Abstract
Several molecules related to antigen presentation, including gamma interferon (IFN-γ) and the major histocompatibility complex (MHC), are encoded by polymorphic genes. Some polymorphisms were found to affect susceptibility to tuberculosis (TB) when they were considered singly in epidemiological studies, but how multiple polymorphisms interact to determine susceptibility to TB in an individual remains an open question. We hypothesized that polymorphisms in some genes may counteract or intensify the effects of polymorphisms in other genes. For example, an increase in IFN-γ expression may counteract the weak binding that a particular MHC variant displays for a peptide from Mycobacterium tuberculosis to establish the same T-cell response as another, more strongly binding MHC variant. To test this hypothesis, we developed a mathematical model of antigen presentation based on experimental data for the known effects of genetic polymorphisms and simulated time courses when multiple polymorphisms were present. We found that polymorphisms in different genes could affect antigen presentation to the same extent and therefore compensate for each other. Furthermore, we defined the conditions under which such relationships could exist. For example, increased IFN-γ expression compensated for decreased peptide-MHC affinity in the model only above a certain threshold of expression. Below this threshold, changes in IFN-γ expression were ineffectual compared to changes in peptide-MHC affinity. The finding that polymorphisms exhibit such relationships could explain discrepancies in the epidemiological literature, where some polymorphisms have been inconsistently associated with susceptibility to TB. Furthermore, the model allows polymorphisms to be ranked by effect, providing a new tool for designing association studies.
Tuberculosis (TB) continues to be a global health problem. An estimated one-third of the human population is infected with the pathogen Mycobacterium tuberculosis, and approximately two million individuals succumb to the disease annually (www.who.int/mediacenter/factsheets/fs104). However, the infections in only a fraction of infected individuals ever progress to disease (62). What distinguishes the people who are able to control the infection from those who are not? In addition to environmental factors, such as nutrition, human immunodeficiency virus coinfection, and differences in bacterial strains, host genetics are likely to play a role, and identifying polymorphisms that predispose individuals to TB continues to be an area of active research (for reviews, see references 3, 15, and 22).
Identification of polymorphisms that affect susceptibility to TB is performed primarily on the basis of epidemiological data from association studies (7). In such studies the frequencies of alleles resulting from polymorphisms in patients and healthy controls are compared. If an allele is found to be overrepresented in patients, it can be hypothesized to encode a protein variant that renders an individual more susceptible to TB.
Association studies do not always yield consistent results, however, and an allele that is found to be correlated with TB in one study may not be found to be correlated with TB in another study (15). Several factors might account for this inconsistency (2). Small sample sizes (i.e., an insufficient number of subjects) may lead to unreliable detection of low-frequency alleles. Also, indirect serological tests may fail to distinguish between closely related alleles.
Even more difficult to account for is the problem of genetic heterogeneity. Because the number of polymorphisms affecting the immune function is vast and many polymorphisms have yet to be discovered, epidemiological studies necessarily fail to assay all polymorphisms. In most studies only a single polymorphism is assayed. Other polymorphisms may compound or counteract the effect of a single polymorphism, and such interactions may go undetected without further testing. For instance, a polymorphism in gene X, X1, may render the human host more susceptible to TB in the presence of one polymorphism in gene Y, Y1, but not in the presence of another polymorphism in Y, Y2. Such interactions underlie the difficulty in comparing the results of studies done with different populations (2), even when the same allele is studied and the study designs are identical.
Antigen presentation requires the contribution of several genes, and many of these genes have polymorphisms that have been associated with TB susceptibility (3, 15, 22) (see Table S1 in the supplemental material). During antigen presentation, receptors known as major histocompatibility complex (MHC) molecules bind peptides from pathogens, and the resulting peptide-MHC (pMHC) complexes are displayed on the surfaces of cells (Fig. 1a). Two classes of MHC molecules exist: class I, primarily for peptides found in the cytoplasm, and class II, primarily for peptides found in endosomal compartments. Cells known as professional antigen-presenting cells (APCs) express both classes of MHC molecules, enabling them to display peptides from a variety of pathogens.
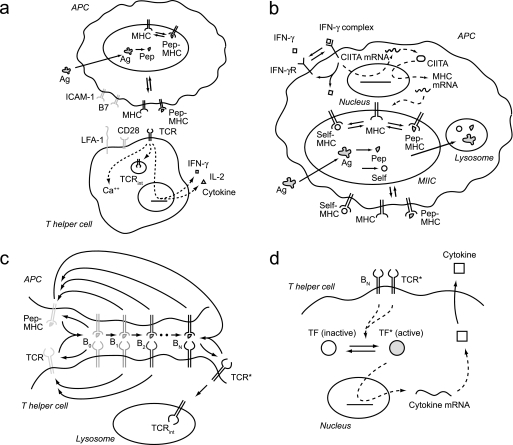
FIG. 1.
Schematic diagram of the multiscale model of antigen presentation. (a) Overview of antigen presentation by APCs and the T-cell response. (b) APC model (input, IFN-γ, exogenous antigen; output, surface pMHCs). (c) T-cell model (input, surface pMHCs from APC model; outputs, activated TCRs and internalized TCRs). (d) Cytokine production model (input, activated TCRs; output, cytokines, particularly IFN-γ). Abbreviations: Ag, antigen; Pep, exogenous peptide; Self, self peptide; B with subscripts 0 through N, pMHC-TCR complexes in different stages of activation; TF, transcription factor; IL-2, interleukin-2. Direct, mechanistic reactions in the model are indicated by solid arrows, while indirect, regulatory interactions in the model are indicated by dashed arrows. The names of cellular compartments are italicized.
A T-cell response to antigen presentation begins when a T cell scans the surface of an APC (Fig. 1a) (for a review, see reference 58). If T-cell receptors (TCRs) expressed by a T cell bind pMHC complexes on the APC surface with sufficient affinity and costimulatory molecules, such as B7 and CD28, are present, a signaling cascade is initiated, which ultimately results in T-cell activation. Within minutes of APC-T-cell contact, the calcium level within the T cell rises and TCRs are internalized from the T-cell surface. Within hours, T cells produce cytokines, including interleukin-2 and gamma interferon (IFN-γ), which result in T-cell proliferation, as well as activation of other cell types. Antigen presentation therefore involves events occurring with timescales ranging from seconds to hours at molecular, cellular, and multicellular scales.
Among the antigen presentation-specific polymorphisms associated with TB, perhaps the best studied are those occurring in genes for human leukocyte antigen (HLA), the human form of MHC. Of the two classes of HLA, HLA class II is particularly relevant to TB because HLA class II molecules bind peptides from antigens in endosomal compartments where M. tuberculosis resides. Over 800 HLA class II alleles have been identified, and several of them have been associated with increased or decreased susceptibility to TB (3, 55). In particular, the DRB1*1501 allele has been associated with increased susceptibility to TB in numerous studies (44, 53, 63, 66) (see Table S1 in the supplemental material). Generally, polymorphisms in HLA map to the peptide-binding regions of the molecule and can therefore be assumed to affect function (52). However, a general mechanism explaining how HLA polymorphisms affect the immune response to pathogens such as M. tuberculosis has not been established. Polymorphisms affecting HLA expression rather than the peptide-binding properties of HLA are also known, but to our knowledge, none of these polymorphisms has been tested yet for TB association (11, 38).
Extracellular signals in the form of cytokines, such as IFN-γ, also affect MHC expression in APCs, and polymorphisms in the IFN-γ gene have been associated with TB. In particular, the +874A allele has been found to be significantly overrepresented in TB patients, suggesting that +874A increases susceptibility to TB (34, 37, 56) (see Table S1 in the supplemental material). Peripheral blood mononuclear cells (PBMCs), which include APCs and T cells, from +874A individuals produce significantly less IFN-γ than PBMCs from +874T individuals produce upon antigenic stimulation in vitro (37, 50). IFN-γ modulates a number of functions in addition to MHC expression, including the activation of macrophages and NK cells and the inhibition of the TH2 phenotype in T cells (40). Exactly which function is undermined in +874A individuals has not been determined yet.
