers. Phylogenetic analysis of single or low-copy nuclear gene sequences offers an effective way to study evolution of allopolyploids (16, 24–26). Homoeologous loci that are contributed by the diploid parents can be cloned and sequenced from an allotetraploid species. Analyses of these sequences, together with the gene sequences of the putative diploid parents, enable one to unravel the reticulate pattern of hybrid speciation. Furthermore, obtaining a phylogeny of the chloroplast genome allows inference of the maternal parent of an allopolyploid. Here, we analyze two nuclear genes, alcohol dehydrogenase gene 1 (Adh1) and 2 (Adh2) (27, 28), and the chloroplast gene matK (29) to reconstruct the phylogeny of the rice genus, with an emphasis on relationships among genome types. Based on this phylogenetic reconstruction, we are able to evaluate the previous circumscription of rice genomes and further recognize another genome type.
Materials and Methods
Total DNA was isolated from 31 accessions representing all of the 23 rice species (Table 1) and 4 closely related genera by using a cetyltrimethylammonium bromide method (30). Seeds and leaf material of the majority of the accessions were provided by the International Rice Genebank at the International Rice Research Institute (Manila, Philippines). The coding region of the chloroplast matK gene was amplified with PCR primers matKF1, 5′-TAATTAAGAGGATTCACCAG, and matKR1, 5′-ATGCAACACCCTGTTCTGAC. PCR products were purified with a GeneClean kit (Bio 101) and sequenced directly.
Adh genes were amplified initially by using primers AdhF1 and AdhR1, which are located on exon 2 and exon 8, respectively (Fig. 1). The primers were designed based on sequences that are conserved across three divergent genera of the grass family, Hordeum, Oryza, and Zea (27, 31), to maximize the chance of amplifying all Adh genes in rice species. The PCR products were then cloned with a TA cloning kit (Invitrogen). For each accession, 12–20 clones were screened by examining restriction sites and/or sequencing with one primer (32). This round of cloning and screening identified two Adh loci, Adh1 and Adh2, for the majority of the rice species. Locus-specific primers, Adh1bR and Adh2RR, were subsequently designed to amplify Adh1 and Adh2 genes, respectively, from certain accessions. The locus-specific primers were also used to amplify each Adh gene from the allotetraploid species. Twelve to fifteen clones were screened for each Adh locus of an allotetraploid species in an attempt to identify all homoeologous loci. Clones with distinct inserts were fully sequenced for both strands and included in the phylogenetic analyses. Sequencing was done on an ABI 373 automated DNA Sequencer with a Dye Terminator Cycle Sequencing Reaction Kit (PE Applied Biosystems).
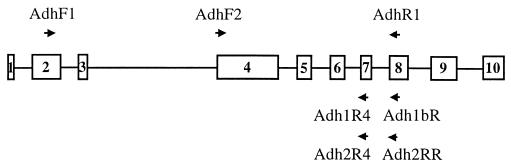
Figure 1.
Diagram of the Adh gene of rice. Boxes represent exons numbered 1–10 from the 5′ to 3′ ends; lines between exons represent introns. Arrows indicate locations and directions of the PCR primers: AdhF1, 5′-CACACCGACGTCTACTTCTG-3′; AdhF2, 5′-AGAGTGTTGGAGAGGGTGTGAC-3′; AdhR1, 5′-ATCATGGCGTTGATGTTGCC-3′; Adh1bR, 5′-TCAGCAAGTACCTAAATTATC-3′; Adh1R4, 5′-TTCWGTGCAACCAAATTTCC-3′; Adh2RR, 5′-CCACCGTTGGTCATCTCAAT-3′; and Adh2R4, 5′-GTCAGTGCAGCCAAACTTCT-3′. Adh1bR and Adh1R4 are Adh1 gene-specific primers, and Adh2RR and Adh2R4 are Adh2 gene-specific primers. Information regarding internal sequencing primers is available from the authors.
