Abstract
Helicobacter pylori VacA is a secreted pore-forming toxin that is comprised of two domains, designated p33 and p55. The p55 domain has an important role in the binding of VacA to eukaryotic cell surfaces. A total of 111 residues at the amino terminus of p55 (residues 312 to 422) are essential for the intracellular activity of VacA, which suggests that this region may constitute a subdomain with an activity distinct from cell binding. To investigate the properties of this subdomain, a small deletion mutation (targeting aspartic acid 346 and glycine 347) was introduced into the H. pylori chromosomal vacA gene. Similar to wild-type VacA, the VacA Δ346-347 mutant protein was proteolytically processed, secreted, and bound to eukaryotic cells. However, VacA Δ346-347 did not cause cell vacuolation or membrane depolarization, and it was impaired in the ability to assemble into large water-soluble oligomeric structures. Interestingly, VacA Δ346-347 was able to physically interact with wild-type VacA to form mixed oligomeric complexes, and VacA Δ346-347 inhibited wild-type vacuolating activity in a dominant-negative manner. These data indicate that the assembly of functional oligomeric VacA complexes is dependent on specific sequences, including amino acids 346 and 347, within the p55 amino-terminal subdomain.
Helicobacter pylori is a gram-negative, microaerophilic bacterium that persistently colonizes the human stomach (10, 36). Most H. pylori-infected patients have no symptoms, but the presence of H. pylori is a risk factor for development of peptic ulcer disease, gastric adenocarcinoma, and gastric lymphoma (10, 36). One of the important virulence factors produced by H. pylori is an exotoxin known as vacuolating toxin (VacA) (3, 13). In a mouse model of infection, a wild-type H. pylori strain expressing VacA has a selective advantage for colonization of the stomach compared to an isogenic vacA-null mutant (31). VacA contributes to gastric damage in animal models (14, 39), and H. pylori strains containing specific allelic forms of vacA are associated with an increased risk of symptomatic disease in humans (2, 12). VacA can cause a wide range of alterations in eukaryotic cells in vitro. One of the most prominent activities of VacA is its capacity to induce the formation of large cytoplasmic vacuoles (21). Other in vitro activities of VacA include permeabilization of the plasma membrane (7, 38), reduction of the mitochondrial transmembrane potential and release of cytochrome c from mitochondria (15, 45), inhibition of the activation and proliferation of T lymphocytes (18, 37), and activation of mitogen-activated protein kinases (28). Many cellular effects of VacA are attributable to the insertion of VacA into membranes and the formation of anion-selective membrane channels (3).
The vacA gene encodes a protein approximately 140 kDa in mass, which undergoes proteolytic processing to yield a 33-amino-acid signal sequence, a mature 88-kDa protein, and a carboxy-terminal domain (6, 32, 39). The secretion of VacA occurs through a classical autotransporter (type Va) pathway (3, 13). In contrast to most proteins that are secreted by the classical autotransporter pathway (8), VacA 88-kDa monomers can assemble into large water-soluble oligomeric complexes. Bilayered flower-shaped complexes (dodecamers and tetradecamers), as well as single-layered complexes (containing six to nine subunits), have been visualized (4, 11). Disassembly of these complexes by treatment at low pH or high pH is required in order for VacA to cause most cellular effects (4). VacA monomers reassemble into oligomeric structures when in contact with membranes (7), and the assembly of VacA into oligomeric structures is likely to be required for membrane channel formation.
The mature 88-kDa VacA passenger domain can undergo proteolytic cleavage to yield an amino-terminal 33-kDa fragment (p33) and a carboxy-terminal 55-kDa fragment (p55), which remain physically associated (39, 41). The p33 and p55 fragments are considered to represent two domains or subunits of VacA (39-41). The p55 domain is comprised predominantly of a β-helical structure, a feature characteristic of autotransporter passenger domains (16). When added individually to the surface of cells, neither p33 nor p55 causes cell vacuolation (40). Similarly, when expressed individually in transiently transfected cells, neither p33 nor p55 causes cell vacuolation (49). However, a mixture of p33 and p55 can cause cell vacuolation when added to the surface of cells, and intracellular coexpression of p33 and p55 results in cell vacuolation (40, 49).
Several putative VacA subdomains with distinct activities have been identified. A hydrophobic region located near the amino terminus of p33 (residues 1 to 32) has an important role in the formation of anion-selective membrane channels (22, 26, 43). A VacA mutant protein with a deletion of amino acid residues 49 to 57 failed to assemble into oligomeric structures, which suggests that specific sequences in this region of the p33 domain are required for VacA oligomerization (19). Amino acid sequences near the carboxy terminus of the p55 domain are required for VacA binding to cells (17, 29, 30, 44). When VacA is expressed in transiently transfected HeLa cells (thus eliminating a requirement for toxin binding to the cell surface and internalization), the minimum portion of VacA required for cell vacuolation comprises a 422-amino-acid protein, corresponding to the entire p33 domain and about 111 amino acids from the amino-terminal portion of p55 (49). This result suggests that the amino-terminal portion of p55 may comprise a subdomain with a functional activity distinct from cell binding.
The goal of the present study was to investigate properties of VacA that are conferred by the p55 amino-terminal subdomain. Thus far, a two-amino-acid deletion mutation (Δ346-347) is the only small alteration within the p55 amino-terminal subdomain that is known to abrogate vacuolating toxin activity (48). Efforts to identify additional small inactivating mutations in the p55 domain using a random mutagenesis approach have not been successful (25). A previous study reported that the VacA Δ346-347 mutant protein lacked vacuolating activity when expressed intracellularly, but the basis for this lack of activity has not yet been investigated (48). Therefore, in the present study we sought to investigate the basis for inactivity of the Δ346-347 mutant protein and to compare the properties of VacA Δ346-347 to those of wild-type VacA. We report that the Δ346-347 mutant protein is proteolytically processed and secreted by H. pylori in a manner similar to that of wild-type VacA. However, the Δ346-347 mutant protein does not cause membrane depolarization and is impaired in the ability to assemble into functional oligomeric VacA complexes. These results provide evidence that assembly of VacA into functional oligomeric complexes is dependent on specific sequences, including amino acids 346 and 347, within the p55 amino-terminal subdomain.
MATERIALS AND METHODS
H. pylori strains and purification of VacA from H. pylori broth culture supernatants.
The vacA gene (GenBank accession number Q48245) from H. pylori 60190 (ATCC 49503) served as the parent DNA for construction of all mutants in the present study. Throughout the present study, we used an amino acid numbering system in which residue 1 refers to alanine-1 of the secreted 88-kDa VacA protein, and the p55 domain corresponds to amino acids 312 to 821. The crystal structure of residues 355 to 811 (within the p55 domain) has recently been determined (16). Wild-type H. pylori strain 60190 and strains that express a VacA Δ6-27 mutant protein or a c-Myc-tagged VacA protein have been described previously (Table 1) (23, 43). An H. pylori strain expressing a Δ346-347 mutant protein was constructed as described below. H. pylori strains were grown in sulfite-free brucella broth containing activated charcoal (20). VacA Δ6-27 and VacA-c-Myc proteins were purified in an oligomeric form from culture supernatants of H. pylori, using gel filtration chromatography (4, 23, 43). It was not possible to purify VacA Δ346-347 from H. pylori broth culture supernatants by using gel filtration. Therefore, for all experiments designed to compare the activity and properties of wild-type VacA and VacA Δ346-347, these proteins were purified from H. pylori culture supernatants by using Cellufine Sulfate Matrex beads (Chisso Corp., Tokyo, Japan) (19), unless otherwise specified. Proteins in H. pylori broth culture supernatants were precipitated with ammonium sulfate, the resuspended proteins were dialyzed in sodium phosphate buffer (20 mM sodium phosphate, 100 mM sodium chloride [pH 7]), and the dialyzed samples were then incubated with Matrex beads at room temperature for 30 min. VacA was eluted from the beads with sequentially increasing concentrations of NaCl. As a negative control, culture supernatant from a vacA-null mutant strain (60190 vacA::km) was processed in the same manner.