Finally, polymorphisms affecting antigen processing (the partial degradation of proteins into peptides) have also been associated with susceptibility to TB, although thus far only in genes affecting MHC class I-mediated antigen presentation. The transporter associated with antigen processing (TAP) translocates peptides from the cytoplasm into the endoplasmic reticulum, where they can be bound by MHC class I molecules. A polymorphism in TAP2, one subunit of TAP, has been found to be overrepresented in TB patients (17, 51) (see Table S1 in the supplemental material). Other enzymes (namely, the cathepsin proteases) perform analogous functions for the MHC class II-mediated pathway, and although polymorphisms in the genes encoding these enzymes are known, to our knowledge none has been tested for TB association (64).
How polymorphisms in HLA, IFN-γ, and other genes interact to ultimately determine genetic susceptibility to TB remains an open question. Mathematical modeling can help provide a unifying framework with which to consider these polymorphisms. A model would ideally have an immunologically relevant readout, such as cytokine production, and allow the effects of different polymorphisms to be simulated and observed, both singly and in combination. Several questions could then be approached using this framework. For example, could a polymorphism that up-regulates IFN-γ expression compensate for a polymorphism that results in deficient HLA-M. tuberculosis peptide binding? How might the T-cell response differ among individuals with different combinations of polymorphisms?
To approach these questions, we developed a multiscale mathematical model for antigen presentation that includes both APCs and T cells and tracks events from the molecular scale to the cellular and multicellular scales. Particular attention was paid to pathways involving MHC and IFN-γ, and experimental data from a variety of sources were used to provide parameter values. Polymorphisms affecting both pathways were then simulated. The extent to which a polymorphism in one gene compounded or counteracted a polymorphism in another gene could be observed, allowing us to determine whether the presence of multiple polymorphisms could be a confounding factor in TB association studies.
MATERIALS AND METHODS
The multiscale model comprises three models that were developed separately (Fig. 1): an APC model representing the events leading to the appearance of pMHC on the APC surface, a T-cell model representing the events leading to TCR internalization, and an intracellular T-cell signaling model representing the events leading to cytokine (IFN-γ) production. We provide an overview of the three models below.
APC model.
Details of the APC model are described elsewhere (1, 8, 59, 60). Briefly, we represented the major events leading to antigen presentation by MHC class II molecules on APCs (e.g., macrophages) using ordinary differential equations (ODEs). These events include de novo synthesis of MHC, the up-regulating effect of IFN-γ on MHC synthesis, uptake and processing of extracellular antigens, formation of pMHC complexes, and trafficking of the pMHC to and from the APC surface (Fig. 1b). Equations are provided in the supplemental material, and parameters are shown in Tables S2 and S3 in the supplemental material.
T-cell model.
To represent the T-cell response to antigen presentation, we developed a T-cell model, which was then linked to the APC model. The T-cell model was based on a model described previously by Coombs et al. (10) and Gonzalez et al. (18) which represents two important features of T-cell signaling, namely, kinetic proofreading (i.e., the requirement for pMHC-TCR engagement to persist for a certain duration before it results in TCR activation) and serial triggering (i.e., the ability of one pMHC to engage multiple TCRs). In the model of Coombs et al., as well as in our model, the following events were represented: engagement of pMHCs by TCRs, progression of pMHC-TCR complexes through various states of activation, and finally internalization of fully activated TCRs as a marker of T-cell activation (Fig. 1c). Parameter values for the model were derived from experiments using T-cell clones or T-cell hybridomas; therefore, the model most closely describes the stimulation of effector T cells (10). CD28 costimulatory molecules, such as B7, ICAM-1, and LFA-1, were assumed to be present in nonlimiting quantities. In order to be internalized, TCRs in the model were required to be fully activated in either free or pMHC-bound forms. The contribution of constitutive recycling of TCRs to the pool of internalized TCRs was not included. Only the contact zone between the APC and the T cell was considered, and the degree of TCR internalization occurring in the contact zone was assumed to be representative of the degree of TCR internalization occurring elsewhere on the T-cell surface (24, 67). The T-cell model comprises a set of ODEs separate from the ODEs of the APC model (equations are provided in the supplemental material, and parameters are shown in Tables S4 and S5 in the supplemental material).
Cytokine production model.
To provide an additional, longer-term readout of T-cell activation, we extended the T-cell model to include signaling events following TCR activation that culminate in the production of cytokines, such as IFN-γ. These events include the recruitment of kinases, such as Lck and ZAP-70, the activation of intermediate signaling molecules, such as phospholipase C and calcineurin, and ultimately the activation of transcription factors NF-AT, NF-κB, and AP-1 (for a review, see reference 36). We developed a simplified model of these events representing transcription factor activation, cytokine gene expression, and cytokine production (Fig. 1d). More detailed features of the T-cell activation pathway, such as intracellular signaling pathways or synthesis and breakdown of transcription factor intermediates, were assumed to have a negligible effect on long-term (>12-h) responses and therefore were not considered in our model. Such features are considered explicitly in other models (16, 23, 30). The cytokine production model comprises a third set of ODEs in addition to the ODEs constituting the APC and T-cell models (equations are provided in the supplemental material, and parameters are shown in Tables S4 and S5 in the supplemental material).
Empirical data for MHC polymorphisms.
Data sets for peptides binding various MHC alleles and their affinities were downloaded from the Immune Epitope Database (IEDB) (http://www.immuneepitope.org) (49). Affinities were generally provided as the 50% inhibitory concentration, the concentration of peptide required to displace 50% of a reporter peptide bound to a given MHC molecule. The 50% inhibitory concentration provides an approximation of the equilibrium dissociation constant (KD) (9). Altogether, four data sets from IEDB were considered, the data sets for the MHC alleles HLA-DR1, -DR2, -DR3, and -DR4. These data sets provided an approximation of the range that could be expected for one parameter in the model, the rate constant for dissociation of the peptide from pMHC.
Solving the multiscale model.
Together, the three models constitute the multiscale model of antigen presentation. The models were run sequentially, as follows: (i) exposure of APCs to IFN-γ in the absence of exogenous antigen for 24 h (APC model alone); (ii) exposure of APCs to exogenous antigen in the absence of IFN-γ for 4 h (APC model alone); and (iii) exposure of APCs to T cells for 24 h (T-cell model and cytokine production model). Information was passed between the APC and T-cell models in the form of the number of pMHCs on the APC surface appearing 4 h after exposure of APCs to antigen (i.e., in a feed-forward manner). Feedback from T cells to APCs, in the form of IFN-γ that could increase MHC expression, was assumed to be negligible for the timescales simulated (≤24 h) and therefore were not represented; such feedback can easily be accommodated by the model when longer timescales are investigated. The three variables serving as outputs were the number of pMHC complexes on the APC surface 4 h after exposure to exogenous antigen (APC model), the fraction of TCRs internalized 5 h after APC—T-cell contact (T-cell model), and the concentration of the cytokine IFN-γ produced 24 h after APC—T-cell contact (cytokine production model), which we considered short-, medium-, and long-term responses, respectively. These outputs have been considered intermediate indicators of the cellular response in the experimental literature and have been quantified (21, 24, 67). Upon exposure to antigen, APCs typically express tens to thousands of pMHC molecules on the surface; a minimum of approximately 200 to 350 pMHC molecules have been found to be necessary to elicit a T-cell response (13, 19). Within hours of APC contact T cells internalize 10 to 90% of the TCRs from the surface depending on the amount of antigen initially present (24, 67). No threshold level of TCR internalization for T-cell activation has been determined, although a correlation with other responses, such as T-cell proliferation, has been observed (24). The amount of the cytokine IFN-γ produced by T cells in response to antigenic stimulation varies over several logs; typically, picomolar amounts are observed in vitro (21, 32, 35). The model was solved using the NDSolve function in Mathematica 4.2 (Wolfram Research, Inc.) and default options, and the outputs were compared to available dose-response data (21, 24, 67). Note that these data are independent of those used to build the model.
Analysis of the sensitivity of the multiscale model.