Sequences of six exons (exons 2–7) and six introns (ones between exons 2 and 8) were aligned with Clustal W (33) and refined manually. A few small regions in introns (ranging from several to 30 bps) that could not be aligned unambiguously were excluded from the analyses. Phylogenetic analyses were conducted with paup* Version 4.0 (34). Parsimony analyses were performed by heuristic search with tree bisection-reconnection (TBR) branch swapping, MULPARS option, ACCTRAN optimization, and 100 random addition replicates. Topological robustness was assessed by bootstrap analysis with 1,000 replicates using simple taxon addition (35). For maximum likelihood analyses, we performed heuristic search with the TBR branch-swapping with the following parameters: empirical base frequencies, a transition–transversion ratio of 2, 10, and equal rates of substitution among sites. The Templeton test, as implemented in paup*, was used to evaluate topological incongruence at the specific nodes between gene trees (36, 37). Leersia, Porteresia, Rhynchoryza, and Zizaniopsis, which are closely related to Oryza based on morphological and molecular evidence (38–40), were used as outgroups. The Adh2 gene of Rhynchoryza was excluded from the phylogenetic analyses because its introns were difficult to align with those of the ingroup species.
Results and Discussion
Adh Gene Phylogenies and Origins of Allotetraploids.
The Adh gene family, comprising usually two to three gene members in a flowering plant (41), has been thoroughly studied in grasses (Poaceae) (31, 42). Two Adh loci, Adh1 and Adh2, have been identified in the cultivated rice, O. sativa, by enzyme electrophoresis, cDNA library screening, and Southern blotting (27, 28). These two loci were duplicated prior to the diversification of the grass family, and their orthologs were found in all members of the grass family studied to date (40, 42). Subsequent gene duplications, occurring at either Adh1 or Adh2 locus, have given rise to additional Adh loci in some grasses, such as barley and maize (42). To amplify all Adh genes from a rice species, we selected the PCR-priming sites (AdhF1 and AdhR1) where sequences are conserved not only between the two Adh loci of O. sativa, but also among Adh genes of Oryza, Hordeum, and Zea. To minimize the chance of omitting an Adh gene member from the PCR products, a relative large number of clones (12–20) were screened for each species. Two types of Adh sequences were identified for every diploid Oryza species. While these two types of sequences could not be aligned with each other in introns, they were aligned well with the previously reported sequences of Adh1 and Adh2 genes of O. sativa, respectively (27, 28). This indicates that every diploid rice species contains these two orthologous Adh loci.
For a diploid species, the majority of clones of the same locus (Adh1 or Adh2) were identical in sequences. Clones that differed from this majority of clones by one or two base pairs were found occasionally. These different clones may represent allelic variation, additional Adh loci, or results of PCR errors. Initial phylogenetic analyses indicated that all clones of the Adh1 or Adh2 gene from the same species formed a monophyletic group, and exclusion of these minor types of clones had no impact on relationships among species. If the sequence polymorphism found in a species represents duplicated Adh genes, the gene duplications must have occurred within the species. Because paralogous relationships can be obtained only when gene duplications precede speciation events (43), it is unlikely that paralogy would cause problems in the present phylogenetic reconstructions.
Two distinct types of sequences were identified at an Adh locus for the majority of tetraploid species. Sequences of the same locus (Adh1 or Adh2) cloned from both diploid and tetraploid species were aligned for phylogenetic analyses. The aligned sequences of the Adh1 gene were 1,955 bp long, of which 781 nucleotide sites were variable and 214 were phylogenetically informative. The Adh2 data set contained 1,779 nucleotide sites, of which 599 sites were variable and 171 were phylogenetically informative. Analysis of the Adh1 data set yielded two equally most parsimonious trees with a consistency index (CI) of 0.75 and a retention index (RI) of 0.87, and the strict consensus tree is shown in Fig. 2A. Analysis of the Adh2 data set yielded four equally most parsimonious trees (CI = 0.76, RI = 0.88), and the strict consensus tree is shown in Fig. 2B. Trees generated by maximum likelihood analyses (data not shown) had topologies identical to the most parsimonious trees.
