Abstract
The trxA gene is regarded as essential in Bacillus subtilis, but the roles of the TrxA protein in this gram-positive bacterium are largely unknown. Inactivation of trxA results in deoxyribonucleoside and cysteine or methionine auxotrophy. This phenotype is expected if the TrxA protein is important for the activity of the class Ib ribonucleotide reductase and adenosine-5′-phosphosulfate/3′-phosphoadenosine-5′-phosphosulfate reductase. We demonstrate here that a TrxA deficiency in addition causes defects in endospore and cytochrome c synthesis. These effects were suppressed by BdbD deficiency, indicating that TrxA in the cytoplasm is the primary electron donor to several different thiol-disulfide oxidoreductases active on the outer side of the B. subtilis cytoplasmic membrane.
Thioredoxin is a well-characterized thiol-disulfide oxidoreductase and an important reducing factor in the cytoplasm of most cells (5, 15, 29). Thiol-disulfide oxidoreductases in the active site contain two cysteine residues that are essential for enzyme function. When a thiol-disulfide oxidoreductase reduces or oxidizes a substrate protein, a disulfide bond is formed between the two proteins as a reaction intermediate. After the reaction is complete, the thiol-disulfide oxidoreductase needs to be reactivated by reduction or oxidation (5, 28). Thioredoxin is reactivated by thioredoxin reductase, with the use of NADPH as the reductant (15).
In bacteria, thioredoxin is involved in the defense against oxidative stress, it is required for the activity of several enzymes, it is a regulatory factor for certain enzymes and receptors, and it functions as an essential component for the assembly of small viruses (15). Some functions of thioredoxin are independent of thiol redox activity and instead depend on the ability of the protein to interact with other proteins to form a transient complex (19).
The trxA gene of the gram-positive bacterium Bacillus subtilis encodes thioredoxin (24) and has been reported to be essential for cell growth on nutrient-rich medium (18, 33). The expression of this gene is induced by various types of stress, and its transcription is initiated from two promoters, one dependent on σA (the main vegetative sigma factor) and the other on σB (a general stress sigma factor) (33). The trxA mRNA has a half-life of about 5 min, which is close to the average half-life for mRNAs in B. subtilis (11). As deduced from the genome sequence (20), B. subtilis contains several cytoplasmic thioredoxin-like proteins in addition to TrxA. The physiological roles of these putative proteins are unknown, but the corresponding genes are not important for growth (37). B. subtilis, like many other gram-positive bacteria, does not contain glutathione; i.e., it lacks genes for glutathione synthesis and glutathione reductase (20, 37). Thioredoxin(s) is seemingly the only proteinaceous disulfide reductant in the cytoplasm of B. subtilis. However, the ytnI gene might encode a glutaredoxin with thiol-disulfide oxidoreductase activity.
Both the primary sequences and the structures of B. subtilis and Escherichia coli TrxA proteins are similar, and both contain the conserved WCGPC active-site sequence (24, 27, 30). B. subtilis arsenate reductase (ArsC) is dependent on TrxA for activity (3, 24). Other enzymes in B. subtilis that are expected to be dependent on the redox activity of thioredoxin are ribonucleotide reductase and adenosine-5′-phosphosulfate (APS)/3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductase (4, 13). It has not been established, however, if the TrxA protein alone or some other thioredoxin functions with these enzymes in vivo.