TABLE 1.
H. pylori strains and plasmids
| H. pylori strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| 60190 | ATCC 49503; encodes wild-type s1/m1 VacA | 6 |
| VT330 | Encodes VacA with c-Myc epitope | 23 |
| AV452 | Encodes VacA Δ6-27 | 43 |
| SI433 | Derived from H. pylori strain VM025, which contains a sacB-kan cassette within vacA; SI433 encodes VacA Δ346-347 | 43; this study |
| 60190 vacA::km | vacA-null mutant | 6 |
| Plasmids | ||
| pMM592 | Encodes wild-type VacA, amino acids 1 to 821 | 24 |
| pSI200 | Derived from pMM592; encodes VacA Δ346-347 | This study |
| pSI201 | Derived from pSI200; encodes VacA Δ346 | This study |
| pSI202 | Derived from pSI200; encodes VacA Δ347 | This study |
| pSI203 | Derived from pSI200; encodes VacA G347A | This study |
| pSI204 | Derived from pSI200; encodes VacA G347R | This study |
| pSI205 | Derived from pSI200; encodes VacA D346L/G347V | This study |
| pSI206 | Derived from pSI200; encodes VacA D346E/G347V | This study |
| pSI207 | Derived from pSI200; encodes VacA D346L/G347R | This study |
| pSI208 | Derived from pSI200; encodes VacA D346R/G347R | This study |
| pSI209 | Encodes VacA His p55 Δ346-347 | This study |
| pET41b VacA p55 | Encodes VacA His p55 | 40 |
| pET41b VacA p33 MH | Encodes VacA p33 Myc-His | 40 |
| pET41b VacA p33 H | Encodes VacA p33 His | 40 |
Expression of VacA Δ346-347 in H. pylori.
A Δ346-347 mutation (encoding a deletion of VacA amino acids 346 and 347) was introduced into the H. pylori chromosomal vacA gene by natural transformation and allelic exchange using a sacB-based counterselection approach, as described previously (23, 26, 43). Sequence analysis of a PCR product was performed to confirm that the desired mutation had been introduced successfully into the chromosomal vacA gene.
Expression of recombinant VacA proteins in Escherichia coli.
pMM592 is a previously described plasmid that allows expression of an 88-kDa VacA protein in E. coli (Table 1) (24). Plasmids for expression of VacA p33 and p55 fragments have been described previously (40); the encoded p33 proteins contain either a His6 tag at the carboxyl terminus of the protein (p33 His) or both c-Myc and His6 tags at the carboxyl terminus of the protein (p33 Myc-His), and p55 contains a His6 tag at the amino terminus of the protein (His p55). To construct pMM592 Δ346-347 (pSI200), the vacA gene from pET20b-1-741 Δ346-347-GFP (48) was digested with EcoRI and KpnI and ligated into EcoRI- and KpnI-digested pMM592. To introduce additional substitution and deletion mutations into the codons for amino acids 346 and 347, we performed inverse PCR, using appropriate primers and pSI200 as template DNA (47). The resulting PCR products were then ligated and transformed into E. coli DH5α. To construct a plasmid encoding the p55 domain of VacA with a Δ346-347 mutation, the corresponding region of vacA (encoding amino acids 312 to 821) was amplified by using primers AND7265 and AND515a (40), which resulted in the insertion of a His6 tag at the amino terminus. The PCR product was digested with SpeI and SalI and ligated into XbaI- and SalI-digested pET41b (Novagen), to yield p55 Δ346-347 (pSI209). In each case, the plasmids were analyzed by sequence analysis to confirm that the desired mutation was present and that no new mutations had been introduced. VacA expression plasmids were transformed into the E. coli expression strain ER2566 (New England Biolabs), which encodes an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible copy of the RNA polymerase gene from bacteriophage T7. VacA-expressing E. coli strains were cultured in Terrific Broth (Invitrogen) supplemented with 25 μg of kanamycin/ml (24, 40), and extracts containing soluble proteins were generated as described previously (24, 25).
Cell culture analysis of VacA proteins expressed in H. pylori or E. coli.
HeLa and AZ-521 cells were grown as described previously (40). In all experiments, preparations of VacA purified from H. pylori culture supernatants were acid-activated by the addition of 100 mM hydrochloric acid, lowering the pH to 3, before VacA was added to cells (9, 27). An equivalent volume of a corresponding preparation from a vacA-null mutant (60190 vacA::km) was used as a negative control. For experiments using multiple recombinant VacA proteins, the relative concentrations of recombinant VacA in different E. coli soluble extracts were assessed by immunoblotting, and the extracts were then normalized so that the relative concentrations of VacA in different preparations were approximately equivalent (40). In comparison to wild-type VacA, none of the mutant proteins exhibited substantial differences in stability or susceptibility to proteolytic degradation. Signals were generated by the enhanced chemiluminescence reaction (Amersham Biosciences) and detected using X-ray film. Recombinant VacA proteins were added to cells as described previously (40). After incubation, cell vacuolation was examined by inverted light microscopy and quantified by a neutral red uptake assay (5). Neutral red uptake data are presented as A540 values (mean ± the standard deviation [SD]). The levels of neutral red uptake produced by negative control samples were subtracted as background.
To test for dominant-negative activity, wild-type VacA (purified by gel filtration) was acid-activated and then mixed with acid-activated preparations of VacA Δ346-347 (purified by Matrex affinity resin), VacA Δ6-27, or a mock sample from a vacA-null mutant strain. The mixtures were then neutralized by diluting with neutral pH medium before addition to HeLa cells (43). The samples were incubated with cells for 1 h at 37°C, the medium overlaying cells was removed, and fresh serum-free medium containing 5 mM ammonium chloride was added to cells for 5 h at 37°C. Cell vacuolation was detected by inverted light microscopy and quantified by a neutral red uptake assay.
Membrane depolarization.
Analysis of membrane potential was performed as described previously (26, 38) except that AZ-521 cells were detached with Accutase. Purified acid-activated VacA, or a mock preparation derived from an H. pylori vacA-null mutant strain, was added to the cells, and the changes in the fluorescence were monitored.