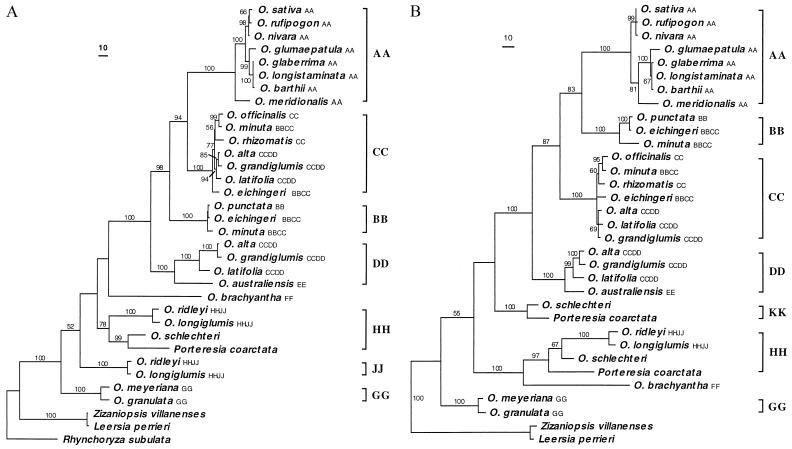
Figure 2.
Phylogenies of Adh1 and Adh2 genes of rice species. (A) Adh1 gene phylogeny. The strict consensus tree of two equally most parsimonious trees (tree length = 1,256, CI = 0.75, RI = 0.87). (B) Adh2 gene phylogeny. The strict consensus tree of four equally most parsimonious trees (tree length = 972, CI = 0.76, RI = 0.88). Numbers are bootstrap percentages above 50%. Branch lengths are proportional to the number of nucleotide substitutions, and scale bars indicate 10 substitutions. Small capital letters following a species name indicate the previously recognized genome type of the species. On each Adh gene tree, the appearance twice of an allotetraploid species represents two distinct types of sequences cloned from the same individual of the species. The genome type of a monophyletic group is indicated.
For allotetraploid species with the BBCC and CCDD genomes, two distinct types of sequences were cloned for each Adh gene. On both Adh gene trees, the types of sequences form monophyletic groups with the diploid species of the BB and CC genomes, respectively (Fig. 2). Two types of sequences cloned from the CCDD genome species form monophyletic groups with the diploid species of the CC and EE genomes, respectively. The congruent relationships between Adh1 and Adh2 gene trees strongly support previous hypotheses of the allotetraploid nature of the BBCC and CCDD genomes. Because gene duplication prior to speciation of the diploid rice species was not detected at either Adh locus, two distinct types of sequences cloned at an Adh locus of a tetraploid individual are much more likely to represent homoeologous loci contributed by the different diploid parents rather than paralogous loci. If the sequence polymorphism is considered to be ancestral polymorphic alleles, lineage sorting has to be invoked to explain each Adh phylogeny. It is, however, very unlikely that lineage sorting, a random process, can result in the identical phylogenetic relationships between the two nuclear loci (25). Therefore, we designate the clade containing the diploid BB genome species and one type of sequence cloned from the BBCC genome species as the clade of the BB genome. The clade that contains the diploid CC genome species and sequences cloned from the BBCC and CCDD genome species is recognized as the CC genome clade (Fig. 2).
The DD type of sequence cloned from the CCDD genome forms a monophyletic group with the diploid EE genome species Oryza australiensis on both Adh gene trees (Fig. 2). Although cytogenetic studies of the F1 hybrids between the CCDD and EE genomes revealed a certain degree of chromosomal pairing, the results were insufficient for recognition of homology between the DD and EE genomes (10). Application of DNA markers, such as nuclear restriction fragment-length polymorphisms, also failed to identify the diploid DD genome (21, 44), which led to speculation that the DD genome species was extinct. Strongly supported monophyly of the DD genome sequence and the EE genome species on both Adh trees suggests that the EE genome is most closely related to the diploid DD genome progenitors giving rise to the CCDD genome. Thus, we designate the clade containing the diploid EE genome and sequences from the CCDD genome as the DD genome clade (Fig. 2).
