Abstract
Recent evidence suggests that deep-sea vestimentiferan tube worms acquire their endosymbiotic bacteria from the environment each generation; thus, free-living symbionts should exist. Here, free-living tube worm symbiont phylotypes were detected in vent seawater and in biofilms at multiple deep-sea vent habitats by PCR amplification, DNA sequence analysis, and fluorescence in situ hybridization. These findings support environmental transmission as a means of symbiont acquisition for deep-sea tube worms.
The mode by which symbionts are passed between successive host generations is a primary question in symbiosis research. Symbiont transmission typically occurs vertically via transfer from parent to offspring, horizontally between cooccurring host individuals, or environmentally via uptake from a free-living population (3). Determining which of these mechanisms operates within a symbiosis is critical, as the transmission mode impacts fundamental ecological and evolutionary processes, including genome evolution, symbiont-host specificity, and coevolution (for examples, see references 7, 19, and 30).
Deep-sea vestimentiferan tube worms, which dominate the fauna at hydrothermal vents and cold seeps, are hypothesized to acquire their bacterial symbionts environmentally from a free-living population. Attempts to detect tube worm symbionts in host eggs and larvae by the use of microscopy and PCR have been unsuccessful (4-6, 12), suggesting that transmission does not occur vertically. Furthermore, most vent vestimentiferan species host symbionts that share identical 16S rRNA sequences, which is also consistent with the hypothesis of environmental transmission (14, 22). Unlike adults, the larvae and small juveniles of vestimentiferan tube worms have a mouth and gut, suggesting environmental acquisition via the ingestion of symbionts during larval development (12, 28). However, Nussbaumer et al. (23) recently demonstrated that bacterial symbionts are found on the developing tubes of settled larvae, entering the host worm through the epidermis and body wall of both larvae and young juveniles (23). These studies strongly suggest that tube worms acquire their symbionts from the surrounding environment and, therefore, that these endosymbionts should be detectable in a free-living form.
Sample collection.
A systematic search for the free-living counterpart to the gammaproteobacterial endosymbiont phylotype shared by three species of vestimentiferan tube worms, Riftia pachyptila, Oasisia alvinae, and Tevnia jerichonana, was conducted at the Tica hydrothermal vent site (∼2,600-m depth) on the East Pacific Rise (EPR) (9°50.447′N, 104°17.493′W) during December 2002 and December 2003. Symbiont-containing tissue was dissected from all three vestimentiferan tube worm hosts (from the trophosome) and from Calyptogena magnifica clams (from the gills) at the Tica vent site for future use as positive and negative controls, respectively. Environmental samples were collected from two distinct habitats: surface-attached biofilms and seawater.
Symbionts in surface-attached biofilms were collected on bacterial settlement devices deployed in four hydrothermal vent environments at increasing distances from tube worm clusters: (i) among tube worms, (ii) adjacent to tube worms, (iii) away from tube worms (∼10 m), and (iv) off-axis (∼100 m outside the axial summit of the caldera) (see Fig. S1 in the supplemental material). Settlement devices were constructed of polyvinyl chloride holders containing three to five basalt pieces (8 by 1 by 1 cm) and 4 to 12 glass microscope slides that were washed, autoclaved, and kept sterile until deployment. Devices were collected within 1 month or after 1 year. Upon collection, the basalt pieces were examined under a dissecting microscope to detect any settled tube worm larvae or juveniles and then immediately stored at −80°C. Pieces with observable tube worms were excluded to eliminate the risk of detecting symbionts living within host tissue. Microscope slides were fixed for fluorescence in situ hybridization (FISH) analysis in 4% paraformaldehyde and stored in 70% ethanol at 4°C.