BN-PAGE.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) (46) was used to investigate the oligomeric state of VacA proteins. In this technique, protein complexes are separated based on molecular size under nondenaturing conditions. Purified wild-type VacA and VacA Δ346-347 proteins were desalted using Zeba Desalt spin columns (Pierce). Approximately 13 μg (5 μl) of each sample was mixed with 1 μl of 1% dodecylmaltoside (a nonionic detergent that is not expected to disrupt protein complexes) and then mixed with 2.5 μl of 50% glycerol and 5% Coomassie blue G-250 dye stock suspension to give a detergent/dye ratio of 1.0 g/g and electrophoresed on a 4 to 13% polyacrylamide gel. Lanes were cut out from the gel, boiled in sodium dodecyl sulfate (SDS) electrophoresis buffer for 10 min, and mounted on top of an 8% SDS-polyacrylamide gel for second-dimension analysis. After transfer to nitrocellulose, the samples were immunoblotted with an anti-VacA polyclonal serum (serum 958), followed by an horseradish peroxidase (HRP)-conjugated secondary antibody. Signals were generated by the enhanced chemiluminescence reaction and detected by using X-ray film.
Modified SDS-PAGE methodology.
The oligomeric state of VacA proteins was also assessed by using a variant of the usual SDS-polyacrylamide gel electrophoresis (PAGE) methodology. VacA preparations were mixed with 4% SDS lysis buffer (containing 1.5% Tris, 20% glycerol, 4% SDS, 10% 2-mercaptoethanol, and 0.002% bromophenol blue), resulting in a final SDS concentration of 2% in each sample. These samples were either boiled or not boiled, electrophoresed on an SDS-polyacrylamide gel (6% separating gel and 4% stacking gel), and analyzed by immunoblotting as described above.
Immunoprecipitation of VacA proteins.
Immunoprecipitations were performed as described previously, with minor modifications (23, 40). Briefly, E. coli soluble extracts containing either c-Myc- or His6-tagged VacA fragments (40) were mixed for 1 h at room temperature. The mixtures tested included the combination of p33 Myc-His with either His p55 or His p55 Δ346-347. Samples were normalized by immunoblotting with an antibody to the His6 epitope (anti-His; Santa Cruz Biotechnology). After 1 h, these samples were diluted in 1 ml of phosphate-buffered saline (pH 7) containing 0.05% Tween 20 and 2% ammonium sulfate. Anti-c-Myc monoclonal antibody (9E10; 3 μg) was added, and the mixture was incubated at 4°C for 2 h. Protein G-Sepharose beads (Amersham Biosciences) were added to the VacA-antibody mixture, followed by incubation for 16 to 18 h at 4°C. The immunoprecipitated proteins were separated from the beads by boiling the beads in SDS-PAGE sample buffer and were analyzed by immunoblotting with an anti-His antibody, followed by the addition of an HRP-conjugated secondary antibody. To analyze a potential interaction between p33 His, His p55 Δ346-347, and full-length 88-kDa VacA, normalized E. coli extracts containing the former two proteins were mixed with acid-activated c-Myc-tagged VacA (Myc-VacA) purified from H. pylori culture supernatant (2 μg/ml) for 1 h at 25°C, and the proteins were immunoprecipitated with an anti-c-Myc antibody as described above. Immunoprecipitated proteins were analyzed by immunoblotting with anti-His and anti-c-Myc antibodies, followed by an HRP-conjugated secondary antibody.
RESULTS
Secretion of VacA Δ346-347 by H. pylori and analysis of binding and vacuolating activity.
To investigate properties of the VacA Δ346-347 protein, we introduced the Δ346-347 mutation into the H. pylori chromosomal vacA gene by natural transformation and allelic exchange as described in Materials and Methods. An 88-kDa VacA protein was detected in broth culture supernatant from the Δ346-347 mutant H. pylori strain, which indicated that the 140-kDa precursor protein (3) containing the Δ346-347 mutation underwent proteolytic processing and secretion, similar to wild-type VacA (Fig. 1). We next investigated the interactions of wild-type VacA and VacA Δ346-347 with HeLa cells. Flow cytometry analysis, performed as described previously (1), revealed that both wild-type VacA and VacA Δ346-347 bound to HeLa cells in a dose-dependent manner (data not shown), thereby indicating that VacA Δ346-347 was not defective in binding to HeLa cells. The capacity of VacA Δ346-347 to be proteolytically processed and secreted by H. pylori, as well its retention of cell-binding activity, suggested that this mutant protein was not grossly misfolded. Notably, wild-type VacA caused cell vacuolation, whereas cell vacuolation was not observed when VacA Δ346-347 was added to HeLa cells (Fig. 2A). Similarly, wild-type VacA caused extensive vacuolation of AZ-521 gastric epithelial cells, whereas VacA Δ346-347 did not cause vacuolation of these cells (data not shown).
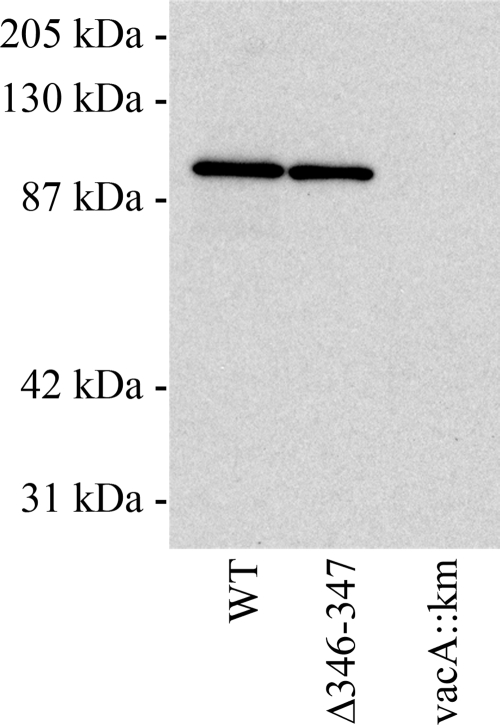
FIG. 1.
Secretion of VacA Δ346-347. Wild-type H. pylori strain 60190, an isogenic mutant strain encoding a VacA Δ346-347 protein, and a vacA-null mutant strain (60190 vacA::km) were cultured in brucella broth containing activated charcoal, and proteins in the broth culture supernatants were precipitated with a 50% saturated solution of ammonium sulfate. Proteins were electrophoresed on a 10% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and immunoblotted with polyclonal anti-VacA serum. Wild-type VacA and VacA Δ346-347 were each proteolytically processed to yield an 88-kDa protein that was secreted by H. pylori into the broth culture supernatant. WT, wild type.
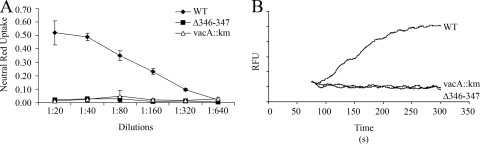
FIG. 2.
Functional analysis of VacA Δ346-347 activity. H. pylori strains expressing wild-type VacA or VacA Δ346-347, and a vacA-null mutant strain (60190 vacA::km) were grown in broth culture, and VacA proteins were purified as described in Materials and Methods. Concentrations of wild-type VacA and VacA Δ346-347 were normalized based on immunoblot assays, and the preparations were tested for vacuolating activity and membrane depolarization. (A) Analysis of vacuolating activity. For VacA-containing samples, a dilution of 1:20 corresponds to a VacA concentration of approximately 15 μg/ml. An equivalent volume of sample from the vacA::km-null mutant strain was tested as a control. Acid-activated samples were added to the medium overlying HeLa cells and vacuolating activity was quantified by using a neutral red uptake assay. The results represent the mean ± the SD from triplicate samples. (B) Analysis of depolarization. AZ-521 cells were loaded with oxonol VI (a probe used to monitor membrane potential). After the addition of acid-activated VacA proteins (10 μg/ml) or a control preparation (vacA::km), changes in fluorescence were monitored. Wild-type VacA induced membrane depolarization, whereas VacA Δ346-347 and the control vacA::km preparation did not. RFU, relative fluorescence units. The results are representative of four experiments. WT, wild type.