For the remaining tetraploid species, Oryza ridleyi, Oryza longiglumis, and Oryza schlechteri, sequence polymorphism was found at one of the Adh loci. Two types of Adh1 sequences were cloned from O. ridleyi and O. longiglumis of the HHJJ genome, while only one type of Adh2 sequence was found. O. schlechteri and Porteresia coarctata are most closely related to each other, according to the two Adh gene and the matK gene trees (Figs. 2 and 3). O. schlechteri is the only rice species with an unknown genome type. P. coarctata was once recognized as a rice species, Oryza coarctata, but later treated as a monotypic genus (45). Two types of Adh2 sequences were cloned from O. schlechteri and P. coarctata, while only one type of Adh1 sequence was identified. Allotetraploid origins of these four species were inferred based on sequence polymorphism at one of the Adh loci, although the hypotheses are not as strong as those concerning the origins of the BBCC and CCDD genomes, which are supported by both Adh genes.
Figure 3.
The single most parsimonious tree generated from matK gene sequences of rice species (tree length = 206, CI = 0.87, RI = 0.90). Numbers are bootstrap percentages above 50%. Branch lengths are proportional to the number of nucleotide substitutions, and the scale bar indicates 10 substitutions. Small capital letters following a species name indicate the previously recognized genome type of the species.
On each Adh gene tree, there is a clade containing all four species, O. ridleyi, O. longiglumis, O. schlechteri, and P. coarctata (Fig. 2). We designate this clade as the HH genome clade. The other clade containing O. ridleyi and O. longiglumis on the Adh1 phylogeny, thus, represents the JJ genome. On the Adh2 phylogeny, the remaining clade containing O. schlechteri and P. coarctata is given a new genome type, KK. Therefore, O. schlechteri and P. coarctata share the same genome type, HHKK. The Adh2 phylogeny suggests a close relationship between the HH genome and the diploid FF genome species, which, however, is not supported by the Adh1 phylogeny. The JJ and KK genomes are not grouped strongly with any diploid species on either Adh phylogeny. These results suggest that the diploid species with the HH, JJ, or KK genome are either extinct or undiscovered.
Failure to uncover one of the homoeologous Adh loci from O. ridleyi, O. longiglumis, O. schlechteri, and P. coarctata may be a result of selective PCR amplification, formation of pseudogenes with mutations occurring at the PCR-priming sites, or gene deletion. To test these alternative hypotheses, we designed a new forward primer (AdhF2) and two new reverse primers (Adh1R4 specific to Adh1, and Adh2R4 specific to Adh2) that are located in the conserved regions of the exons different from those on which the original primers (AdhF1, AdhR1, Adh1bR, and Adh2RR) are located (Fig. 1). Two combinations of a new and an original primer were tried to amplify the missing homoeologous loci; they are AdhF1–Adh1R4 and AdhF2–Adh1bR for amplifying the Adh1 gene from O. schlechteri and P. coarctata, and AdhF1–Adh2R4 and AdhF2–Adh2RR for amplifying the Adh2 gene from O. ridleyi and O. longiglumis. For each of the primer combinations, more than 10 clones were screened for every species. All of the clones belong to the same homoeologous loci identified previously with the original primer combinations (AdhF1–AdhR1 for both Adh loci, AdhF1–Adh1bR for the Adh1 locus, and Adh1F-Adh2RR for the Adh2 locus). The failure to uncover the Adh1 or Adh2 homoeologous locus is very unlikely to have resulted from selective PCR amplification or mutations occurring at the PCR-priming sites given that four combinations of five primers have been attempted for each gene. These results thus support the hypothesis of gene deletion, i.e., deletion of the Adh1 gene from the KK genome of O. schlechteri and P. coarctata, and deletion of the Adh2 gene from the JJ genome of O. ridleyi and O. longiglumis.