We used sampling-based sensitivity analysis to determine how variability in processes represented in the model affected model outputs. Briefly, we varied the values for different parameters in the model, generated an output for each set of parameter values, and then determined the degree of correlation between each parameter and the output. A particular parameter was varied if a genetic polymorphism was known to affect the corresponding process or to ensure that each of three constituent models was represented by approximately the same number of varying parameters during the analysis; a total of 16 parameters were varied (see Table S6 in the supplemental material). Parameters were assigned log-uniform distributions; that is, minimum and maximum values were assigned to each parameter, and sampling was done uniformly for a range defined by the log transform of these values. When several biological values were available in the literature, the approximate ranges of the values used were as follows: for the pMHC off-rate constant, 10−6 to 10−2 s−1 (42, 57); for the pMHC-TCR off-rate constant, 10−3 to 100 s−1 (12); for the IFN-γ dose, 10−12 to 10−6 M (33); and for the antigen dose, 10−9 to 10−4 M. In all other cases, a range of 1 order of magnitude above the baseline value to 1 order of magnitude below the baseline value was used (see the supplemental material). Five hundred values for each parameter were generated by a Latin hypercube sampling scheme (5, 20), resulting in 500 sets of parameter values. An equivalent number of output values were then derived, and correlations between output values and parameter values were quantified using partial rank correlation coefficients (5). Significance was assigned based on a Bonferroni-corrected α value of 0.05 (4).
Experimental scenarios simulated.
Using the multiscale model of antigen presentation, we were able to simulate in vitro protocols intended to test the responsiveness of host cells to particular antigens (for an example of such a protocol, see reference 28). In such protocols PBMCs are isolated from patient blood, stimulated by antigens, such as purified protein derivative from M. tuberculosis, and then assayed for the response either indirectly (e.g., by measuring tritiated thymidine uptake as a marker of proliferation) or directly (by an enzyme-linked immunosorbent assay for a cytokine). Because monocytes, which are precursors of macrophages, serve as APCs in PBMCs and significant quantities of IFN-γ are unlikely to be present in the blood, the model simulates PBMC protocols when the amount of IFN-γ initially present is set to zero. The model can also simulate an in vivo scenario of antigen presentation at a site of infection when macrophages and activated T cells are present; in this case, the amount of IFN-γ initially present in the model is set at a nonzero value. Both of these scenarios were examined during sensitivity analysis.
Trade-off plots.
In addition to performing a sensitivity analysis using various multiple parameters concurrently, we also examined the relationship between model processes in a pairwise manner by varying two parameters at a time. Pairs of parameter values that yielded approximately the same target output value were compiled and plotted. Because such plots show how a change in one parameter is able to compensate for a change in another parameter, we refer to such plots as trade-off plots. When values for both parameters are first log transformed and then plotted, regions in which the curves are diagonal (slope, approximately 1 or −1) indicate that there is a compensatory relationship; that is, a 1-log change in one parameter is able to compensate for a 1-log change in another parameter to maintain a given output value. In contrast, regions in which the curves are horizontal or vertical identify conditions under which one parameter has a dominant effect on the output of the other parameter. In such regions the output is relatively insensitive to changes in one parameter, the parameter represented on the axis parallel to the curve. To generate trade-off plots, parameters were chosen from either the same-scale submodel (the intramodel case) or different-scale submodels (the intermodel case). A sufficiently wide range of values was assigned to each parameter during the generation of each trade-off plot to capture the full range of behaviors in each curve. Biologically realistic values were then overlaid on each plot as boxes whose edges represented the range of values observed experimentally, usually in vitro. When in vitro measurements were not available, a range of 1 order of magnitude above the baseline value to 1 order of magnitude below the baseline value was indicated (see Tables S2 to S5 in the supplemental material). When pMHC or pMHC-TCR off-rate constants were varied, parameter values were plotted as the KD values, assuming invariant on-rate constants (27). We chose different values for the three outputs to serve as target output values, generally accepting pairs of parameter values that resulted in values between 80 and 120% of the target output values. The target output values were 100, 500, or 1,000 pMHCs on the APC surface; 10, 40, or 80% internalization of the total TCRs; and production of 0.1, 1, or 5 pM IFN-γ, corresponding to ∼2, ∼20, and ∼200 pg/ml IFN-γ. To assist visualization of plots, curve fitting was done using the SplineFit function (Bezier option) of the NumericalMath library in Mathematica 4.2 (Wolfram Research, Inc.), except in cases where more than one y value mapped to the same x value (as in the plots of pMHC-TCR affinity versus pMHC affinity and the plots of TCR internalization versus pMHC affinity); in these cases, curves were drawn by hand.
RESULTS
To relate genetic polymorphisms to changes in APC and T-cell responses, we developed a multiscale model of antigen presentation that traverses several biological and temporal scales (from molecular to multicellular and from seconds to hours) (Fig. 1). This model represents different immunological processes that may vary due to genetic polymorphisms and allows us to examine the effect of multiple polymorphisms occurring simultaneously.
Empirical data quantify effects of genetic polymorphisms on pMHC binding involving M. tuberculosis peptides.
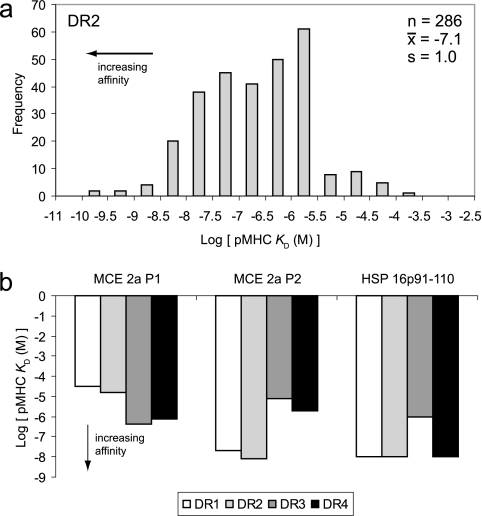
Data from a variety of previous studies were used to obtain values for parameters in the multiscale model, as described in the supplemental material. In some cases, the effects of genetic variability could be observed directly. For example, variability within MHC sequences results in MHC molecules that have different affinities for the same peptide (e.g., peptides from M. tuberculosis proteins), as well as for a broad range of peptides; these affinities in turn inform pMHC on- and off-rate constants in the model. Many of these affinities are usually measured at equilibrium binding, the concentration at which binding is inhibited by 50% (which approximates KD), have been measured and compiled in databases, such as the IEDB (Fig. 2a). Using the IEDB, we found that MHC molecules exhibit affinities (KD) for peptides that vary greatly, from 10−9 to 10−4 M (Fig. 2a; see Fig. S1 in the supplemental material), with the majority of affinities less than 10−6 M. In these data are the affinities of different MHC molecules for an identical set of M. tuberculosis peptides (Fig. 2b). This subset of the data shows that the range of affinities that different MHC molecules exhibit for a given peptide is approximately the same as the range of affinities that a given MHC molecule exhibits for different peptides. For example, different MHC molecules bind the P2 peptide from the M. tuberculosis protein MCE2A with affinities that vary from 10−5 to 10−8 M (Fig. 2b), which is approximately the same as the range described above.
FIG. 2.
Experimentally quantified effects of MHC polymorphisms on peptide-binding affinities. (a) Survey of pMHC affinities involving HLA-DR1 as found in the IEDB (49). n, number of peptides; x̄, mean value; s, standard deviation. (b) Affinities of different MHC molecules for a common set of M. tuberculosis peptides. MCE 2a P1 and MCE 2a P2 refer to M. tuberculosis MCE family protein MCE 2 peptides P1 and P2 (47), respectively, and HSP 16p91-110 refers to a peptide comprising amino acids 91 to 110 from M. tuberculosis heat shock protein 16.3 (26).
Output from the multiscale model agrees with experimental data.
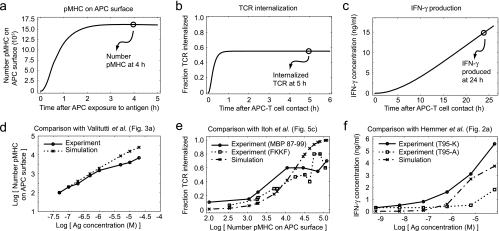
Initially, we tested the model by comparing three model outputs to behaviors expected from experimental data; these data were independent from those used to develop the model. As a negative control we checked baseline results of the model. In the absence of exogenous antigen, no exogenous pMHC complexes were formed, no TCRs were internalized, and no IFN-γ was produced (data not shown). As a positive control, we examined outputs of the model when exogenous antigen was present. The dynamics of pMHC display, TCR internalization, and IFN-γ production approximated experimentally observed time courses (Fig. 3a to c). Specifically, the number of pMHCs on the APC surface peaked within 4 h after antigen exposure (Fig. 3a) (19); the majority of TCR internalization occurred within the first 2 h of exposure of T cells to APCs (Fig. 3b) (67); and IFN-γ production continued to increase during the first 24 h of exposure of T cells to APCs (Fig. 3c) (35). The model also recapitulated dose-response data available for the various outputs (Fig. 3d to f).