Analysis of cellular depolarization.
The addition of wild-type VacA to cells results in depolarization of the resting membrane potential, a phenomenon attributed to insertion of VacA into the plasma membrane to form anion-selective channels (33, 38). In the next experiments, we compared the capacity of wild-type VacA and VacA Δ346-347 to cause depolarization of AZ-521 cells. Consistent with previously published results, we found that addition of wild-type VacA to cells induced membrane depolarization (Fig. 2B), and a mock preparation derived from an H. pylori vacA-null mutant strain did not induce depolarization. When VacA Δ346-347 was added to AZ-521 cells, membrane depolarization was not detected (Fig. 2B). Similar results were obtained when we used HeLa cells instead of AZ-521 cells (data not shown). The failure of the VacA Δ346-347 mutant protein to depolarize AZ-521 cells suggests that this mutant toxin is defective in membrane channel formation.
Oligomerization of VacA Δ346-347.
VacA 88-kDa monomers produced by H. pylori are known to assemble into large water-soluble, flower-shaped structures (4). The assembly of VacA monomers into oligomeric structures is likely to be required for membrane channel formation and membrane depolarization. To test whether VacA Δ346-347 formed oligomers similar to those formed by wild-type VacA, we initially used gel filtration followed by analysis of fractions by immunoblotting with anti-VacA serum. Consistent with previous studies (4, 11), wild-type VacA was detected in fractions corresponding to a molecular mass of about 1,000 kDa. When VacA Δ346-347 was analyzed in the same manner, only trace amounts of VacA Δ346-347 were detected in these high-molecular-mass fractions (data not shown). Trace amounts of VacA Δ346-347 were detected in a broad range of lower molecular mass fractions, without evidence of a well-defined peak. The gel filtration elution properties of VacA Δ346-347 are similar to those reported for a mutant VacA protein containing a deletion of the p33 domain (30) and similar to several mutant VacA proteins containing large deletions within the p33 domain (43). The gel filtration properties of VacA Δ346-347 differ markedly from those of several previously described inactive VacA proteins with mutations in the p33 domain, which formed large oligomeric structures similar to wild-type VacA (25, 26, 43).
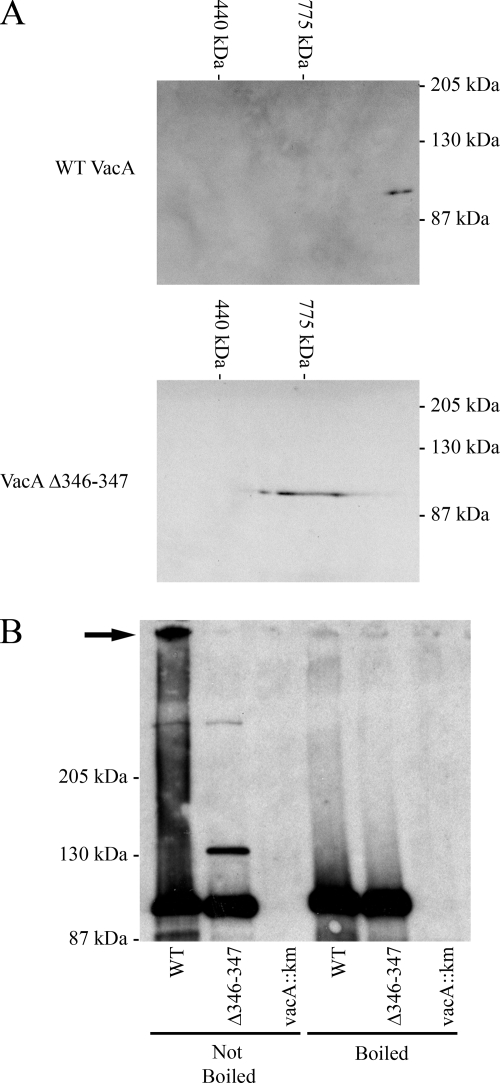
We next used gel electrophoresis methods to analyze the oligomeric state of VacA Δ346-347. Efforts to detect wild-type VacA oligomers using native gel electrophoresis were unsuccessful, because VacA did not enter the gel. Therefore, we analyzed the wild-type and mutant VacA proteins by using BN-PAGE, as described in Materials and Methods. Wild-type VacA was detected as a complex substantially larger than 775 kDa (Fig. 3A, top panel). In this analysis, the molecular mass of VacA Δ346-347 was substantially lower than that of wild-type VacA (Fig. 3A, bottom panel). VacA Δ346-347 appeared as a horizontal streak instead of a well-circumscribed spot, which suggested that this proteinaceous spot may be comprised of a heterogeneous mixture of oligomeric structures.
FIG. 3.
Analysis of oligomer formation by wild-type VacA and VacA Δ346-347. (A) BN-PAGE. Wild-type VacA (top panel) and VacA Δ346-347 (bottom panel) were purified and then analyzed by BN-PAGE, followed by immunoblotting with an anti-VacA serum. (B) Analysis by modified SDS-PAGE. VacA was precipitated from H. pylori broth culture supernatants with ammonium sulfate and VacA protein concentrations were normalized based on immunoblot analysis. Equivalent amounts of precipitated proteins were suspended in an SDS-containing buffer. One set of preparations (lanes 1 to 3, left) was not boiled, and a duplicate set of preparations (lanes 4 to 6, right) was boiled prior to SDS-PAGE. Samples were run on a 6% SDS gel, followed by immunoblotting with an anti-VacA serum. The arrow indicates a large oligomeric VacA complex. WT, wild type.
As another approach to compare the oligomeric state of VacA Δ346-347 and wild-type VacA, we used a modification of the SDS-PAGE procedure in which samples were suspended in loading buffer containing SDS and then either boiled or not boiled prior to electrophoresis. As expected, both wild-type VacA and VacA Δ346-347 yielded 88-kDa bands if the proteins were boiled prior to SDS-PAGE (Fig. 3B). In the absence of boiling, wild-type VacA was detected as both a high-molecular-mass complex (>250 kDa) and an 88-kDa band (Fig. 3B). When unboiled VacA Δ346-347 was analyzed in the same manner, the high-molecular-mass complex was not detected, but a smaller complex was detected (Fig. 3B). This modified SDS-PAGE assay does not permit an accurate determination of the molecular mass of nondenatured proteins or protein complexes, based on comparison with molecular mass markers. However, the results suggest that VacA Δ346-347 can form a complex with a mass larger than that of the 88-kDa VacA monomer, but smaller than that of wild-type VacA oligomers. Collectively, the gel filtration results, BN-PAGE experiments, and modified SDS-PAGE results all suggest that VacA Δ346-347 and wild-type VacA differ in the ability to assemble into large oligomeric complexes. In addition, it is possible that complexes formed by VacA Δ346-347 are less stable in the presence of detergent than are the complexes formed by wild-type VacA.