One of the homoeologous loci derived from the diploid parents may undergo gene silencing because of genetic redundance in a tetraploid genome (46). It has been found in allotetraploid peony species that one of the homoeologous Adh loci has become a pseudogene (25). Results in this study suggest that deletion of one of the homoeologous loci may be a mechanism of reduction of genetic redundance in these allotetraploid rice species. On both Adh phylogenies, sequences of the HHJJ and HHKK genomes occupy the basal positions relative to those of the BBCC and CCDD genomes (Fig. 2), suggesting that the former had more ancient origins, which may have allowed more extensive genomic rearrangement, including possible deletions of some homoeologous loci. Southern blotting experiments can be employed to further test the hypothesis of gene deletion.
matK Gene Phylogeny—The Maternal Genealogy.
Because the chloroplast genome is maternally inherited in rice (47), the matK gene tree represents a maternal genealogy of rice species that offers an opportunity to identify the maternal parents of allotetraploid species. The aligned matK sequences were 1,552 bp in length, of which 166 nucleotide sites were variable and 48 were phylogenetically informative. Analysis of the matK sequences resulted in a single most parsimonious tree with a CI of 0.87 and an RI of 0.90 (Fig. 3). The CCDD genome species form a monophyletic group with the CC genome species on the matK phylogeny, suggesting that the CC genome served as the maternal parent of the CCDD genome species. On the Adh and matK gene trees, three species of the CCDD genome form monophyletic groups in which Oryza alta and Oryza grandiglumis are sister species, implying that these three allotetraploid species originated from a single hybridization event. In contrast, two species with the BBCC genome, Oryza minuta and Oryza eichingeri, may have had different origins. The former had a maternal parent of the BB genome, and the latter had a maternal parent of the CC genome, according to the matK phylogeny. The position of O. schlechteri and P. coarctata on the matK gene tree is apparently the same as that of the KK genome on the Adh2 tree, suggesting that the KK genome was the maternal parent of the HHKK genome. The maternal parent of the HHJJ genome, however, seems to be less clear.
Phylogeny of Rice Genomes Inferred from Three Gene Trees.
Understanding congruence and incongruence between gene trees has attracted considerable theoretical discussion (43, 48–51). Congruence between gene trees provides a strong indication of orthology, and thus enhances robustness of phylogenetic reconstructions. Here, we compare the two Adh gene trees and infer phylogenetic relationships of rice genomes based on congruence between them. On both Adh1 and Adh2 gene trees, each clade of a previously recognized genome type (including sequences from both diploid and tetraploid species) is supported by a high bootstrap value, mostly 100% (Fig. 2). Phylogenetic relationships among the genomes are largely congruent between the two Adh gene trees except at two places. One is the relationship of the BB and CC genomes relative to the AA genome, and the other involves the position of the FF genome species, Oryza brachyantha (Fig. 2). The AA and CC genomes are sister groups on the Adh1 tree, whereas the AA and BB genomes are sister groups on the Adh2 tree. To evaluate this incongruence, the Templeton test was conducted in both ways, i.e., using relationships on the Adh1 tree as a constraint to test the Adh2 data set and vice versa. The test results are significant when either the Adh1 (P < 0.05) or Adh2 (P < 0.0001) data set was tested against the constraint topology. The FF genome is grouped strongly with the HH genome on the Adh2 phylogeny, but does not form a strongly supported group with any genome type on the Adh1 phylogeny. When the FF and HH genomes were forced to form a monophyletic group on the Adh1 phylogeny, the resulting tree was significantly worse (P < 0.0001).
Therefore, we did not combine the two data sets (48, 49), but rather generated a consensus tree in which the congruent relationships between the two gene trees were maintained and the incongruent relationships were treated as unresolved clades (52). Reasons for the topological incongruence need to be explored, and relationships among AA, BB, and CC genomes and the position of the FF genome should be further clarified with additional gene phylogenies (51). The topology of the matK gene tree is largely congruent with the consensus tree of diploid genomes inferred from the Adh gene trees, except for the position of the GG genome. The Templeton test indicated that this topological incongruence is not significant (P > 0.1). An overall hypothesis of the phylogeny of the rice genomes based on the three gene phylogenies is presented in Fig. 4.