Seawater samples were collected 1 m away from an R. pachyptila tube worm cluster using a McLane large-volume water transfer system water pump attached to the deep submergence vehicle Alvin. Samples (200 liters each) were filtered in situ through a 1-μm Petex prefilter (Sefar) and then through a 0.45-μm mixed-cellulose ester filter (Millipore). Control seawater samples (80 liters each) were collected from the ocean surface above the EPR and from the Atlantic Ocean in Nahant, MA. All filters were stored at −80°C until DNA extraction.
16S rRNA gene sequence analyses.
PCR amplification and DNA sequence analyses were used to test for the presence of the vestimentiferan symbiont in biofilm and seawater samples. DNA was extracted by standard methods (27). The vestimentiferan symbiont 16S rRNA gene (a 401-bp fragment) was PCR amplified using primers specific for the shared 16S phylotype: RifTO44 (5′-GGCCTAGATTGACGCTGCGGTA-3′) (this study) and RifTO445 (23). To detect contamination by host tissue, primers specific for the genes encoding the vestimentiferan host exoskeleton protein RP43 (GenBank accession no. AF233595), RifTOExoF (5′-CTAAAGGCAGTGTCAAGAGCGGGAC-3′) and RifTOExoR (5′-TTCCTCGAAGTTGCCGTATGCCG-3′), were used. PCR products were cloned into a pCR2 cloning vector (Invitrogen) and sequenced by standard methods using BigDye Terminator cycle sequencing reaction kits (PE Biosystems) with M13 forward and reverse primers. Symbiont- and host-specific primers amplified their target genes in the control symbiont-containing tissue samples from R. pachyptila, T. jerichonana, and O. alvinae worms, while vestimentiferan symbionts were not amplified from C. magnifica gill tissue, the negative control.
The free-living vestimentiferan symbiont 16S rRNA phylotype was detected in both biofilm and seawater samples collected at the Tica vent site. The symbiont phylotype (GenBank accession no. U77478) (9) was amplified from all basalt pieces retrieved after 1 month and after 1 year, including those from the off-axis site, away from active venting, and those from vent seawater samples on both 0.45- and 1-μm-pore-size water filters (Table 1). Host tissue was detected only on a single prefilter (1 μm) water sample. PCR amplifications from surface seawater control samples yielded positive PCR results with universal Bacteria primers (27f and 1492r) (13) but yielded negative results when either the vestimentiferan symbiont- or host-specific primers were used, suggesting that symbiont phylotypes were present only in deep-seawater samples. Little is yet known about the metabolic state or energy source for symbionts outside of their tube worm hosts, but it is possible that free-living symbionts may be cystic or quiescent while awaiting the inoculation of larval or juvenile tube worms.
TABLE 1.
Detection of free-living symbiont phylotype of vent vestimentiferan tube worms via PCR and sequence analyses of biofilmsa
| Time of collection and type of deployment | Total no. of blocksd | No. of blocks with positive result by:
|
|||
|---|---|---|---|---|---|
| PCR amplificationb
|
Sequence verificationc | ||||
| Bacteria positive control | Tube worm symbiont | Tube worm host | |||
| ∼1 mo | |||||
| Among R. pachyptila | 3 | 3 | 3 | NDe | 3 |
| Near R. pachyptila | 4 | 4 | 4 | ND | 3 |
| Away from R. pachyptila | 5 | 5 | 5 | ND | 3 |
| Off-axis | 3 | 2 | 2 | ND | 1 |
| 1 yr | |||||
| Among R. pachyptila | 1 | 1 | 1 | ND | 1 |
| Near R. pachyptila | 2 | 2 | 2 | ND | NAf |
| Away from R. pachyptila | 3 | 3 | 3 | ND | 3 |
| Off-axis | 3 | 3 | 3 | ND | 3 |
Basalt blocks, deployed at various distances from R. pachyptila tube worm clusters at the Tica vent site along the EPR, were collected and analyzed after ∼1 month and after 1 year.
PCR amplification was performed using primers for universal Bacteria (27f and 1492r), vestimentiferan host-specific primers (for RP43, RifTOExoF and RifTOExoR), and symbiont-specific primers (RifTO44 and RifTO445).