In this work, we have analyzed the effects of inactivation of the trxA gene in B. subtilis. The focus is on the role of TrxA in endospore and cytochrome c maturation, which are processes involving the extracytoplasmic membrane-bound thiol-disulfide oxidoreductases CcdA, ResA, and StoA (7, 8, 34, 35). The CcdA protein is a polytopic membrane protein of 26 kDa that is homologous to the central (β) domain of DsbD of, e.g., Escherichia coli (26, 29). It is thought to be reduced by a thioredoxin in the cytoplasm and to transfer reducing equivalents to ResA and StoA on the outer side of the cytoplasmic membrane. ResA (20 kDa) functions to reduce apocytochrome c during cytochrome c maturation (7), whereas StoA (19 kDa) is important for efficient endospore cortex synthesis (8).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli cells were grown at 37°C in LB medium or on LA plates (31). B. subtilis cells were grown in baffled E-flasks, on a rotary shaker (at 200 rpm), in LB medium, in nutrient sporulation medium with phosphate (NSMP) (9) or in Spizizen minimal medium (38) supplemented with 0.5% glucose and 50 mg/liter tryptophan. Tryptose blood agar base (TBAB; Difco Co.) was used as the solid medium for B. subtilis. Deoxyribonucleosides (deoxyadenosine, deoxycytidine, deoxyguanosine, and deoxythymidine) were each added to a final concentration of 25 mg/liter. Cysteine and methionine were each added to a final concentration of 20 mg/liter. For the growth of strain LUW327, the medium was supplemented with 50 μg/ml isopropyl-β-d-thiogalactopyranoside (IPTG) to sustain wild-type growth (21, 37). Antibiotics were used at the following concentrations: for E. coli, ampicillin was used at 100 mg/liter, chloramphenicol at 12.5 mg/liter, and kanamycin at 50 mg/liter; and for B. subtilis, chloramphenicol was used at 4 mg/liter, erythromycin at 0.3 or 0.5 mg/liter, lincomycin at 10 or 12.5 mg/liter, and kanamycin at 10 mg/liter.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Descriptiona | Reference or sourceb |
|---|---|---|
| E. coli strains | ||
| MM294 | supE44 hsdR endA thi | 1 |
| TUNER(DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm lacY1(DE3)/pLysS; Cmr | Novagen |
| B. subtilis strains | ||
| 1A1 | trpC2 | BGSCc |
| LU60A1 | trpC2 ΔccdA::ble; Pmr | 34 |
| LUL10 | trpC2 bdbD::Tn10; Spr | 6 |
| LUW327 | trpC2 pMUTIN2mcs insertion in trxA promoter; Emr | 21 |
| LMM1 | trpC2 trxA::kan; Kmr | pLMC6-kan → 1A1; this study |
| LMM5 | trpC2 bdbD::Tn10 pMUTIN2mcs insertion in trxA promoter; Emr Spr | LUW327 + LUL10 →1A1; this study |
| LMM24 | trpC2 trxA::kan; Kmr | LMM1 → 1A1; this study |
| LMM82 | trpC2 bdbD::Tn10 trxA::kan; Kmr Spr | LMM24 + LUL10 → 1A1; this study |
| LMM83 | trpC2 bdbD::Tn10 trxA::kan; Kmr Spr | LMM24 + LUL10 → 1A1; this study |
| LMM241 | trpC2 trxA::kan; Kmr | LMM24 → 1A1; this study |
| Plasmids | ||
| pBluescript (SK−) | Cloning vector; Amr | Stratagene |
| pBAD-HisB | Expression vector; Amr | Invitrogen |
| pRSETB | Expression vector; Amr | Invitrogen |
| pDG780 | Carries antibiotic resistance cassette; Kmr Amr | 10 |
| pLMC1 | pBluescript with a 339-bp KpnI-HindIII fragment containing B. subtilis trxAwt Amr | This study |
| pLMC2 | pBAD-HisB with a 339-bp KpnI-HindIII fragment containing trxAwt from pLMC1; Amr | This study |
| pLMC6 | pRSETB with a 339-bp KpnI-HindIII fragment containing a trxAwt fragment from pLMC2; Amr | This study |
| pLMC6-kan | pLMC6 with trxA disrupted by insertion of a 1,486-bp ClaI fragment from pDG780; Amr Kmr | This study |
Amr, ampicillin; Cmr, chloramphenicol; Emr, erythromycin; Kmr, kanamycin; Pmr, phleomycin; Spr, spectinomycin; wt, wild type.
Arrows indicate transformation of the indicated strain with the indicated plasmid or chromosomal DNA.
Bacillus Genetic Stock Center, Ohio.
DNA techniques.