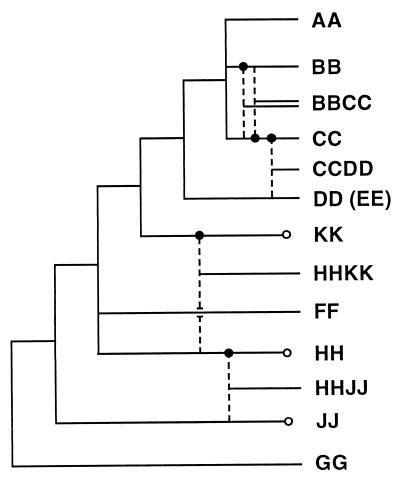
Figure 4.
Evolutionary relationships of the rice genomes inferred from Adh1, Adh2, and matK gene phylogenies. Dashed lines indicate origins of allotetraploids; ● indicate maternal parents. ○ indicate unidentified diploid genomes.
Implications of the Phylogenetic Reconstruction.
Monophyletic groups revealed by the phylogenetic reconstruction (Fig. 4) are either concordant or discordant with taxonomic sections recognized in the most recent classification of the genus (8) (Table 1). The AA, BB, and CC genomes are most closely related and together form a sister group with the DD (EE) genome. This monophyletic group, containing the AA through EE genomes, corresponds to section Oryza. The GG genome, which occupies the most basal position of the genus, constitutes section Granulata. The remaining genome types that are included in section Ridleyanae, however, form a paraphyletic group in the phylogenetic hypothesis (Fig. 4).
According to the crossability between O. sativa and other rice species, the wild species have been categorized as the primary, secondary, and tertiary gene pools for the cultivars (9). Species of the AA genome are easily crossed with O. sativa and are regarded as the primary gene pool. The BB through EE genomes constitute the secondary gene pool, and the remaining genomes constitute the tertiary gene pool. Clearly, crossability correlates well with the phylogenetic relationships of the rice genomes (Fig. 4). A variety of beneficial traits, such as high yield, disease and pest resistance, and tolerance to environmental stresses, have been incorporated into rice cultivars from these gene pools (53, 54). The tertiary gene pool contributes less than one-third of the species diversity, but nearly half of the genomic diversity (genomes FF through KK). These genomes are distantly related to the cultivated species and have great potential to provide novel beneficial genes.
The AA genome, which contains the cultivated rice, is one of the most recently diverged lineages within the rice genus (Fig. 4). It contains the largest number of diploid species and has the widest geographic distribution of any rice genome. Apparently, the AA genome is a recently diversified, rapidly radiated, and well adapted group. According to the Adh phylogenies that resolve relationships within the AA genome, the widely cultivated species O. sativa is most closely related to two wild species distributed in Asia, Oryza nivara and Oryza rufipogon, supporting the previous hypothesis of an Asian origin of O. sativa (9, 18). The African cultivated species, Oryza glaberrima, is most closely related to two African wild species, Oryza barthii and Oryza longistaminata, and also to Oryza glumaepatula, which occurs in Central and South America.
Conclusions
The present study demonstrates that phylogenetic analyses of sequences of low-copy nuclear genes together with chloroplast genes offer an effective approach to study genomic composition and relationships of rice species. Particularly, comparison of the two Adh gene phylogenies has led to a robust reconstruction of origins of allotetraploid species, including those whose diploid parents may be extinct. Genome types of the maternal parents of the allotetraploid species were inferred based on the chloroplast matK gene phylogeny. Beyond the previous understanding of rice genomic composition, this phylogenetic study revealed that the EE genome is most closely related to the DD genome progenitor that gave rise to the CCDD genome. Based on the phylogenetic reconstruction, we were able to recognize the additional genome type HHKK for O. schlechteri and P. coarctata, which suggests that P. coarctata should be treated as an Oryza species. A relatively robust phylogeny of diploid and tetraploid rice genomes was inferred by synthesizing two nuclear and one chloroplast gene phylogenies. The remaining ambiguous relationships among genome types resulting from topological incongruence between the Adh gene trees need to be further clarified with additional gene phylogenies.