A subset of basalt biofilm samples was analyzed to verify the presence of the free-living tube worm symbiont phylotype.
Number of basalt blocks analyzed for each deployment location and time.
Tube worm hosts were not detected (ND) on any analyzed samples; samples with tube worm hosts were discarded.
Only one sample was not analyzed (NA) because sequencing reactions did not work (including positive controls), perhaps due to inhibitory substrates in the DNA.
FISH.
FISH was used to provide direct visual evidence of the tube worm symbiont on glass slides recovered from bacterial settlement devices. For each slide, a universal Bacteria probe, Eub338 (1), either 5′end labeled with fluorescein or stained with the DNA-binding fluorescent dye 4′,6′-diamidino-2-phenylindole (DAPI), was used as a positive control along with the symbiont-specific probe RifTO147, RifTO445, or RifTO830 that was 5′end labeled with Cy3 (23). The images from the control and symbiont-specific probes were then overlaid. The probe specificity was tested on R. pachyptila trophosome tissue, and the formamide concentration was increased until no probe remained hybridized (probe dependent, 20% [for Fig. S2 in the supplemental material] or 35% [for Fig. 1]). On each slide, either a nonsense probe, NON338 (23), or a 1-base-mismatch probe was used as a negative control. Hybridized slides were viewed and digitally photographed using a Leica model DMRB fluorescence microscope.
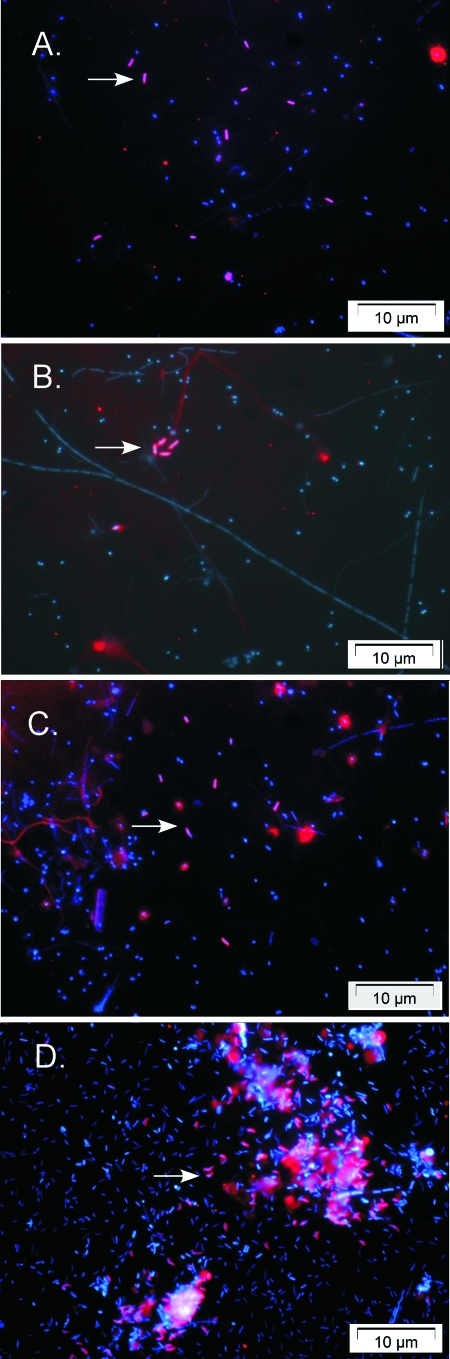
FIG. 1.
FISH detection of free-living vestimentiferan bacterial symbionts. Representative slides deployed at the Tica hydrothermal vent site on the EPR for ∼1 month among Riftia pachyptila tube worms (A), near tube worms (B and C), and 10 m away from tube worms (D) are shown. The overlay of two images with symbiont-specific probes (red [Cy3]) and DAPI (blue) shows the free-living symbionts (arrows) labeled with the symbiont-specific probes RifTO830 (A and C), RifTO147 (B), and RifTO445 (D).