FIG. 3.
Comparison of multiscale model predictions with experimental data. (a) Time course for the number of pMHCs on the APC surface in the model. (b) Time course for TCR internalization in T cells in the model. (c) Time course for IFN-γ production in the model. (d) Dose-response curve for pMHCs as the antigen concentration is varied in the model, along with experimental data. (e) Dose-response curve for TCR internalization as the number of pMHCs on the APC surface is varied in the model, along with experimental data. (f) Dose-response curve for IFN-γ production as the antigen concentration is varied in the model, along with experimental data. When more than one curve was available from the experimental data (e and f), the highest and lowest nonzero experimental curves were selected and are shown. Model parameter values are provided in the supplemental material. Ag, antigen.
T-cell response is sensitive to multiple genetically variable processes.
To determine how biological variability due to genetic polymorphisms might affect APC and T-cell responses, we simulated variability in the multiscale model and correlated changes in output to changes in input parameters (Table 1). The outputs occurred either in the same submodel as the parameters being varied (the intramodel case) or in different submodels (the intermodel case). Two scenarios were simulated, the absence and presence of IFN-γ initially, which represented antigen presentation during PBMC protocols and at the site of infection, respectively.
TABLE 1.
Influence of various processes on APC and T-cell responses as quantified by using partial rank correlation coefficients generated from sensitivity analysis of the multiscale modela
| Biological process or factor | No IFN-γ initially present
|
IFN-γ initially present
|
||||
|---|---|---|---|---|---|---|
| No. of pMHCsb | No. of TCRsc | Amt of IFN-γd | No. of pMHCsb | No. of TCRsc | Amt of IFN-γd | |
| IFN-γ dosee | NA | NA | NA | 0.64 | 0.14 | 0.15 |
| MHC expressionf | 0.41 | 0.19 | 0.15 | 0.29 | (0.07) | (0.05) |
| pMHC affinityg | −0.80 | −0.44 | −0.40 | −0.65 | −0.29 | −0.28 |
| Antigen dose | 0.97 | 0.70 | 0.68 | 0.97 | 0.71 | 0.72 |
| Antigen processingh | 0.66 | 0.17 | 0.16 | 0.62 | 0.21 | 0.24 |
| TCR expression | NA | 0.55 | 0.42 | NA | 0.55 | 0.34 |
| pMHC-TCR affinityi | NA | −0.58 | −0.60 | NA | −0.56 | −0.60 |
| pMHC-TCR activationj | NA | 0.51 | 0.49 | NA | 0.46 | 0.46 |
| Activated free TCR internalizationk | NA | (0.08) | −0.24 | NA | (0.07) | −0.23 |
| IFN-γ signalingl | NA | NA | 0.56 | NA | NA | 0.66 |
Results for 10 of 16 parameters that were varied are shown (see Table S6 in the supplemental material for a complete table). Parameters corresponding to processes in which genetic polymorphisms have been observed are indicated by bold type (see Table S1 in the supplemental material and references therein). Nonsignificant partial rank correlation coefficient values (α = 0.05, Bonferroni adjusted) are in parentheses. NA indicates that the parameter for a process occurs subsequent to the output of interest in the model and therefore does not affect the output value.
Number of pMHCs on the APC surface 4 h after antigen exposure.
Number of TCRs internalized by the T cell 5 h after APC—T-cell contact.
Amount of IFN-γ produced by the T cell 24 h after APC—T-cell contact.
Amount of IFN-γ to which APCs are exposed 24 h prior to antigen exposure.
Number of MHC molecules initially expressed on an APC.
Expressed as pMHC KD when the peptide-MHC dissociation rate constant was varied.
Rate constant for antigen processing.
Expressed as pMHC-TCR KD when the pMHC-TCR dissociation rate constant was varied.
Rate constant for progressive activation of pMHC-TCR complexes.
Rate constant for internalization of free, activated TCR.
Rate constant for TCR-induced IFN-γ transcription.
Multiple parameters were found to correlate significantly with model outputs, identifying biological processes that may positively or negatively govern antigen presentation and the T-cell response (Table 1). Genetic variability in a number of these processes is known to exist and in some cases has been associated with susceptibility to TB. For example, the model showed that pMHC affinity and IFN-γ dose (which may vary in humans due to polymorphisms in MHC and IFN-γ) correlated significantly with all three outputs: number of pMHC molecules on the APC surface, the extent of TCR internalization, and the level of cytokine production (Table 1). Other processes in the model are associated with polymorphisms that are likely functional but have not previously been associated with TB susceptibility. For example, antigen processing, for which polymorphisms are also known (17, 51), correlated significantly with all three outputs but more strongly at early time points than at later time points (Table 1). Likewise, MHC expression also correlated significantly with all three outputs more strongly at early time points than at later time points. However, this correlation was observed only in the absence of IFN-γ, a scenario resembling PBMC protocols rather than infection in vivo, illustrating the overlapping effects of changes in IFN-γ expression and MHC expression (Table 1).
Most parameters displayed similar degrees of correlation (either positive or negative) for the two T-cell responses, TCR internalization and IFN-γ production (Table 1). One exception was the rate constant for the internalization of free, activated TCR, which correlated positively with TCR internalization and negatively with IFN-γ production. In the model, internalized TCRs are not capable of initiating signal transduction and do not contribute to cytokine production. Internalization, therefore, decouples one indicator of T-cell activation (T-cell internalization) from another indicator (cytokine production), resulting in the oppositely signed correlations observed during this analysis.
Trade-off plots depict potentially confounding effects of multiple polymorphisms.
Sensitivity analysis demonstrated that multiple processes, including several processes that may vary due to genetic polymorphisms, govern the dynamics of antigen presentation and subsequent T-cell responses. To examine interactions between polymorphisms in more detail, we varied parameters in a pairwise manner and determined the extent to which one parameter could compensate for another parameter in governing subsequent dynamics in output variables.
(i) IFN-γ expression and HLA binding polymorphisms can be compensatory.
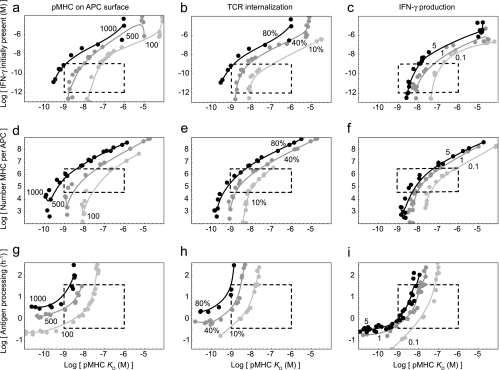
The polymorphisms in the antigen presentation pathway most commonly associated with TB susceptibility affect the level of IFN-γ expressed by T cells and pMHC binding in APCs (3). The consequences of two polymorphisms in different genes acting simultaneously on antigen presentation have not been examined either experimentally or theoretically to our knowledge. To simulate these polymorphisms, we varied parameters for IFN-γ levels and pMHC binding affinity in the APC model and plotted the pairs of parameter values resulting in approximately the same values for our three outputs: number of pMHC molecules on the APC surface, TCR internalization, and cytokine production (Fig. 4a to c). In the case of the pMHC output, the minimum levels known to elicit a T-cell response were chosen (13, 19).
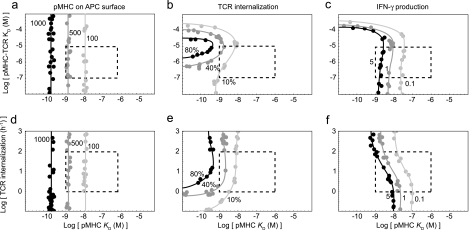
FIG. 4.
Trade-off plots showing that polymorphisms in different genes affecting APCs may compensate for deficiencies in pMHC binding to maintain a given response. Values for pairs of parameters were varied, and pairs resulting in the same target output value were plotted. (a to c) IFN-γ expression (expressed as the amount initially available to APCs) versus pMHC binding. (d to f) MHC expression versus pMHC binding. (g to i) Antigen processing versus pMHC binding. The target output values were 100, 500, or 1,000 pMHCs on the APC surface; 10, 40, or 80% internalization of total TCRs; and production of 0.1, 1, or 5 pM IFN-γ, corresponding to ∼2, ∼20, and ∼200 pg/ml IFN-γ. Dashed boxes indicate biologically plausible values (see Materials and Methods). Model parameter values are provided in the supplemental material.