Plasmid DNA was isolated by using a Quantum Prep plasmid miniprep kit (Bio-Rad) or by CsCl density gradient centrifugation. E. coli strains were transformed by electroporation (12), and B. subtilis was grown to competence essentially as described by Hoch (14). PCR was carried out using Taq DNA polymerase (Roche) or Phusion polymerase (Finnzymes). DNA ligation was performed using T4 DNA ligase (New England Biolabs, Inc.). Plasmid and strain constructs were confirmed by DNA sequencing (BM labbet, Lund, Sweden; or MWG Biotech AG, Martinsried, Germany).
Construction of plasmids and strains.
Plasmid pLMC1 was constructed by amplifying the trxA gene of B. subtilis strain 1A1, using the primers MM001 (5′-GGCCATGGTACCATGGCTATCGTAAAGCAAC-3′) and MM002 (5′-GGCCATAAGCTTCTGGCATGTAAGCAGCG-3′) in a PCR (KpnI-HindIII sites are underlined). The PCR product was cut with KpnI and HindIII and cloned into pBluescript (SK−). The fragment was excised from the obtained plasmid pLMC1, using the same enzymes, and ligated into pBAD-HisB, creating the pLMC2 plasmid. pLMC6 was constructed by cutting out the trxA fragment of pLMC2, using KpnI and HindIII, and ligating it into pRSETB cut with the same enzymes. Sequencing of the trxA gene of B. subtilis strain 1A1 showed that base 243 in the open reading frame was a C, whereas it is an A in the published genome sequence (20). This nucleotide difference, however, does not change the predicted protein sequence.
To obtain plasmid pLMC6-kan, pLMC6 was cut with ClaI and ligated with a kanamycin resistance cassette on a ClaI fragment obtained from plasmid pDG780. B. subtilis LMM1 was obtained by transformation of the 1A1 strain with ScaI-cleaved pLMC6-kan and the selection of colonies on TBAB plates supplemented with kanamycin, cysteine, methionine, and deoxyribonucleosides, as described in Results and Discussion. Strain LMM24 was obtained by the transformation of 1A1, using LMM1 chromosomal DNA. Similarly, strain LMM241 was obtained by the transformation of 1A1 with LMM24 chromosomal DNA. BdbD TrxA double-deficient strains were obtained by transformation of 1A1 with a mixture of DNAs including LUL10 chromosomal DNA and either LUW327 or LMM1 chromosomal DNA.
Production and affinity purification of His-tagged TrxA.
Hexahistidinyl-tagged wild-type B. subtilis TrxA was produced in E. coli TUNER(DE3)/pLysS containing pLMC6. Cells were grown in LB medium at 37°C at 200 rpm. At an optical density at 600 nm of 0.5, the expression of trxA was induced, using 0.4 mM IPTG. Ninety minutes later, the cells were collected by centrifugation and frozen at −20°C. After cells were thawed, they were resuspended in 40 ml of buffer A (50 mM sodium phosphate buffer, 300 mM NaCl [pH 8.0]) containing 10 mM imidazole and DNase. The cells were disrupted by using a French pressure cell operated at 18,000 lb/in2. The lysate obtained was centrifuged at 48,000 × g for 30 min at 4°C, and the supernatant was mixed with Ni-nitrilotriacetic acid agarose (Qiagen). After it was mixed for 1 h at 4°C, the resin was washed with 10 mM imidazole in buffer A, and TrxA was eluted from the resin by using 100 mM imidazole in buffer A. The purity of the isolated His-tagged proteins was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. The authenticity of the purified His-tagged wild-type TrxA was verified by mass spectrometry (Swegene Proteomics Platform, Lund, Sweden), giving a value of 15,859.7 Da, which is in reasonable agreement with the expected mass of 15,850.7 Da. Isolated B. subtilis His6-TrxA protein and E. coli thioredoxin (TrxA), used as a reference, showed similar thiol-disulfide oxidoreductase activity, as determined by using the insulin reduction assay (16). Insulin and E. coli thioredoxin were purchased from Sigma Chem. Antiserum against TrxA was obtained by immunizing rabbits with isolated His-tagged TrxA. Antisera were produced at PickCell Laboratories (Amsterdam, The Netherlands).