The tube worm symbiont phylotype was detected using FISH on all slides tested (Fig. 1; see Fig. S2 in the supplemental material) with the exception of the off-axis samples that were collected from devices deployed for less than 1 month. Although not directly quantified, the overall bacterial abundance appeared to be greatest on slides deployed for 1 year among, adjacent to, or away from the tube worms. The direct detection of the tube worm symbiont in biofilms supports the hypothesis that these bacteria exist in the free-living vent environment. Indeed, in a coastal marine endosymbiosis, the 16S phylotype of bacterial symbionts of Codakia orbicularis clams is readily found in the sea grass sediment surrounding their hosts (11).
Endosymbiont ITS diversity.
If vestimentiferan tube worms acquire their symbionts from a diverse environmental source population, it can be hypothesized that the symbiont population within a host may consist of multiple closely related phylotypes (8, 31). The symbiont internal transcribed spacer (ITS), which is under relaxed selection relative to the 16S and has been used extensively to assess strain-level variation in bacteria (29), was cloned and sequenced to test for the presence of multiple symbiont phylotypes within individual tube worms. The ITS, located between the 16S and 23S rRNA genes in the bacterial rRNA operon, occurs as a single copy in the vestimentiferan symbiont genome (16, 26). By using symbiont-specific primers embedded in the 16S and 23S rRNA genes (Sym-ITS-1322F and Sym-ITS-23SR) (31), the ITS was PCR amplified (30 cycles with Taq polymerase) from DNA extracted from the trophosomes of three adult R. pachyptila worms. PCR products were cloned and sequenced (96 clones per specimen; 288 in total).
Analysis of the ITS sequences from the three R. pachyptila symbiont clone libraries revealed high levels of genetic homogeneity in intracellular symbiont populations. Sequence analysis revealed one dominant symbiont phylotype within each of the three host specimens (accounting for 65, 77, and 41% of the sequences, respectively), and the third specimen hosted a second phylotype (27%), which consistently differed by the same two nucleotides. The majority of the remaining ITS sequences were singletons that cannot be distinguished from errors resulting from PCR or Taq analyses. The detection of diverse ITS sequences in R. pachyptila worms further supports the acquisition of bacteria from the environment, but the diversity of free-living symbionts has not yet been investigated.
Evidence for environmental symbiont acquisition.
Detection of the free-living tube worm symbiont phylotype supports the hypothesis that newly settled tube worms obtain their bacteria from the vent environment. Along a spatial gradient, free-living symbionts were present among, adjacent to, and away from (within 10 m) tube worms and were also detected 100 m outside the areas of hydrothermal activity. The presence of free-living symbiotic bacteria at multiple spatial scales within a vent site suggests a potentially large environmental pool of symbionts. During host larval development and the colonization of new vents (17, 20, 21), an abundant free-living bacterial population would facilitate the initiation of the symbiosis. The environmental transmission of symbionts seems to be a risky strategy for obligate tube worm symbioses, as the survival of the mouthless and gutless adult host requires that developing larvae or juveniles successfully acquire their symbionts from a potentially unstable free-living source population. However, this developmental mode might be beneficial if it provides the host with opportunities to acquire specific, locally adapted symbiont genotypes.
Influence of symbiotic bacteria on free-living microbial diversity.
Symbioses, notably those that are facultative, clearly have an impact on and may be a driving force of local microbial diversity in varied ecosystems (2, 10). Indeed, the bacterial symbionts of the shrimp Rimicaris exoculata make up a major component of the surrounding microbial community at hydrothermal vents in the Atlantic Ocean (25). Likewise, a free-living counterpart to the bioluminescent symbiotic bacterium Vibrio fischeri of squid has been identified in coastal environments, revealing a connection between the symbiotic relationship and microbial abundance and distribution (15). The same situation appears to be true in legume-rhizobium symbioses; the host species is thought to be a major factor in determining the characteristics of the soil microbial community (18). Endosymbiont and free-living populations may affect each other via positive feedback cycles, whereby the host inoculates the free-living population, and the free-living population inoculates the host (24). This study serves as the basis for future investigations of the biodiversity and biogeography of free-living marine symbionts at multiple spatial scales.