In the trade-off plots, three distinct regions can be discerned (described here for TCR internalization [Fig. 4b]). First, at low IFN-γ concentrations (<10−10 M), TCR internalization is determined almost entirely by pMHC affinity and is invariant when there are small changes in the IFN-γ concentration, which is apparent from the nearly vertical lines on the plots. Under these conditions few of the IFN-γ receptors are bound, and small changes in IFN-γ concentrations do not alter MHC expression. Second, at intermediate IFN-γ concentrations (between 10−10 and 10−6 M), changes in IFN-γ have an effect on TCR internalization nearly equal to that of changes in pMHC affinity, which is apparent from the diagonal curves on the plots. In this region, for example, 80% TCR internalization can be achieved by pairing either 10−9 M IFN-γ and a pMHC binding affinity (expressed as KD) of 10−9 M or 10−8 M IFN-γ and a pMHC binding affinity of 10−8 M. Finally, at high IFN-γ concentrations (>10−6 M), TCR internalization is again determined almost entirely by pMHC affinity, which is apparent from the nearly vertical curves on the plots. Under these conditions most of the IFN-γ receptors are bound, and small changes in IFN-γ concentrations do not affect near-maximal increases in MHC expression.
Superimposing experimental data onto these plots allows regions to be defined which encompass all possible two-polymorphism combinations in an individual and the effects that these combinations have on APCs and the T-cell response. For example, IFN-γ expression in PBMCs from individuals with the +874A and +874T alleles has been measured and found to differ as much as threefold, in the range from 10−10 to 10−11 M (37, 50; I. Aguilar-Delfin, unpublished data). A wider range of IFN-γ concentrations (between 10−9 and 10−12 M) is typically applied in vitro (Fig. 4a to c). The affinities of different pMHC class II complexes have also been measured and found to vary greatly between 10−6 and 10−9 M (49, 57) (Fig. 2a; see Fig. S1 in the supplemental material). When these values are superimposed on the relevant trade-off plot, the plot shows that at realistic levels of IFN-γ expression, variability in pMHC affinity has a stronger effect on all outputs, from the number of pMHCs displayed by APCs to the amount of cytokine produced by T cells (Fig. 4a to c).
(ii) HLA expression and HLA binding polymorphisms can be compensatory.
Although polymorphisms in HLA promoters have been identified, no such polymorphism has yet been associated with susceptibility to TB (11, 38). One reason for this may be the difficulty involved in measuring the total level of expression of a particular HLA class II variant both within an APC and on the APC surface simultaneously. Another reason may be the difficulty involved in attributing an association with TB to the HLA promoter rather than the HLA coding sequence with which it is likely in linkage disequilibrium. In the model, HLA expression and binding affinity are separate parameters and were found to have nearly equivalent effects on output values (Fig. 4d to f). For instance, when 105 MHC molecules were expressed by the APCs, a pMHC affinity of 10−9 M resulted in nearly the same degree of TCR internalization (∼80%) as a weaker pMHC affinity (10−8 M) when more MHC molecules (106 molecules) were expressed (Fig. 4e). Tenfold differences in expression from different HLA promoters have been observed previously (such as the differences between HLA-DRB3 and HLA-DRB5 [38]). At lower levels of MHC expression (<105 MHC molecules per APC), however, pMHC affinity became more strongly determinative of the T-cell response. The possibility that higher levels of expression might compensate for lower binding affinity has been raised previously in animal studies (29, 68).
(iii) Antigen processing and HLA binding polymorphisms can be compensatory.
Like polymorphisms affecting MHC expression, polymorphisms affecting antigen processing have been identified, although none have been associated with susceptibility to TB yet (64). Polymorphisms affecting antigen processing can be expected to either increase or decrease the availability of antigenic peptides available to bind MHC and thereby affect antigen presentation and subsequent T-cell responses. In the model, variability in antigen processing was found to compensate for variability in pMHC affinity in a nearly 1:1 manner (Fig. 4g to i). For example, to maintain 5 pM IFN-γ production from T cells, an increase in the rate constant for antigen processing (from 100 to 101 h−1) could compensate for a decrease in pMHC binding affinity (from 10−9 to 10−8 M) (Fig. 4i). The extent to which polymorphisms affect the activity of cathepsin proteases responsible for processing antigen for MHC class II is not known (64), but within a 1-log range of the level of activity observed in vitro, the trade-off plots show that variability in cathepsin activity and subsequent antigen processing may affect the T-cell response to the same extent as variability in pMHC affinity. While the cathepsin proteases generally exhibit a high degree of redundancy, in some cases a particular cathepsin has been found to be necessary for processing a particular antigen (e.g., cathepsin D for antigen 85B from M. tuberculosis [61]). Polymorphisms are known to exist in this cathepsin (e.g., +224T, which has been associated with an increased risk of Alzheimer's disease [48]), but no association studies have been done yet with TB to our knowledge, nor is the precise effect of these polymorphisms on enzymatic function known.
(iv) Optimal pMHC-TCR affinity affects TCR internalization, but not IFN-γ production.
The binding affinity of the pMHC-TCR trimolecular complex has been shown to be an important quantity in determining the T-cell response (41). We examined trade-offs between pMHC and pMHC-TCR affinities in eliciting different responses (Fig. 5a to c). Because pMHC-TCR binding occurs outside the scope of the APC model, variability in pMHC-TCR affinity does not affect the number of pMHCs. This lack of an effect is apparent from the vertical lines on the trade-off plot for this output (Fig. 5a). Coombs et al. (10) and Gonzalez et al. (18) showed that under certain conditions there is an optimal half-life for pMHC-TCR interaction, resulting in maximal TCR internalization. Because our model of the T cell was based on the model of Coombs et al. (10), it was not surprising to observe an optimal binding affinity for pMHC-TCR on trade-off plots for which pMHC-TCR affinity was varied and TCR internalization was selected as the output (Fig. 5b). However, the peak representing the optimal affinity was less prominent at lower pMHC affinities, particularly when IFN-γ production was the output (Fig. 5c). Indeed, at biological values (pMHC KD, 10−9 to 10−6), pMHC affinity was more determinative of the T-cell response than pMHC-TCR affinity, which was apparent from vertical lines on the plots (Fig. 5b and c).
FIG. 5.
Trade-off plots showing that polymorphisms in different genes affecting a T cell may compensate for deficiencies in pMHC binding to maintain a given response. (a to c) Plots of pMHC-TCR affinity versus pMHC affinity, showing that an optimal value for one parameter (in this case pMHC-TCR affinity) appears as a peak on such plots. (d to f) Plots of the internalization rate constant for free, activated TCRs versus pMHC affinity, showing that parameters may affect two outputs differently, resulting in curves with different slopes. The target output values were 100, 500, or 1,000 pMHCs on the APC surface; 10, 40, or 80% internalization of the total TCRs; and production of 0.1, 1, or 5 pM IFN-γ, corresponding to ∼2, ∼20, and ∼200 pg/ml IFN-γ. Dashed boxes indicate biologically plausible values (see Materials and Methods). Model parameter values are provided in the supplemental material.
(v) Internalization of activated TCR is inversely correlated with different T-cell responses.
We also examined trade-offs between pMHC affinity and the rate constant for TCR internalization (Fig. 5d to f). While most parameters in the model correlated consistently (either positively or negatively) with the three different responses, the parameter for internalization of free, activated TCR differed in that it was positively correlated with one response, TCR internalization, and negatively correlated with another response, IFN-γ production (Fig. 5e to f and Table 1). These correlations persisted up to a certain value for the internalization rate constant (∼1 h−1), above which other processes, such as pMHC binding, became limiting (vertical lines in Fig. 5e to f). These results were obtained under the assumption that TCRs do not continue to signal after internalization, an assumption that has been challenged for TCRs, as well as for other receptors (6, 39). If internalized TCRs were assumed to continue signaling in the model, vertical trade-off plots with pMHC affinity were observed and TCR internalization had little effect on IFN-γ production (data not shown).