Interaction between p33 and p55 Δ346-347.
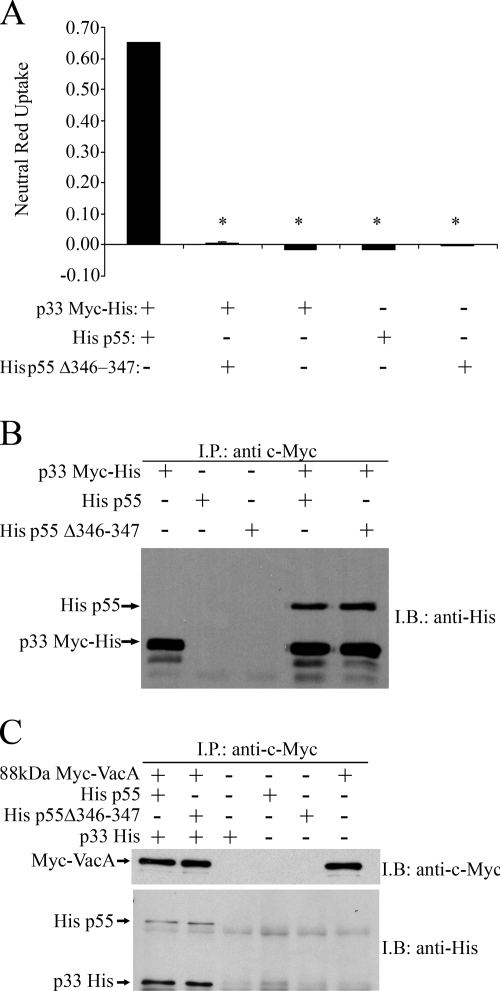
When E. coli extracts containing VacA p33 and p55 fragments are mixed and added to HeLa cells, extensive cell vacuolation is seen, whereas when added to cells individually, the p33 and p55 proteins do not induce cell vacuolation (40). To further investigate the effect of the Δ346-347 mutation on VacA activity and oligomerization, we expressed an isolated p55 VacA fragment containing the Δ346-347 mutation in E. coli. As expected, a mixture of p33 plus wild-type p55 proteins induced vacuolation of HeLa cells (Fig. 4A). When a mixture of p33 plus p55 Δ346-347 was added to HeLa cells, vacuolation was not detected. To determine whether p55 Δ346-347 could physically interact with p33, we performed immunoprecipitation experiments. Different combinations of epitope-tagged recombinant proteins were mixed, and immunoprecipitation was performed as described in Materials and Methods. As shown in Fig. 4B, p33 and p55 Δ346-347 interacted in solution, similar to the interaction of p33 with wild-type p55. The inclusion of two negative controls excluded nonspecific interactions between p55 and the antibody or beads (Fig. 4B). These data indicate that the Δ346-347 mutation does not abrogate interactions between the p33 and p55 VacA domains.
FIG. 4.
Analysis of recombinant p33, p55, and p55 Δ346-347 VacA domains. (A) Vacuolating toxin activity. E. coli soluble extracts containing the indicated recombinant VacA proteins were normalized and added to HeLa cells as described in Materials and Methods. Vacuolating activity was quantified by using a neutral red uptake assay. The results represent the mean ± the SD from triplicate samples. *, P ≤ 0.05 as determined by using analysis of variance (ANOVA), followed by Dunnett's post hoc test compared to p33 Myc-His combined with His p55. (B) Interaction of p55 Δ346-347 with p33. E. coli soluble extracts containing normalized concentrations of p33 Myc-His, His p55, or His p55 Δ346-347 were mixed and proteins were immunoprecipitated (I.P.) with an anti-c-Myc antibody. Immunoprecipitated proteins were electrophoresed on a 10% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and immunoblotted (I.B.) with an anti-His antibody. (C) Interaction of p33-p55 Δ346-347 with 88-kDa VacA. E. coli extracts containing normalized concentrations of p33 His, His p55, and His p55 Δ346-347 were mixed with acid-activated c-Myc-tagged 88-kDa VacA protein (Myc-VacA). Proteins were immunoprecipitated with an anti-c-Myc antibody and then immunoblotted with an anti-c-Myc antibody (top panel) or an anti-His antibody (bottom panel).
Interactions of p33 and p55 domains with full-length 88-kDa VacA.
We have previously shown that a mixture of p33 and p55 VacA fragments can physically interact with wild-type full-length VacA (40, 42). Therefore, we next investigated whether a mixture of p33 and p55 Δ346-347 could interact with 88-kDa VacA. For these experiments, we used a c-Myc-tagged 88-kDa VacA protein (Myc-VacA) purified from H. pylori culture supernatant and recombinantly expressed p33 and p55 Δ346-347. After mixing Myc-VacA with p33 and p55 fragments, proteins were immunoprecipitated with an anti-c-Myc antibody. As expected, when the wild-type p33-p55 mixture was incubated with 88-kDa Myc-VacA, all three proteins were immunoprecipitated (Fig. 4C). Similarly, when the p33-p55 Δ346-347 mixture was incubated with 88-kDa Myc-VacA, both fragments interacted with full-length VacA (Fig. 4C).
Inhibition of wild-type VacA cytotoxic activity by VacA Δ346-347.
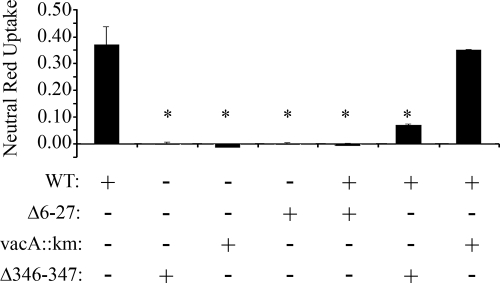
Certain inactive mutant forms of VacA can act as dominant-negative inhibitors of wild-type VacA activity (19, 23, 42, 43). Previous studies have suggested that the dominant-negative activity of mutant VacA proteins requires protein-protein interactions between wild-type VacA and the mutant proteins (23, 42, 43). The observation that VacA Δ346-347 can form mixed oligomeric complexes suggested that this mutant toxin might be capable of acting in a dominant-negative manner. Therefore, we next investigated whether VacA Δ346-347 could inhibit wild-type VacA activity. As a control, we tested another mutant toxin (VacA Δ6-27) previously shown to act as a dominant-negative inhibitor (43). Wild-type VacA and mutant toxins (VacA Δ346-347 or VacA Δ6-27) were mixed together, and then the mixtures were added to HeLa cells. As shown in Fig. 5, VacA Δ346-347 inhibited the cell-vacuolating activity of wild-type VacA, even when the concentration of VacA Δ346-347 was lower than that of wild-type VacA. These results indicate that VacA Δ346-347 acts as a dominant-negative inhibitor of wild-type VacA activity.
FIG. 5.