Supplementary Material
Acknowledgments
We thank Chief Scientists Charles Fisher and Craig Cary; George Silva, Gary Chiljean, and the crew of the research vessel Atlantis; and the deep submergence vehicle Alvin group for collaborative expeditions, expert sample deployment, and collection. We thank Alan Fleer for instruction and use of the McLane pumps, Ansel Payne for technical assistance, and Thomas Auchtung, Stephanie Huff, and Irene Garcia Newton for assistance both on land and at sea. The paper was greatly improved thanks to Frank Stewart and four anonymous reviewers.
This work was supported by grants from the Austria Science Foundation (FWF H00087 and P13762) and the Austrian Academy of Science to M.B., the National Science Foundation (NSF DBI-0400591) to E.G.D, and the NOAA National Undersea Research Center for the West Coast and Polar Regions (UAF 03-0092) and the NSF (OCE-0453901) to C.M.C., which we gratefully acknowledge.
Footnotes
Published ahead of print on 11 April 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, A. C. 2003. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34:661-689. [Google Scholar]
- 3.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, Inc., New York, NY.
- 4.Cary, S. C., H. Felbeck, and N. D. Holland. 1989. Observations on the reproductive biology of the hydrothermal vent tubeworm Riftia pachyptila. Mar. Ecol. Prog. Ser. 52:89-94. [Google Scholar]
- 5.Cary, S. C., W. Warren, E. Anderson, and S. J. Giovannoni. 1993. Identification and localization of bacterial endosymbionts in hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol. Mar. Biol. Biotechnol. 2:51-62. [PubMed] [Google Scholar]
- 6.Cavanaugh, C. M., S. L. Gardiner, M. L. Jones, H. W. Jannasch, and J. B. Waterbury. 1981. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213:340-342. [DOI] [PubMed] [Google Scholar]
- 7.Dale, C., and N. A. Moran. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126:453-465. [DOI] [PubMed] [Google Scholar]
- 8.DeChaine, E. G., A. E. Bates, T. M. Shank, and C. M. Cavanaugh. 2006. Off-axis symbiosis found: characterization and biogeography of bacterial symbionts of Bathymodiolus mussels from Lost City hydrothermal vents. Environ. Microbiol. 8:1902-1912. [DOI] [PubMed] [Google Scholar]
- 9.Feldman, R., M. Black, C. Cary, R. Lutz, and R. Vrijenhoek. 1997. Molecular phylogenetics of bacterial endosymbionts and their vestimentiferan hosts. Mol. Mar. Biol. Biotechnol. 6:268-277. [PubMed] [Google Scholar]
- 10.Finlay, R. D. 2005. Mycorrhizal symbiosis: myths, misconceptions, new perspectives and future research priorities. Mycologist 19:90-95. [Google Scholar]
- 11.Gros, O., M. Liberge, A. Heddi, C. Khatchadourian, and H. Felbeck. 2003. Detection of the free-living forms of sulfide-oxidizing gill endosymbionts in the lucinid habitat (Thalassia testudinum environment). Appl. Environ. Microbiol. 69:6264-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, M. L., and S. L. Gardiner. 1988. Evidence for a transient digestive tract in Vestimentifera. Proc. Biol. Soc. Wash. 101:423-433. [Google Scholar]
- 13.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 14.Laue, B. E., and D. C. Nelson. 1997. Sulfur-oxidizing symbionts have not co-evolved with their hydrothermal vent tubeworm hosts: RFLP analysis. Mol. Mar. Biol. Biotechnol. 6:180-188. [PubMed] [Google Scholar]