Acknowledgments
We thank F. Wang, S.-Z. Zhang, and Y. Zhou for lab assistance and R. Allison, D. Ferguson, R. Lenski, D. Tank, and anonymous reviewers for valuable comments on the manuscript. We also thank the International Rice Research Institute (Manila, Philippines) for providing plant material for this study. This research was supported by Michigan State University, Michigan Agricultural Experiment Station, and the Chinese Academy of Sciences (KZ951-B1-102).
Abbreviations
- CI
consistency index
- RI
retention index
Footnotes
References
- 1.Mann C C. Science. 1999;283:310–313. doi: 10.1126/science.283.5400.314. [DOI] [PubMed] [Google Scholar]
- 2.Tanksley S D, Nelson J C. Theor Appl Genet. 1996;92:191–203. doi: 10.1007/BF00223376. [DOI] [PubMed] [Google Scholar]
- 3.Tanksley S D, McCouch S R. Science. 1997;277:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- 4.Xiao J H, Grandillo S, Ahn S N, McCouch S R, Tanksley S D, Li J M, Yuan L P. Nature (London) 1996;384:223–224. [Google Scholar]
- 5.Sasaki T. Proc Natl Acad Sci USA. 1998;95:2027–2028. doi: 10.1073/pnas.95.5.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kellogg E A. Proc Natl Acad Sci USA. 1998;95:2005–2010. doi: 10.1073/pnas.95.5.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCouch S. Proc Natl Acad Sci USA. 1998;95:1983–1985. doi: 10.1073/pnas.95.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaughan D A. The Wild Relatives of Rice: A Genetic Resources Handbook. Philippines: International Rice Research Institute; 1994. [Google Scholar]
- 9.Khush G S. Plant Mol Biol. 1997;35:25–34. [PubMed] [Google Scholar]
- 10.Li H W, Chen C C, Wu H K, Lu K C L. In: Rice Genetics and Cytogenetics. Tsunoda S, Takahashi N, editors. Amsterdam: Elsevier; 1964. pp. 118–131. [Google Scholar]
- 11.Morinaga T. In: Rice Genetics and Cytogenetics. Tsunoda S, Takahashi N, editors. Amsterdam: Elsevier; 1964. pp. 91–102. [Google Scholar]
- 12.Aggarwal R K, Brar D S, Khush G S. Mol Gen Genet. 1997;254:1–12. doi: 10.1007/s004380050384. [DOI] [PubMed] [Google Scholar]
- 13.Gaut B S. Plant Species Biol. 1996;11:1–11. [Google Scholar]
- 14.Doyle J J, Kanazin V, Shoemaker R C. Mol Phylogenet Evol. 1996;6:438–447. doi: 10.1006/mpev.1996.0092. [DOI] [PubMed] [Google Scholar]
- 15.Wendel J F, Albert V A. Syst Bot. 1992;17:115–143. [Google Scholar]
- 16.Small R L, Ryburn J A, Cronn R C, Seelanan T, Wendel J F. Am J Bot. 1998;85:1301–1315. [PubMed] [Google Scholar]
- 17.Morishima H K, Oka H I. Evolution. 1960;14:153–165. [Google Scholar]
- 18.Second G. Jpn J Genet. 1982;57:25–57. [Google Scholar]
- 19.Dally A, Second G. Theor Appl Genet. 1989;80:209–222. doi: 10.1007/BF00224389. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar R, Raina S N. Theor Appl Genet. 1992;85:127–132. doi: 10.1007/BF00223855. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z Y, Second G, Tanksley S D. Theor Appl Genet. 1992;83:565–581. doi: 10.1007/BF00226900. [DOI] [PubMed] [Google Scholar]
- 22.Grant V. Plant Speciation. 2nd Ed. New York: Columbia Univ. Press; 1981. [Google Scholar]