Inhibition of wild-type VacA cytotoxic activity by VacA Δ346-347. Preparations of acid-activated wild-type VacA (WT; 15 μg/ml) were incubated with 15 μg of acid-activated VacA Δ6-27/ml, 8 μg of acid-activated VacA Δ346-347/ml, or an equivalent volume of an acidified preparation from a vacA-null mutant strain (vacA::km) as described in Materials and Methods, and the mixtures were then added to the medium overlying HeLa cells for 1 h at 37°C. The toxins were removed, and fresh medium containing 5 mM ammonium chloride was added to HeLa cells for 5 h at 37°C. The vacuolating activity was quantified by using a neutral red uptake assay. The results represent the mean ± the SD from triplicate samples. *, P ≤ 0.05 as determined by using ANOVA, followed by Dunnett's post hoc test compared to WT.
Mutational analysis of residues 346 and 347.
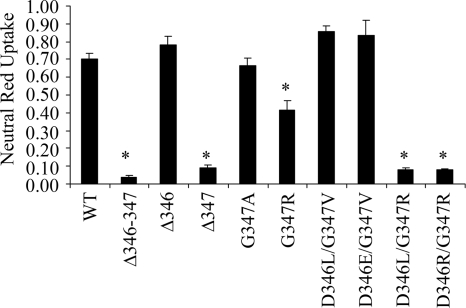
To undertake a more detailed mutational analysis, we expressed VacA Δ346-347 using a system that allows expression of a functional 88-kDa cytotoxic form of VacA in E. coli (24). It has not been possible to purify well-defined dodecameric structures when VacA is expressed in E. coli (M. S. McClain and T. L. Cover, unpublished results), but this system nevertheless permits analysis of vacuolating toxic activity (24). Soluble E. coli extracts containing VacA Δ346-347 or wild-type VacA were generated as described in Materials and Methods. Both recombinant proteins were successfully expressed based on immunoblotting analysis (data not shown). As expected, when extracts containing wild-type VacA were added to cells, extensive cell vacuolation was detected. In contrast, when E. coli extracts containing VacA Δ346-347 were added to HeLa cells, no vacuolation was detected (Fig. 6). To further investigate the role of VacA amino acids 346 (aspartic acid) and 347 (glycine) in VacA activity, we introduced several additional mutations into these sites. E. coli soluble extracts containing the mutant proteins were added to HeLa cells, and the vacuolating activity was measured by neutral red uptake. A mutant protein containing a deletion of amino acid 346 caused cell vacuolation similar to that caused by wild-type VacA (Fig. 6). In contrast, when amino acid 347 was deleted, vacuolating activity was not detected (Fig. 6).
FIG. 6.
Mutational analysis of VacA residues 346 and 347. Full-length WT VacA and a panel of VacA proteins containing mutations in residues 346 and/or 347 were expressed in E. coli. E. coli soluble extracts containing the indicated recombinant proteins were normalized based on immunoblotting so that they contained equivalent concentrations of VacA and were then added to the medium overlying HeLa cells. Vacuolating activity was quantified by using a neutral red uptake assay. The results represent the mean ± the SD from triplicate samples. *, P ≤ 0.05 as determined by using ANOVA, followed by Dunnett's post hoc test compared to WT VacA.
The introduction of specific pairs of substitution mutations at position 346 and 347 (D346L/G347R or D346R/G347R) abrogated VacA activity (Fig. 6). In contrast, other pairs of substitution mutations at these positions (D346L/G347V or D346E/G347V) did not abrogate VacA activity. Introduction of the G347R mutation alone (without any change at position 346) resulted in a partial loss of VacA activity. Thus, the loss of activity resulting from the Δ346-347 mutation was recapitulated by deletion of a single residue (amino acid 347) or by specific pairs of substitution mutations.
DISCUSSION
In this study, we sought to elucidate the properties of VacA that are conferred by an amino-terminal p55 subdomain. A recent analysis of the p55 VacA crystal structure (residues 355 to 811) showed that a substantial portion of the p55 domain comprises a beta-helical fold (16). The crystal structure reveals that the amino-terminal portion of p55 is spatially separated from carboxy-terminal portions of p55, which is consistent with the concept of an amino-terminal subdomain. Our analysis focused on alterations in VacA that result from a small deletion mutation (Δ346-347, corresponding to the deletion of contiguous aspartic acid and glycine residues, respectively). High-resolution structural data are not available for the portion of VacA comprising residues 346 and 347 (16), but the presence of adjacent glycine and proline residues at positions 347 and 348, respectively, suggests that this segment will represent part of a loop or turn.
As described in the present study, VacA Δ346-347 lacked vacuolating activity when added to the surface of cells and, in contrast to wild-type VacA, VacA Δ346-347 did not cause cell depolarization. The failure of this mutant toxin to induce cell depolarization suggests that it is unable to form membrane channels (26, 38). Multiple biochemical analyses provided evidence that the Δ346-347 mutation disrupts or weakens intermolecular VacA interactions. Defective VacA intermolecular interactions could result in impaired assembly or impaired stability of oligomeric complexes required for membrane channel activity or could result in an impaired ability of oligomers to undergo a conformational transition necessary for channel formation.
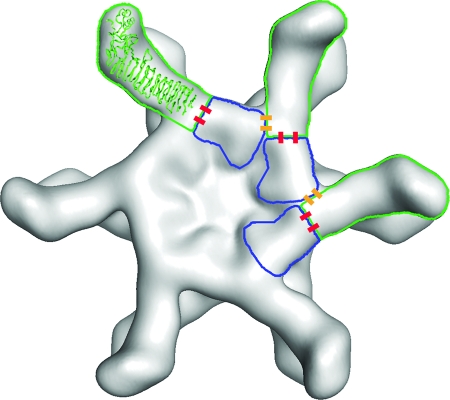
Thus far, very little is known about which amino acid sequences in VacA contribute to protein-protein interactions and oligomerization. Prior to the present study, the smallest mutation known to disrupt VacA oligomerization was a deletion of residues 49 to 57, located within the p33 domain (19). The current data indicate that residues 346 to 347 (located within the p55 domain) contribute to VacA oligomerization. Thus, amino acid sequences in both the p33 domain and the p55 domain are required for assembly of VacA into large oligomeric structures. This conclusion is consistent with a model for VacA oligomerization that is based on docking of the p55 crystal structure into a 19-Å cryo-EM map of a VacA dodecamer (16) (Fig. 7).
FIG. 7.
Model depicting how the Δ346-347 mutation interferes with oligomerization of VacA. The model is based on docking the p55 crystal structure into a 19-Å cryo-EM map of a VacA dodecamer (11, 16). p55 subunits are outlined in green, and p33 subunits are outlined in blue. Intramolecular p33-p55 interactions are depicted as red bars, and intermolecular p33-p55 interactions are depicted as yellow bars. We predict that residues 346 to 347 are located at or near the sites of yellow bars, and thus a Δ346-347 mutation would disrupt intermolecular p33-p55 interactions.