- 15.Lee, K.-H., and E. G. Ruby. 1992. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl. Environ. Microbiol. 58:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markert, S., C. Arndt, H. Felbeck, D. Becher, S. M. Sievert, M. Hugler, D. Albrecht, J. Robidart, S. Bench, R. A. Feldman, M. Hecker, and T. Schweder. 2007. Physiological proteomics of the uncultured symbiont of Riftia pachyptila. Science 315:247-250. [DOI] [PubMed] [Google Scholar]
- 17.Marsh, A. G., L. S. Mullineaux, C. M. Young, and D. T. Manahan. 2001. Larval dispersal potential of the tubeworm Riftia pachyptila at deep-sea hydrothermal vents. Nature 411:77-80. [DOI] [PubMed] [Google Scholar]
- 18.Miethling, R., G. Wieland, H. Backhaus, and G. C. Tebbe. 2000. Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb. Ecol. 40:43-56. [DOI] [PubMed] [Google Scholar]
- 19.Moya, A., J. Pereto, R. Gil, and A. Latorre. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9:218-229. [DOI] [PubMed] [Google Scholar]
- 20.Mullineaux, L. S., C. R. Fisher, C. H. Peterson, and S. W. Schaeffer. 2000. Tubeworm succession at hydrothermal vents: use of biogenic cues to reduce habitat selection error? Oecologia 123:275-284. [DOI] [PubMed] [Google Scholar]
- 21.Mullineaux, L. S., S. W. Mills, A. K. Sweetman, A. H. Beaudreau, A. Metaxas, and H. L. Hunt. 2005. Vertical, lateral and temporal structure in larval distributions at hydrothermal vents. Mar. Ecol. Prog. Ser. 293:1-16. [Google Scholar]
- 22.Nelson, K., and C. R. Fisher. 2000. Absence of cospeciation in deep-sea vestimentiferan tube worms and their bacterial endosymbionts. Symbiosis 28:1-15. [Google Scholar]
- 23.Nussbaumer, A. D., C. R. Fisher, and M. Bright. 2006. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441:345-348. [DOI] [PubMed] [Google Scholar]
- 24.Polz, M. F., J. A. Ott, M. Bright, and C. M. Cavanaugh. 2000. When bacteria hitch a ride: associations between sulfur-oxidizing bacteria and eukaryotes represent spectacular adaptations to environmental gradients. ASM News 66:531-539. [Google Scholar]
- 25.Polz, M. F., and C. M. Cavanaugh. 1995. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc. Natl. Acad. Sci. USA 92:7232-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robidart, J. C., S. R. Bench, R. A. Feldman, A. Novoradovsky, S. B. Podell, T. Gaasterland, E. E. Allen, and H. Felbeck. 2008. Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ. Microbiol. 10:727-737. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 28.Southward, E. C. 1988. Development of the gut and segmentation of newly settled stages of Ridgeia (Vestimentifera): implications for relationships between Vestimentifera and Pogonophora. J. Mar. Biol. Assoc. U.K. 68:465-487. [Google Scholar]
- 29.Stewart, F. J., and C. M. Cavanaugh. 2007. Intragenomic variation and evolution of the internal transcribed spacer of the rRNA operon in bacteria. J. Mol. Evol. 65:44-67. [DOI] [PubMed] [Google Scholar]
- 30.Stewart, F. J., I. L. Newton, and C. M. Cavanaugh. 2005. Chemosynthetic endosymbioses: adaptations to oxic-anoxic interfaces. Trends Microbiol. 13:439-448. [DOI] [PubMed] [Google Scholar]
- 31.Won, Y.-J., S. J. Hallam, G. D. O'Mullan, I. L. Pan, K. R. Buck, and R. C. Vrijenhoek. 2003. Environmental acquisition of thiotrophic endosymbionts by deep-sea mussels of the genus Bathymodiolus. Appl. Environ. Microbiol. 69:6785-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.