Immunoblotting.
Proteins were fractionated by SDS-PAGE and transferred to a polyvinylidene difluoride blotting membrane (Immobilon P; Millipore), using a wet blot (Bio-Rad). Transfer buffer was 20 mM Tris and 150 mM glycine buffer containing 20% (vol/vol) methanol. Immunodetection was carried out by chemiluminescence, using horseradish peroxidase-labeled anti-rabbit immunoglobulin G antibodies (GE Healthcare) and the SuperSignal West Pico chemiluminescent substrate (Pierce Chem Co.). Luminescence was recorded using a Kodak Image station.
Sporulation assay.
Cells were grown for 2 days at 30°C or at 37°C at 200 rpm in NSMP supplemented with deoxyribonucleosides and 50 mg/ml cysteine and methionine. Sporulation efficiency of the strains was analyzed by heating 5 ml of culture at 80°C for 15 min, as described previously (8). Serial dilutions of treated and untreated samples were spread on TBAB plates supplemented with deoxyribonucleosides. After plates were incubated at 37°C, colonies were counted.
Other methods.
Protein concentrations were determined by using the bicinchoninic acid protein assay (Pierce Chemical Co.) with bovine serum albumin as the standard or by measuring the absorbance at 280 nm. SDS-PAGE was carried out using the Schägger and von Jagow buffer system (32). Isolation of membranes, N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) oxidation activity staining of colonies on plates, and determination of cytochrome c oxidase activity were done as described previously (2). Staining of proteins for covalently bound heme after SDS-PAGE was carried out as described previously (36).
RESULTS AND DISCUSSION
Growth of the trxA conditional mutant B. subtilis strain LUW327.
E. coli thioredoxin was first identified as a hydrogen donor to ribonucleotide reductase, required for the synthesis of deoxyribonucleotides (22). Thioredoxin in E. coli is also involved in the biosynthesis of cysteine by donating electrons to APS/PAPS reductase (39). Using transcriptome analysis, Smits et al. showed that some B. subtilis genes for proteins that function in sulfur assimilation and cysteine biosynthesis and the genes for the type Ib ribonucleotide reductase are up-regulated in TrxA-deprived cells (37). It was also shown that the addition of cysteine to the growth medium partly restores the growth of a TrxA-deprived strain.
To explore the roles of TrxA in B. subtilis, we initially made use of strain LUW327 (Table 1). In this strain, the trxA gene in the chromosome is transcribed from the pSpac promoter whose activity is negatively controlled by the binding of LacI. Wild-type cellular levels of thioredoxin are obtained in strains containing this conditional trxA expression system if the liquid growth medium is supplemented with 100 μM IPTG. In the presence of only 25 μM IPTG, the TrxA protein is not detectable in cell extracts, using immunoblotting analysis (37). LUW327 was found to require ≥25 μM IPTG for wild-type-like levels of growth in liquid rich media, i.e., NSMP and LB. On solid rich medium, i.e., TBAB plates, 50 μM IPTG was needed to support wild-type-like levels of growth of the strain. When LUW327 cells were spread onto solid rich medium without IPTG, colonies of better-growing clones containing suppressor mutations appeared at a high frequency. These suppressor mutations are presumably located in the pSpac promoter region or in lacI, making the trxA gene expression IPTG independent.
Cysteine requirement of LUW327.
To investigate the importance of TrxA in deoxyribonucleotide and cysteine biosynthesis in B. subtilis, we grew LUW327 without IPTG in minimal-glucose medium supplemented with 20 mg/liter cysteine or 20 mg/liter methionine. Cysteine or methionine essentially restored growth to the same level as that obtained by adding 100 μM IPTG to the medium. We ruled out the possibility that growth in the absence of IPTG was due to suppressor mutations by plating cells from the culture onto plates without IPTG, where no growth was detected.