The Δ346-347 mutation could potentially interfere with VacA oligomerization by disrupting p33-55 interactions. It is also theoretically possible that the Δ346-347 mutation may disrupt p55-p55 interactions; however, at present there is no convincing evidence indicating that p55-p55 interactions are required for VacA oligomer formation, and interactions between isolated p55 fragments have not been readily detectable by immunoprecipitation or yeast two-hybrid methods (40, 41). Immunoprecipitation experiments indicated that p55 Δ346-347 can interact in solution with p33 (Fig. 4B), and a mixture of p33 plus p55 Δ346-347 can interact with full-length VacA (Fig. 4C). Thus, the Δ346-347 mutation does not completely abrogate p33-p55 interactions. Our model predicts that the assembly of VacA proteins into large oligomeric structures requires multiple types of p33-p55 interactions, including interaction of a single p55 domain with p33 domains from one or two adjacent molecules (intermolecular interactions), as well as an interaction with p33 from the same molecule (intramolecular interaction) (Fig. 7). These multiple types of p33-p55 interactions presumably are mediated by multiple different contact points on the surface of p55 (Fig. 7). Based on this model, the Δ346-347 mutation could interfere with VacA oligomerization, despite failure of this mutation to completely abrogate p33-p55 interactions.
An interesting property of VacA Δ346-347 is its ability to inhibit the activity of wild-type toxin in a dominant-negative manner (Fig. 5). Several dominant-negative mutant forms of VacA have been described in previous studies (19, 23, 42, 43), and at least one of these mutants (VacA Δ6-27) is able to block membrane channel formation by wild-type VacA (43). It has been hypothesized that the inhibitory activity of these mutants is dependent on their ability to physically interact with wild-type VacA, thereby forming mixed oligomeric complexes that are defective in functional activity (23, 43). Similarly, the dominant-negative phenotype of several Bacillus anthracis protective antigen mutants is also due to the formation of mixed oligomeric complexes containing wild-type and mutant proteins (34, 35). In contrast to two previously described dominant-negative mutant proteins (VacA Δ6-27 and VacA s2/m1) (23, 43), the mutant protein described in the present study (VacA Δ346-347) failed to assemble into large oligomeric structures. Similarly, a recent study reported that another mutant protein, VacA Δ49-57, failed to cause cell vacuolation and did not form large oligomeric structures but was able to inhibit the activity of wild-type VacA in a dominant-negative manner (19). One possibility is that the mechanisms of dominant-negative inhibition are different for one group of mutants (VacA Δ6-27 and VacA s2/m1) compared to the second group of mutants (VacA Δ346-347 and VacA Δ49-57). Alternatively, it is likely that the latter mutants, although defective in assembly into large oligomeric structures, are still able to physically interact with wild-type VacA. Specifically, the VacA Δ346-347 protein has a defective oligomerization site within the p55 domain, but it has an intact p33 domain. As shown in Fig. 7, each subunit within a dodecamer makes contact with other subunits via multiple p33-p55 intermolecular interactions. Based on this model, it is predicted that VacA Δ346-347 (via its p33 domain) would be able to interact with wild-type VacA. We speculate that oligomers containing both wild-type and mutant components would be defective in the ability to undergo conformational changes required for channel formation and therefore would be defective in vacuolating activity.
In summary, the present study provides new insights into properties of VacA that are conferred by the p55 amino-terminal subdomain. Ongoing structure-function studies of VacA should lead to a better understanding of how VacA forms membrane channels and causes alterations in human cells.
Acknowledgments
This study was supported by National Institutes of Health (NIH) grant R01 AI39657 and the Department of Veterans Affairs (T.L.C.) and NIH grant R01 AI45928, the Robert A. Welch Foundation (E-1311), and the American Heart Association (98BG472) (S.R.B.).
We thank Beverly Hosse for assistance with VacA purification.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 28 April 2008.
REFERENCES
- 1.Algood, H. M., V. J. Torres, D. Unutmaz, and T. L. Cover. 2007. Resistance of primary murine CD4+ T cells to Helicobacter pylori vacuolating cytotoxin. Infect. Immun. 75334-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 27017771-17777. [DOI] [PubMed] [Google Scholar]
- 3.Cover, T. L., and S. R. Blanke. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3320-332. [DOI] [PubMed] [Google Scholar]
- 4.Cover, T. L., P. I. Hanson, and J. E. Heuser. 1997. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J. Cell Biol. 138759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cover, T. L., W. Puryear, G. I. Perez-Perez, and M. J. Blaser. 1991. Effect of urease on HeLa cell vacuolation induced by Helicobacter pylori cytotoxin. Infect. Immun. 591264-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cover, T. L., M. K. Tummuru, P. Cao, S. A. Thompson, and M. J. Blaser. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 26910566-10573. [PubMed] [Google Scholar]
- 7.Czajkowsky, D. M., H. Iwamoto, T. L. Cover, and Z. Shao. 1999. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc. Natl. Acad. Sci. USA 962001-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dautin, N., and H. D. Bernstein. 2007. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu. Rev. Microbiol. 6189-112. [DOI] [PubMed] [Google Scholar]
- 9.de Bernard, M., E. Papini, V. de Filippis, E. Gottardi, J. Telford, R. Manetti, A. Fontana, R. Rappuoli, and C. Montecucco. 1995. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. J. Biol. Chem. 27023937-23940. [DOI] [PubMed] [Google Scholar]
- 10.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Bez, C., M. Adrian, J. Dubochet, and T. L. Cover. 2005. High resolution structural analysis of Helicobacter pylori VacA toxin oligomers by cryo-negative staining electron microscopy. J. Struct. Biol. 151215-228. [DOI] [PubMed] [Google Scholar]
- 12.Figueiredo, C., J. C. Machado, P. Pharoah, R. Seruca, S. Sousa, R. Carvalho, A. F. Capelinha, W. Quint, C. Caldas, L. J. van Doorn, F. Carneiro, and M. Sobrinho-Simoes. 2002. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J. Natl. Cancer Inst. 941680-1687. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, W., R. Buhrdorf, E. Gerland, and R. Haas. 2001. Outer membrane targeting of passenger proteins by the vacuolating cytotoxin autotransporter of Helicobacter pylori. Infect. Immun. 696769-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujikawa, A., D. Shirasaka, S. Yamamoto, H. Ota, K. Yahiro, M. Fukada, T. Shintani, A. Wada, N. Aoyama, T. Hirayama, H. Fukamachi, and M. Noda. 2003. Mice deficient in protein tyrosine phosphatase receptor type Z are resistant to gastric ulcer induction by VacA of Helicobacter pylori. Nat. Genet. 33375-381. [DOI] [PubMed] [Google Scholar]