- 23.Masterson J. Science. 1994;264:421–423. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 24.Gaut B S, Doebley J F. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sang T, Zhang D. Syst Bot. 1999;24:148–163. [Google Scholar]
- 26.Small R L, Ryburn J A, Wendel J F. Mol Biol Evol. 1999;16:491–501. doi: 10.1093/oxfordjournals.molbev.a026131. [DOI] [PubMed] [Google Scholar]
- 27.Xie Y, Wu R. Plant Mol Biol. 1989;13:53–68. doi: 10.1007/BF00027335. [DOI] [PubMed] [Google Scholar]
- 28.Xie Y, Wu R. Gene. 1990;87:185–191. doi: 10.1016/0378-1119(90)90300-g. [DOI] [PubMed] [Google Scholar]
- 29.Olmstead R G, Palmer J D. Am J Bot. 1994;81:1205–1224. [Google Scholar]
- 30.Doyle J J, Doyle J L. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 31.Gaut B S, Morton B R, Mcgaig B C, Clegg M T. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sang T, Donoghue M J, Zhang D. Mol Biol Evol. 1997;14:994–1007. doi: 10.1093/oxfordjournals.molbev.a025716. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 1998. , Version 4. [Google Scholar]
- 35.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 36.Templeton A R. Evolution. 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 37.Mason-Gamer R J, Kellogg E A. Syst Biol. 1996;45:524–545. [Google Scholar]
- 38.Terrell E, Robinson H. Bull Torr Bot Club. 1974;101:235–245. [Google Scholar]
- 39.Duvall M R, Peterson P M, Terrell E E, Christensen A H. Am J Bot. 1993;80:83–88. [Google Scholar]
- 40.Morton B R, Gaut B S, Clegg M T. Proc Natl Acad Sci USA. 1996;93:11735–11739. doi: 10.1073/pnas.93.21.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gottlieb L D. Science. 1982;216:373–380. doi: 10.1126/science.216.4544.373. [DOI] [PubMed] [Google Scholar]
- 42.Gaut B S, Peek A S, Morton B R, Clegg M T. Mol Biol Evol. 1999;16:1086–1097. doi: 10.1093/oxfordjournals.molbev.a026198. [DOI] [PubMed] [Google Scholar]
- 43.Maddison W P. Syst Biol. 1997;46:523–536. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- 44.Kiefer-Meyer M C, Reddy A S, Delseny M. Genome. 1995;38:681–694. doi: 10.1139/g95-086. [DOI] [PubMed] [Google Scholar]
- 45.Tateoka T. Bull Natl Sci Mus Ser B (Tokyo) 1965;8:405–406. [Google Scholar]
- 46.Leitch I J, Bennett M D. Trends Plant Sci. 1997;2:470–476. [Google Scholar]
- 47.Corriveau J L, Coleman A W. Am J Bot. 1988;75:1443–1458. [Google Scholar]
- 48.de Queiroz A, Donoghue M J, Kim J. Annu Rev Ecol Syst. 1995;26:657–681. [Google Scholar]
- 49.Huelsenbeck J P, Bull J J, Cunningham C W. Trends Ecol Evol. 1996;11:152–158. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 50.Doyle J J. Syst Biol. 1997;46:537–553. doi: 10.1093/sysbio/46.3.537. [DOI] [PubMed] [Google Scholar]
- 51.Wendel J F, Doyle J J. In: Molecular Systematics of Plants II: DNA Sequencing. Soltis D E, Soltis P S, Doyle J J, editors. Boston: Kluwer; 1998. pp. 265–296. [Google Scholar]
- 52.Swofford D L. In: Phylogenetic Analysis of DNA Sequences. Miyamoto M M, Cracraft J, editors. New York: Oxford Univ. Press; 1991. pp. 295–333. [Google Scholar]
- 53.Amante-Bordeos A, Sitch L A, Nelson R, Dalmacio R D, Oliva N P, Aswidinnoor H, Leung H. Theor Appl Genet. 1992;84:345–354. doi: 10.1007/BF00229493. [DOI] [PubMed] [Google Scholar]
- 54.Brar D S, Khush G S. Plant Mol Biol. 1997;35:25–34. [PubMed] [Google Scholar]