DISCUSSION
A large body of epidemiological data links polymorphisms in various host genes to increased susceptibility to TB (3, 15, 22). However, there are no mechanistic explanations for how polymorphisms that have been identified increase susceptibility to TB. We posed a fundamental question: How do polymorphisms in multiple genes acting simultaneously affect immune functions, such as antigen presentation? For example, considering that IFN-γ up-regulates MHC expression, could an allele of IFN-γ increase the number of MHC molecules per APC enough to offset deficiencies exhibited by some HLA alleles in binding epitopes from M. tuberculosis and elicit the same T-cell response?
To examine these questions, we developed a multiscale model of antigen presentation that links molecular and intracellular events to cellular and multicellular outcomes. By varying parameters for IFN-γ expression, pMHC binding, and other processes, we were able to simulate changes in different processes that result from genetic variation and to analyze the sensitivity of antigen presentation and T-cell responses to these changes. Sensitivity analysis showed that many of the processes in the model had strong and comparable effects on the outputs. For instance, both IFN-γ expression (as represented by the amount of IFN-γ to which APCs were initially exposed) and pMHC binding were found to significantly affect all outputs in the model, both at the same spatial scale (i.e., intrascale, within a single APC) and at different scales (i.e., interscale, at the level of the T cell). These outputs included the number of pMHC molecules appearing on the APC surface, the degree of TCR internalization, and the amount of cytokine produced by T cells (Table 1).
We then analyzed interactions between genetically variable processes in more detail using trade-off plots which showed how different pairs of values for two parameters may result in the same output. We found that changes in some processes may compensate for changes in other processes under certain conditions that we were also able to determine from the trade-off plots. For instance, within a certain range of concentrations (10−10 to 10−6 M), variations in the amount of IFN-γ to which APCs were exposed exerted the same degree of influence as variations in pMHC binding affinity in determining the T-cell response (Fig. 4b and c). Outside this range, however, pMHC affinity had a more dominant effect on the T-cell response, minimizing the contribution of IFN-γ. In primary cultures of PBMCs restimulated with antigen, IFN-γ has been detected at concentrations of 10−11 to 10−10 M. At these concentrations polymorphisms in MHC may mask the effect of polymorphisms in IFN-γ. This interaction may account for inconsistencies in epidemiological studies that attempted to link IFN-γ polymorphisms and TB susceptibility. In particular, the +874A polymorphism in the gene for IFN-γ results in decreased IFN-γ expression (relative to the +874T variant) and has been associated with susceptibility to TB in some but not all studies (45). Could variability in peptide binding exhibited by different HLA alleles mask the effect of IFN-γ polymorphisms in such studies? Jepson et al. (25) found that variability in the immune response to TB antigens was the result of variability in both non-HLA genes (such as the IFN-γ gene) and HLA genes. Given the significant presence of HLA polymorphisms in human populations, our study suggests that the accuracy and consistency of association studies could be increased by comparing the frequencies of concurrent pairs of polymorphisms (such as IFN-γ +874A/HLA-DRB1*1501) in TB patients rather single polymorphisms alone. Thus, our analysis described here provides a tool to not only rank the effects of individual polymorphisms but also identify combinations of polymorphisms of interest for association studies. For example, one can construct, before genotyping, hypotheses about the combined effects of specific polymorphisms.
We also found that polymorphisms need not affect the same cell or even the same timescale (i.e., occur intrascale) to be compensatory. Parameters affecting different scales (i.e., occurring interscale) may be compensatory as well. For instance, pMHC affinity and pMHC-TCR affinity exhibit a compensatory relationship, although the former process affects APCs, while the latter affects the interface between APCs and T cells. Because TCRs are generated by somatic recombination, TCR alleles do not exist in the human population, although an individual can be expected to express a diverse set of TCRs, each with a different affinity for a given pMHC ligand (12). The importance of pMHC-TCR affinity in determining the T-cell response has been demonstrated experimentally (41, 43). Previous models have suggested that an increase in pMHC-TCR affinity can compensate for a decrease in pMHC affinity, but the conditions under which this relationship exists have not been defined previously (1, 14). These conditions can be defined using our plots, although verification of our results requires experimental data that are more detailed than the data currently available, namely, measurements of pMHC affinities, pMHC-TCR affinities, and T-cell responses for the same set of peptide, MHC, and TCR molecules.
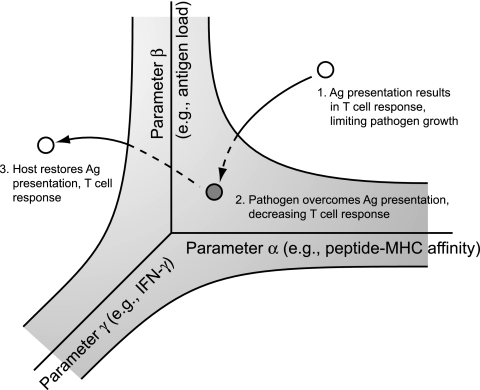
In the future we hope to consider additional questions regarding genetic variability and the dynamic interplay between host and pathogen. In addition to other polymorphisms in the human host, including polymorphisms that may exist at larger scales (e.g., polymorphisms affecting T-cell-dendritic cell contact in the lymph node [31, 54]), the model may also be amended to incorporate polymorphisms in M. tuberculosis genes, a number of which have been observed (e.g., in the PE-PGRS genes of M. tuberculosis, whose products are antigenic [65]). During the course of an infection, all of these factors may play a role in determining whether the host is able to respond successfully (for example, by displaying a sufficient number of pMHCs to elicit a T-cell response) (13, 19). Plotting all combinations of parameter values that result in this threshold would yield a surface on a multidimensional trade-off plot (shown on the conceptualized plot in Fig. 6 with two host-specific parameters and one pathogen-specific parameter). Above this surface all points would represent a successful immune response (Fig. 6, point 1). During the course of an infection, measurements could be obtained for these parameters and then overlaid on the plot to reveal whether the host or pathogen occupied a position of advantage. Given a series of measurements, one possibility is that the host and the pathogen would appear to trade positions of advantage, similar to what has been called a “cycle of antigen frustration” (46). Over the course of such a cycle antigen production by M. tuberculosis and antigen presentation by the human host are hypothesized to expand and contract in response to each other. A series of measurements would then trace a path on a multidimensional trade-off plot, dipping above and below the threshold surface (Fig. 6, points 2 and 3). In this way, models of the immune response and trade-off plots made using these models, together with experimental measurements of the relevant parameters obtained during an infection, may help to test hypotheses related to TB, reveal the nature of latency, and suggest new strategies for vaccine and therapy development. For example, modulating two different steps during antigen presentation, such as antigen processing and TCR expression, even by only moderate amounts, may be sufficient to produce a dramatic effect on T-cell activation.
FIG. 6.
Conceptualized multidimensional trade-off plot showing how the host and pathogen may respond to each other during the course of an infection. The gray area represents all parameters that lead to a threshold number of pMHCs on the APC surface or a corresponding T-cell response. Points represent values measured for the three parameters at different time points during an infection, with the points above and below the surface representing successful and unsuccessful immune responses, respectively. Ag, antigen.
In summary, the multiscale modeling approach taken here offers a new means for both understanding the biology underlying genotype-phenotype associations and developing mechanistic hypotheses for experimental validation. In addition, the model provides a new method for ranking polymorphisms for genotyping and for determining combinations of polymorphisms for genotyping in association studies.
Supplementary Material
Acknowledgments
We express our gratitude to Irma Aguilar-Delfin, Dan Coombs, and Zhenhua Yang for helpful discussions. We also thank anonymous reviewers for insightful comments.
We acknowledge support from NIH grants R01 LM 009027-01, R01 HL 072682, and R01 HL 68526 and from Merck Research Laboratories to J.J.L. S.T.C. was partially supported by a Rackham Predoctoral Fellowship from the University of Michigan.