The results suggested that LUW327 cells growing in minimal medium contain TrxA, but only very small amounts, due to incomplete blockage of the pSpac promoter activity. This small amount of TrxA in the cell does not support the required APS/PAPS reductase activity but is large enough to support sufficient deoxyribonucleotide synthesis. It should be noted that cysteine can be synthesized from methionine (17). Methionine supplementation therefore supports the growth of LUW327 cells in minimal medium. Cysteine, a low-molecular-weight thiol reagent, can act as a redox buffer in the cytoplasm, and it is possible that this makes a contribution when the TrxA concentration is low (23).
Inactivation of trxA.
With the knowledge that cysteine or methionine can support the growth of cells containing little TrxA, we tested to see whether B. subtilis cells with the trxA gene inactivated can grow if the medium is supplemented with cysteine, methionine, and deoxyribonucleosides. To inactivate trxA, plasmid pLMC6-kan (which contains trxA disrupted by the insertion of a kanamycin resistance cassette) in linear form was used to transform B. subtilis strain 1A1. TBAB plates supplemented with kanamycin, cysteine, methionine, and deoxyribonucleosides were used for selection of transformants. Small colonies were obtained after several days of incubation at 37°C. One clone that did not grow without supplementation of the medium with the four deoxyribonucleosides was named LMM1. The disruption of the trxA gene in the chromosomal DNA of LMM1 was verified by using PCR with primers MM001 and MM002, followed by DNA sequence analysis of the PCR product.
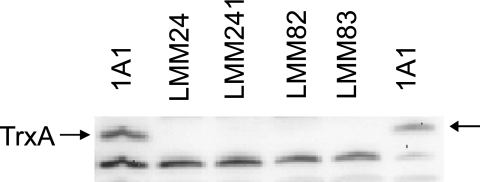
Isolated LMM1 chromosomal DNA was used to transform B. subtilis 1A1. Kanamycin-resistant transformants were selected on TBAB plates supplemented with cysteine, methionine, and deoxyribonucleosides. Transformants appeared as small colonies after incubation at 37°C overnight, and all were found to require deoxyribonucleosides for growth. The trxA insertion mutation was confirmed as before in two of the backcrossed clones. One clone, LMM24 (Table 1), was kept for further analysis. The lack of the TrxA polypeptide in the mutant cells grown in NSMP supplemented with cysteine, methionine, and deoxynucleosides was confirmed by immunoblotting analysis (Fig. 1).
FIG. 1.
Immunoblotting analysis for TrxA in different B. subtilis strains. Proteins in cell lysates were separated by SDS-PAGE, electroblotted to a filter, and probed with rabbit antiserum obtained by using recombinant B. subtilis TrxA produced in E. coli as the immunogen. 1A1 is the parental strain. The trxA gene is inactivated in the other strains (Table 1). Protein (10 μg) was loaded in each lane, except in the case of the lane with 1A1 on the right side, where only 5 μg of protein was loaded. The position of TrxA polypeptide in the gel is indicated by arrows. A polypeptide smaller than TrxA and cross-reacting with the antiserum is seen in all lanes.
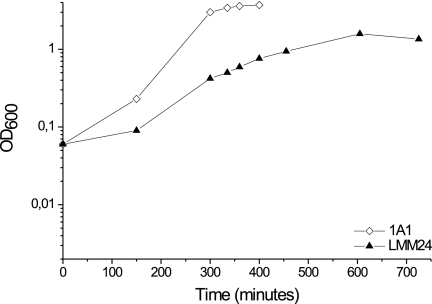
The growth of LMM24 colonies on TBAB plates containing the four deoxyribonucleosides was independent of the addition of cysteine or methionine (these amino acids are available in TBAB). Concentrations higher than 25 mg/liter of deoxyribonucleosides in the TBAB plates did not promote better growth. As judged from the use of combinations of deoxyribonucleosides, deoxycytidine was absolutely required for growth. The growth of strain LMM24 compared to that of the parental strain 1A1 in NSMP supplemented with deoxyribonucleosides, cysteine, and methionine is shown in Fig. 2.
FIG. 2.
Growth curves for B. subtilis 1A1 and the TrxA-deficient strain LMM24 in NSMP supplemented with the four deoxynucleosides, cysteine, and methionine. See Materials and Methods for details.
TrxA-deficient strains are defective in cytochrome c synthesis.