- 15.Galmiche, A., J. Rassow, A. Doye, S. Cagnol, J. C. Chambard, S. Contamin, V. de Thillot, I. Just, V. Ricci, E. Solcia, E. Van Obberghen, and P. Boquet. 2000. The N-terminal 34-kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J. 196361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangwer, K. A., D. J. Mushrush, D. L. Stauff, B. Spiller, M. S. McClain, T. L. Cover, and D. B. Lacy. 2007. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc. Natl. Acad. Sci. USA 10416293-16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garner, J. A., and T. L. Cover. 1996. Binding and internalization of the Helicobacter pylori vacuolating cytotoxin by epithelial cells. Infect. Immun. 644197-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebert, B., W. Fischer, E. Weiss, R. Hoffmann, and R. Haas. 2003. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science 3011099-1102. [DOI] [PubMed] [Google Scholar]
- 19.Genisset, C., C. L. Galeotti, P. Lupetti, D. Mercati, D. A. Skibinski, S. Barone, R. Battistutta, M. de Bernard, and J. L. Telford. 2006. A Helicobacter pylori vacuolating toxin mutant that fails to oligomerize has a dominant negative phenotype. Infect. Immun. 741786-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawrylik, S. J., D. J. Wasilko, S. L. Haskell, T. D. Gootz, and S. E. Lee. 1994. Bisulfite or sulfite inhibits growth of Helicobacter pylori. J. Clin. Microbiol. 32790-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leunk, R. D., P. T. Johnson, B. C. David, W. G. Kraft, and D. R. Morgan. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 2693-99. [DOI] [PubMed] [Google Scholar]
- 22.McClain, M. S., P. Cao, and T. L. Cover. 2001. Amino-terminal hydrophobic region of Helicobacter pylori vacuolating cytotoxin (VacA) mediates transmembrane protein dimerization. Infect. Immun. 691181-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClain, M. S., P. Cao, H. Iwamoto, A. D. Vinion-Dubiel, G. Szabo, Z. Shao, and T. L. Cover. 2001. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J. Bacteriol. 1836499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClain, M. S., and T. L. Cover. 2003. Expression of Helicobacter pylori vacuolating toxin in Escherichia coli. Infect. Immun. 712266-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClain, M. S., D. M. Czajkowsky, V. J. Torres, G. Szabo, Z. Shao, and T. L. Cover. 2006. Random mutagenesis of Helicobacter pylori vacA to identify amino acids essential for vacuolating cytotoxic activity. Infect. Immun. 746188-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClain, M. S., H. Iwamoto, P. Cao, A. D. Vinion-Dubiel, Y. Li, G. Szabo, Z. Shao, and T. L. Cover. 2003. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. J. Biol. Chem. 27812101-12108. [DOI] [PubMed] [Google Scholar]
- 27.McClain, M. S., W. Schraw, V. Ricci, P. Boquet, and T. L. Cover. 2000. Acid activation of Helicobacter pylori vacuolating cytotoxin (VacA) results in toxin internalization by eukaryotic cells. Mol. Microbiol. 37433-442. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama, M., M. Kimura, A. Wada, K. Yahiro, K. Ogushi, T. Niidome, A. Fujikawa, D. Shirasaka, N. Aoyama, H. Kurazono, M. Noda, J. Moss, and T. Hirayama. 2004. Helicobacter pylori VacA activates the p38/activating transcription factor 2-mediated signal pathway in AZ-521 cells. J. Biol. Chem. 2797024-7028. [DOI] [PubMed] [Google Scholar]
- 29.Pagliaccia, C., M. de Bernard, P. Lupetti, X. Ji, D. Burroni, T. L. Cover, E. Papini, R. Rappuoli, J. L. Telford, and J. M. Reyrat. 1998. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc. Natl. Acad. Sci. USA 9510212-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyrat, J. M., S. Lanzavecchia, P. Lupetti, M. de Bernard, C. Pagliaccia, V. Pelicic, M. Charrel, C. Ulivieri, N. Norais, X. Ji, V. Cabiaux, E. Papini, R. Rappuoli, and J. L. Telford. 1999. 3D imaging of the 58-kDa cell binding subunit of the Helicobacter pylori cytotoxin. J. Mol. Biol. 290459-470. [DOI] [PubMed] [Google Scholar]
- 31.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect. Immun. 69730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt, W., and R. Haas. 1994. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol. Microbiol. 12307-319. [DOI] [PubMed] [Google Scholar]
- 33.Schraw, W., Y. Li, M. S. McClain, F. G. van der Goot, and T. L. Cover. 2002. Association of Helicobacter pylori vacuolating toxin (VacA) with lipid rafts. J. Biol. Chem. 27734642-34650. [DOI] [PubMed] [Google Scholar]
- 34.Sellman, B. R., M. Mourez, and R. J. Collier. 2001. Dominant-negative mutants of a toxin subunit: an approach to therapy of anthrax. Science 292695-697. [DOI] [PubMed] [Google Scholar]
- 35.Singh, Y., H. Khanna, A. P. Chopra, and V. Mehra. 2001. A dominant-negative mutant of Bacillus anthracis protective antigen inhibits anthrax toxin action in vivo. J. Biol. Chem. 27622090-22094. [DOI] [PubMed] [Google Scholar]
- 36.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 3471175-1186. [DOI] [PubMed] [Google Scholar]
- 37.Sundrud, M. S., V. J. Torres, D. Unutmaz, and T. L. Cover. 2004. Inhibition of primary human T-cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc. Natl. Acad. Sci. USA 1017727-7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szabo, I., S. Brutsche, F. Tombola, M. Moschioni, B. Satin, J. L. Telford, R. Rappuoli, C. Montecucco, E. Papini, and M. Zoratti. 1999. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 185517-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Telford, J. L., P. Ghiara, M. Dell'Orco, M. Comanducci, D. Burroni, M. Bugnoli, M. F. Tecce, S. Censini, A. Covacci, Z. Xiang, et al. 1994. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J. Exp. Med. 1791653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres, V. J., S. E. Ivie, M. S. McClain, and T. L. Cover. 2005. Functional properties of the p33 and p55 domains of the Helicobacter pylori vacuolating cytotoxin. J. Biol. Chem. 28021107-21114. [DOI] [PubMed] [Google Scholar]
- 41.Torres, V. J., M. S. McClain, and T. L. Cover. 2004. Interactions between p-33 and p-55 domains of the Helicobacter pylori vacuolating cytotoxin (VacA). J. Biol. Chem. 2792324-2331. [DOI] [PubMed] [Google Scholar]
- 42.Torres, V. J., M. S. McClain, and T. L. Cover. 2006. Mapping of a domain required for protein-protein interactions and inhibitory activity of a Helicobacter pylori dominant-negative VacA mutant protein. Infect. Immun. 742093-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinion-Dubiel, A. D., M. S. McClain, D. M. Czajkowsky, H. Iwamoto, D. Ye, P. Cao, W. Schraw, G. Szabo, S. R. Blanke, Z. Shao, and T. L. Cover. 1999. A dominant-negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. J. Biol. Chem. 27437736-37742. [DOI] [PubMed] [Google Scholar]
- 44.Wang, H. J., and W. C. Wang. 2000. Expression and binding analysis of GST-VacA fusions reveals that the C-terminal approximately 100-residue segment of exotoxin is crucial for binding in HeLa cells. Biochem. Biophys. Res. Commun. 278449-454. [DOI] [PubMed] [Google Scholar]
- 45.Willhite, D. C., and S. R. Blanke. 2004. Helicobacter pylori vacuolating cytotoxin enters cells, localizes to the mitochondria, and induces mitochondrial membrane permeability changes correlated to toxin channel activity. Cell Microbiol. 6143-154. [DOI] [PubMed] [Google Scholar]
- 46.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1418-428. [DOI] [PubMed] [Google Scholar]
- 47.Wren, B. W., J. Henderson, and J. M. Ketley. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16994-996. [PubMed] [Google Scholar]
- 48.Ye, D., and S. R. Blanke. 2002. Functional complementation reveals the importance of intermolecular monomer interactions for Helicobacter pylori VacA vacuolating activity. Mol. Microbiol. 431243-1253. [DOI] [PubMed] [Google Scholar]
- 49.Ye, D., D. C. Willhite, and S. R. Blanke. 1999. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. J. Biol. Chem. 2749277-9282. [DOI] [PubMed] [Google Scholar]