Editor: F. C. Fang
Footnotes
Published ahead of print on 28 April 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Agrawal, N. G., and J. J. Linderman. 1996. Mathematical modeling of helper T lymphocyte/antigen-presenting cell interactions: analysis of methods for modifying antigen processing and presentation. J. Theor. Biol. 182487-504. [DOI] [PubMed] [Google Scholar]
- 2.Alcais, A., N. Remus, L. Abel, and J. L. Casanova. 2001. Genetic susceptibility to tuberculosis: from monogenic to polygenic inheritance. Sepsis 4237-246. [Google Scholar]
- 3.Bellamy, R. 2005. Genetic susceptibility to tuberculosis. Clin. Chest Med. 26233-246. [DOI] [PubMed] [Google Scholar]
- 4.Bland, J. M., and D. G. Altman. 1995. Multiple significance tests: the Bonferroni method. BMJ 310170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blower, S. M., and H. Dowlatabadi. 1994. Sensitivity and uncertainty analysis of complex models of disease transmission: an HIV model, as an example. Int. Stat. Rev. 2229-243. [Google Scholar]
- 6.Burke, P., K. Schooler, and H. S. Wiley. 2001. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell 121897-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casanova, J. L., and L. Abel. 2002. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 20581-620. [DOI] [PubMed] [Google Scholar]
- 8.Chang, S. T., J. J. Linderman, and D. E. Kirschner. 2005. Multiple mechanisms allow Mycobacterium tuberculosis to continuously inhibit MHC class II-mediated antigen presentation by macrophages. Proc. Natl. Acad. Sci. USA 1024530-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, Y., and W. H. Prusoff. 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 223099-3108. [DOI] [PubMed] [Google Scholar]
- 10.Coombs, D., A. M. Kalergis, S. G. Nathenson, C. Wofsy, and B. Goldstein. 2002. Activated TCRs remain marked for internalization after dissociation from pMHC. Nat. Immunol. 3926-931. [DOI] [PubMed] [Google Scholar]
- 11.Cowell, L. G., T. B. Kepler, M. Janitz, R. Lauster, and N. A. Mitchison. 1998. The distribution of variation in regulatory gene segments, as present in MHC class II promoters. Genome Res. 8124-134. [DOI] [PubMed] [Google Scholar]
- 12.Davis, M. M., J. J. Boniface, Z. Reich, D. Lyons, J. Hampl, B. Arden, and Y. Chien. 1998. Ligand recognition by alpha beta T cell receptors. Annu. Rev. Immunol. 16523-544. [DOI] [PubMed] [Google Scholar]
- 13.Demotz, S., H. M. Grey, and A. Sette. 1990. The minimal number of class II MHC-antigen complexes needed for T cell activation. Science 2491028-1030. [DOI] [PubMed] [Google Scholar]
- 14.Eberl, G., M. A. Roggero, and G. Corradin. 1995. A simple mathematical model for the functional peptide/MHC/TCR interactions. J. Immunol. 154219-225. [PubMed] [Google Scholar]
- 15.Fernando, S. L., and W. J. Britton. 2006. Genetic susceptibility to mycobacterial disease in humans. Immunol. Cell Biol. 84125-137. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, W. G., P. C. Yang, R. K. Medikonduri, and M. S. Jafri. 2006. NFAT and NFκB activation in T lymphocytes: a model of differential activation of gene expression. Ann. Biomed. Eng. 341712-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez, L. M., J. F. Camargo, J. Castiblanco, E. A. Ruiz-Narvaez, J. Cadena, and J. M. Anaya. 2006. TAP1 analysis of IL1B, TAP2 and IKBL polymorphisms on susceptibility to tuberculosis. Tissue Antigens 67290-296. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, P. A., L. J. Carreno, D. Coombs, J. E. Mora, E. Palmieri, B. Goldstein, S. G. Nathenson, and A. M. Kalergis. 2005. T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proc. Natl. Acad. Sci. USA 1024824-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding, C. V., and E. R. Unanue. 1990. Quantitation of antigen-presenting cell MHC class II/peptide complexes necessary for T-cell stimulation. Nature 346574-576. [DOI] [PubMed] [Google Scholar]
- 20.Helton, J. C., and F. J. Davis. 2002. Illustration of sampling-based methods for uncertainty and sensitivity analysis. Risk Anal. 22591-622. [DOI] [PubMed] [Google Scholar]
- 21.Hemmer, B., I. Stefanova, M. Vergelli, R. N. Germain, and R. Martin. 1998. Relationships among TCR ligand potency, thresholds for effector function elicitation, and the quality of early signaling events in human T cells. J. Immunol. 1605807-5814. [PubMed] [Google Scholar]
- 22.Hill, A. V. 2006. Aspects of genetic susceptibility to human infectious diseases. Annu. Rev. Genet. 40469-486. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann, A., A. Levchenko, M. L. Scott, and D. Baltimore. 2002. The IκB-NF-κB signaling module: temporal control and selective gene activation. Science 2981241-1245. [DOI] [PubMed] [Google Scholar]
- 24.Itoh, Y., B. Hemmer, R. Martin, and R. N. Germain. 1999. Serial TCR engagement and down-modulation by peptide:MHC molecule ligands: relationship to the quality of individual TCR signaling events. J. Immunol. 1622073-2080. [PubMed] [Google Scholar]
- 25.Jepson, A., W. Banya, F. Sisay-Joof, M. Hassan-King, C. Nunes, S. Bennett, and H. Whittle. 1997. Quantification of the relative contribution of major histocompatibility complex (MHC) and non-MHC genes to human immune responses to foreign antigens. Infect. Immun. 65872-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jurcevic, S., A. Hills, G. Pasvol, R. N. Davidson, J. Ivanyi, and R. J. Wilkinson. 1996. T cell responses to a mixture of Mycobacterium tuberculosis peptides with complementary HLA-DR binding profiles. Clin. Exp. Immunol. 105416-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasson, P. M., J. D. Rabinowitz, L. Schmitt, M. M. Davis, and H. M. McConnell. 2000. Kinetics of peptide binding to the class II MHC protein I-Ek. Biochemistry 391048-1058. [DOI] [PubMed] [Google Scholar]
- 28.Katial, R. K., D. Sachanandani, C. Pinney, and M. M. Lieberman. 1998. Cytokine production in cell culture by peripheral blood mononuclear cells from immunocompetent hosts. Clin. Diagn. Lab. Immunol. 578-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufman, J., and J. Salomonsen. 1997. The “minimal essential MHC” revisited: both peptide-binding and cell surface expression level of MHC molecules are polymorphisms selected by pathogens in chickens. Hereditas 12767-73. [DOI] [PubMed] [Google Scholar]
- 30.Kemp, M. L., L. Wille, C. L. Lewis, L. B. Nicholson, and D. A. Lauffenburger. 2007. Quantitative network signal combinations downstream of TCR activation can predict IL-2 production response. J. Immunol. 1784984-4992. [DOI] [PubMed] [Google Scholar]
- 31.Kirschner, D. E., S. T. Chang, T. W. Riggs, N. Perry, and J. J. Linderman. 2007. Toward a multi-scale model of antigen presentation in immunity. Immunol. Rev. 21693-118. [DOI] [PubMed] [Google Scholar]
- 32.Laaksonen, K., M. Waris, M. J. Makela, E. O. Terho, and J. Savolainen. 2003. In vitro kinetics of allergen- and microbe-induced IL-4 and IFN-γ mRNA expression in PBMC of pollen-allergic patients. Allergy 5862-66. [DOI] [PubMed] [Google Scholar]
- 33.Lin, Y., M. Zhang, F. M. Hofman, J. Gong, and P. F. Barnes. 1996. Absence of a prominent Th2 cytokine response in human tuberculosis. Infect. Immun. 641351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lio, D., V. Marino, A. Serauto, V. Gioia, L. Scola, A. Crivello, G. I. Forte, G. Colonna-Romano, G. Candore, and C. Caruso. 2002. Genotype frequencies of the +874T→A single nucleotide polymorphism in the first intron of the interferon-gamma gene in a sample of Sicilian patients affected by tuberculosis. Eur. J. Immunogenet. 29371-374. [DOI] [PubMed] [Google Scholar]
- 35.Listvanova, S., S. Temmerman, P. Stordeur, V. Verscheure, S. Place, L. Zhou, C. Locht, and F. Mascart. 2003. Optimal kinetics for quantification of antigen-induced cytokines in human peripheral blood mononuclear cells by real-time PCR and by ELISA. J. Immunol. Methods 28127-35. [DOI] [PubMed] [Google Scholar]