B. subtilis contains four different types of cytochrome c, and they are all membrane bound and functional on the outer side of the cytoplasmic membrane (40). CcdA-deficient mutants lack all four types of cytochrome c and are also deficient in endospore synthesis, but vegetative cells show normal growth in both minimal and rich media (34, 35).
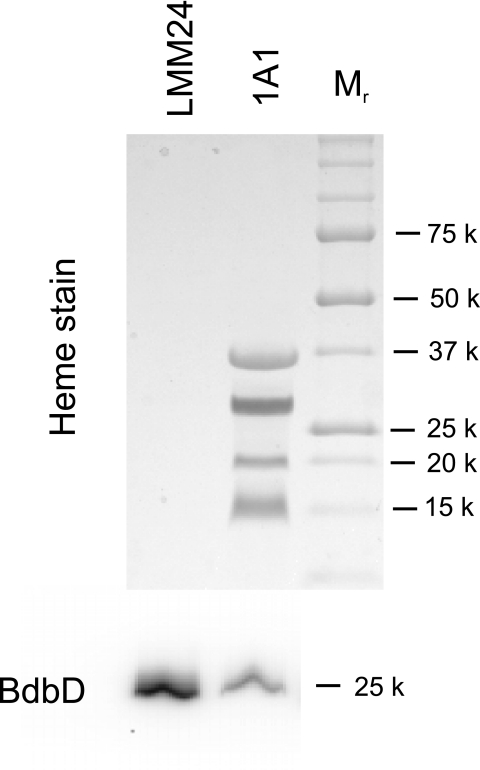
Relatively high production of cytochrome c is obtained when wild-type B. subtilis cells are grown in NSMP. Cytochrome c synthesis in TrxA-deficient cells was examined by TMPD oxidation staining of colonies and cytochrome c oxidase activity of isolated membranes. Both of these activities rely on the presence of a functional cytochrome caa3 oxidase, which in turn depends on the presence of cytochrome c (2). The parental strains 1A1 and LU60A1 (a CcdA-deficient strain) were used as controls in the experiments. Colonies of LMM1, LMM24, and LU60A1 grown on NSMP agar plates supplemented with the four deoxyribonucleosides did not oxidize TMPD, whereas 1A1 colonies on such plates showed strong oxidation activity. Also, strain LUW327 grown on NSMP agar plates without added IPTG lacked TMPD oxidation activity but showed such activity when the medium was supplemented with 25 μM IPTG. Membranes of strains LMM24 and LMM241 (obtained by a backcross to 1A1, using LMM24 chromosomal DNA; Table 1) completely lacked all four cytochrome c proteins, as determined by heme staining of SDS-polyacrylamide gels (Fig. 3 and gel not shown), and contained only a background level of cytochrome c oxidase activity (Table 2). LUW327 grown without IPTG added showed about 4% cytochrome c activity compared to that of the wild type (Table 2). This low oxidase activity can be explained by the incomplete inhibition of pSpac promoter activity, resulting in low amounts of TrxA in the cell. The combined findings show that TrxA-deficient strains lack cytochrome c.
FIG. 3.
Analysis of membrane proteins for covalently bound heme. Isolated membranes of the TrxA-deficient strain LMM24 and the parental strain 1A1 were subjected to SDS-PAGE. The upper part of the figure shows the result of staining the gel for heme after electrophoresis. The four bands seen with 1A1 correspond to heme in CtaC, QcrC, QcrB, and CccA/CccB (40). Lane Mr contained prestained molecular size markers (k, thousand). The lower part of the figure shows, as a positive control, a Western blot with the same bacterial membranes, using anti-BdbD antiserum. For the heme stain and Western blot, 100 μg and 5 μg membrane protein, respectively, were loaded per lane.
TABLE 2.