- 36.Liu, J. O. 2005. The yins of T cell activation. Sci. STKE 2651-8. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Maderuelo, D., F. Arnalich, R. Serantes, A. Gonzalez, R. Codoceo, R. Madero, J. J. Vazquez, and C. Montiel. 2003. Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 167970-975. [DOI] [PubMed] [Google Scholar]
- 38.Louis, P., R. Vincent, P. Cavadore, J. Clot, and J.-F. Eliaou. 1994. Differential transcriptional activities of HLA-DR genes in the various haplotypes. J. Immunol. 1535059-5067. [PubMed] [Google Scholar]
- 39.Luton, F., V. Legendre, J. P. Gorvel, A. M. Schmitt-Verhulst, and C. Boyer. 1997. Tyrosine and serine protein kinase activities associated with ligand-induced internalized TCR/CD3 complexes. J. Immunol. 1583140-3147. [PubMed] [Google Scholar]
- 40.Maher, S. G., A. L. Romero-Weaver, A. J. Scarzello, and A. M. Gamero. 2007. Interferon: cellular executioner or white knight? Curr. Med. Chem. 141279-1289. [DOI] [PubMed] [Google Scholar]
- 41.Matsui, K., J. J. Boniface, P. Steffner, P. A. Reay, and M. M. Davis. 1994. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc. Natl. Acad. Sci. USA 9112862-12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McFarland, B. J., and C. Beeson. 2002. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med. Res. Rev. 22168-203. [DOI] [PubMed] [Google Scholar]
- 43.McMahan, R. H., J. A. McWilliams, K. R. Jordan, S. W. Dow, D. B. Wilson, and J. E. Slansky. 2006. Relating TCR-peptide-MHC affinity to immunogenicity for the design of tumor vaccines. J. Clin. Investig. 1162543-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehra, N. K., R. Rajalingam, D. K. Mitra, V. Taneja, and M. J. Giphart. 1995. Variants of HLA-DR2/DR51 group haplotypes and susceptibility to tuberculoid leprosy and pulmonary tuberculosis in Asian Indians. Int. J. Lepr. Other Mycobact. Dis. 63241-248. [PubMed] [Google Scholar]
- 45.Moran, A., X. Ma, R. A. Reich, and E. A. Graviss. 2007. No association between the +874T/A single nucleotide polymorphism in the IFN-gamma gene and susceptibility to TB. Int. J. Tuberc. Lung Dis. 11113-115. [PubMed] [Google Scholar]
- 46.Murray, P. J.. 1999. Defining the requirements for immunological control of mycobacterial infections. Trends Microbiol. 7366-372. [DOI] [PubMed] [Google Scholar]
- 47.Panigada, M., T. Sturniolo, G. Besozzi, M. G. Boccieri, F. Sinigaglia, G. G. Grassi, and F. Grassi. 2002. Identification of a promiscuous T-cell epitope in Mycobacterium tuberculosis Mce proteins. Infect. Immun. 7079-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papassotiropoulos, A., M. Bagli, A. Kurz, J. Kornhuber, H. Forstl, W. Maier, J. Pauls, N. Lautenschlager, and R. Heun. 2000. A genetic variation of cathepsin D is a major risk factor for Alzheimer's disease. Ann. Neurol. 47399-403. [PubMed] [Google Scholar]
- 49.Peters, B., J. Sidney, P. Bourne, H. H. Bui, S. Buus, G. Doh, W. Fleri, M. Kronenberg, R. Kubo, O. Lund, D. Nemazee, J. V. Ponomarenko, M. Sathiamurthy, S. Schoenberger, S. Stewart, P. Surko, S. Way, S. Wilson, and A. Sette. 2005. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 3e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pravica, V., A. Asderakis, C. Perrey, A. Hajeer, P. J. Sinnott, and I. V. Hutchinson. 1999. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-gamma gene. Eur. J. Immunogenet. 261-3. [DOI] [PubMed] [Google Scholar]
- 51.Rajalingam, R., D. P. Singal, and N. K. Mehra. 1997. Transporter associated with antigen-processing (TAP) genes and susceptibility to tuberculoid leprosy and pulmonary tuberculosis. Tissue Antigens 49168-172. [DOI] [PubMed] [Google Scholar]
- 52.Rammensee, H. 1995. Chemistry of peptides associated with MHC class I and class II molecules. Curr. Opin. Immunol. 785-96. [DOI] [PubMed] [Google Scholar]
- 53.Ravikumar, M., V. Dheenadhayalan, K. Rajaram, S. S. Lakshmi, P. P. Kumaran, C. N. Paramasivan, K. Balakrishnan, and R. M. Pitchappan. 1999. Associations of HLA-DRB1, DQB1 and DPB1 alleles with pulmonary tuberculosis in south India. Tuber. Lung Dis. 79309-317. [DOI] [PubMed] [Google Scholar]
- 54.Riggs, T., A. Walts, N. Perry, L. Bickle, J. N. Lynch, A. Myers, J. Flynn, J. J. Linderman, M. J. Miller, and D. E. Kirschner. 2008. A comparison of random vs. chemotaxis driven contacts of T cells with dendritic cells during repertoire scanning. J. Theor. Biol. 250732-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson, J., M. J. Waller, P. Parham, N. de Groot, R. Bontrop, L. J. Kennedy, P. Stoehr, and S. G. Marsh. 2003. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 31311-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossouw, M., H. J. Nel, G. S. Cooke, P. D. van Helden, and E. G. Hoal. 2003. Association between tuberculosis and a polymorphic NFκB binding site in the interferon gamma gene. Lancet 3611871-1872. [DOI] [PubMed] [Google Scholar]
- 57.Rothbard, J. B., and M. L. Gefter. 1991. Interactions between immunogenic peptides and MHC proteins. Annu. Rev. Immunol. 9527-565. [DOI] [PubMed] [Google Scholar]
- 58.Santana, M. A., and F. Esquivel-Guadarrama. 2006. Cell biology of T cell activation and differentiation. Int. Rev. Cytol. 250217-274. [DOI] [PubMed] [Google Scholar]
- 59.Singer, D. F., and J. J. Linderman. 1990. The relationship between antigen concentration, antigen internalization, and antigenic complexes: modeling insights into antigen processing and presentation. J. Cell Biol. 11155-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singer, D. F., and J. J. Linderman. 1991. Antigen processing and presentation: how can a foreign antigen be recognized in a sea of self proteins? J. Theor. Biol. 151385-404. [DOI] [PubMed] [Google Scholar]
- 61.Singh, C. R., R. A. Moulton, L. Y. Armitige, A. Bidani, M. Snuggs, S. Dhandayuthapani, R. L. Hunter, and C. Jagannath. 2006. Processing and presentation of a mycobacterial antigen 85B epitope by murine macrophages is dependent on the phagosomal acquisition of vacuolar proton ATPase and in situ activation of cathepsin D. J. Immunol. 1773250-3259. [DOI] [PubMed] [Google Scholar]
- 62.Small, P. M., and P. I. Fujiwara. 2001. Management of tuberculosis in the United States. N. Engl. J. Med. 345189-200. [DOI] [PubMed] [Google Scholar]
- 63.Sriram, U., P. Selvaraj, S. M. Kurian, A. M. Reetha, and P. R. Narayanan. 2001. HLA-DR2 subtypes and immune responses in pulmonary tuberculosis. Indian J. Med. Res. 113117-124. [PubMed] [Google Scholar]
- 64.Taggart, R. T. 1992. Genetic variation of human aspartic proteinases. Scand. J. Clin. Lab. Investig. Suppl. 210111-119. [PubMed] [Google Scholar]
- 65.Talarico, S., M. D. Cave, B. Foxman, C. F. Marrs, L. Zhang, J. H. Bates, and Z. Yang. 2007. Association of Mycobacterium tuberculosis PE PGRS33 polymorphism with clinical and epidemiological characteristics. Tuberculosis 87338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teran-Escandon, D., L. Teran-Ortiz, A. Camarena-Olvera, G. Gonzalez-Avila, M. A. Vaca-Marin, J. Granados, and M. Selman. 1999. Human leukocyte antigen-associated susceptibility to pulmonary tuberculosis: molecular analysis of class II alleles by DNA amplification and oligonucleotide hybridization in Mexican patients. Chest 115428-433. [DOI] [PubMed] [Google Scholar]
- 67.Valitutti, S., S. Muller, M. Cella, E. Padovan, and A. Lanzavecchia. 1995. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature 375148-151. [DOI] [PubMed] [Google Scholar]
- 68.Wegner, K. M., M. Kalbe, G. Rauch, J. Kurtz, H. Schaschl, and T. B. Reusch. 2006. Genetic variation in MHC class II expression and interactions with MHC sequence polymorphism in three-spined sticklebacks. Mol. Ecol. 151153-1164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.