Cytochrome c oxidase activities in membranes of B. subtilis strainsa
| Strain | Enzyme deficiency | Cytochrome c oxidase activity (% of normal)b |
|---|---|---|
| 1A1 | None (parental strain) | 100 |
| LMM24 | TrxA | <1 |
| LMM241 | TrxA | <1 |
| LMM82 | TrxA BdbD | 10 |
| LUW327c | TrxA | 4 |
| LMM5c | TrxA BdbD | 31 |
Cytochrome c oxidase activities in membranes isolated from B. subtilis strains grown in NSMP supplemented with deoxyribonucleosides, cysteine, and methionine.
The cytochrome c activity of strain 1A1 was 180 nmol cytochrome c oxidized per min per mg of membrane protein.
Membranes of strains LUW327 and LMM5 were isolated from cells grown without the addition of IPTG.
Endospore synthesis depends on TrxA.
Strains lacking TrxA, i.e., LMM24 and LMM241, grown for sporulation in NSMP medium supplemented with cysteine and deoxyribonucleosides, produced few heat-resistant cells compared to the parental strain (Table 3). The cultures did not contain any phase-contrast bright cells as observed using a light microscope. These results indicated defective endospore synthesis in TrxA-deficient cells.
TABLE 3.
Heat resistance of B. subtilis strainsa
| Strain | Enzyme deficiency | Cell titer (CFU/ml)b | Spore titer (CFU/ml)c | Sporulation efficiency (%)d |
|---|---|---|---|---|
| 1A1 | None (parental strain) | 1.3 × 108 | 5.5 × 107 | 42 |
| LMM24 | TrxA | 6.2 × 107 | <1 × 102 | <0.0002 |
| LMM241 | TrxA | 6.7 × 107 | 2 × 102 | <0.0003 |
| LMM82 | TrxA BdbD | 1.2 × 106 | 2.8 × 105 | 23 |
Data indicate the heat resistance of B. subtilis strains grown for sporulation in NSMP supplemented with deoxyribonucleosides, cysteine, and methionine.
Cell titer values are CFU/ml before treatment.
Spore titers are CFU/ml after heat treatment.
Sporulation efficiency was calculated as the spore titer divided by the cell titer.
BdbD deficiency suppresses the effects of the trxA gene inactivation on cytochrome c and endospore synthesis.
BdbD is an extracytoplasmic membrane-bound oxidative thiol-disulfide oxidoreductase that catalyzes the formation of disulfide bonds in proteins on the outer side of the membrane (6, 25). The defects in cytochrome c and endospore maturation caused by deficiencies of ResA and StoA, respectively, and those caused by CcdA deficiency are suppressed by BdbD deficiency (6, 7, 8). Suppression is explained by the fact that BdbD catalyzes disulfide bond formation in extracytoplasmic proteins, and some proteins are thereby inactivated. ResA counteracts this process by specifically disrupting disulfide bonds in apocytochrome c. Similarly, StoA breaks disulfide bonds in hitherto-unidentified protein substrates present in the intermembrane space of the forespore during endospore maturation (6, 7, 26).
If TrxA is the electron donor to CcdA, and thereby indirectly to both ResA and StoA, we predicted that the negative effects of TrxA deficiency on cytochrome c and endospore maturation should be suppressed by BdbD deficiency. Therefore, we constructed strains deficient in both TrxA and BdbD, LMM82 and LMM83 (Table 1 and Fig. 1). These strains showed about 10% cytochrome c oxidase activity compared to that of the parental strain (Table 2) and almost normal production of heat-resistant endospores (Table 3). Suppression of the defect in cytochrome c maturation caused by TrxA deficiency was also observed with a constructed BdbD-deficient derivative of LUW327, LMM5 (Table 2). The extent of suppression obtained by the absence of BdbD in TrxA-deficient cells is similar to that seen with strains lacking ResA, CcdA, or StoA (6, 7, 8).
Our findings with the TrxA BdbD doubly deficient cells demonstrate that the effects on cytochrome c and endospore maturation obtained as a result of the trxA gene inactivation are manifested on the extracytoplasmic side of the membrane.
Conclusion.
We have successfully inactivated the “essential” trxA gene in B. subtilis cells and have shown that this results in deoxyribonucleoside auxotrophy, most likely because the TrxA protein is required for ribonucleotide reductase activity. It has previously been demonstrated that ribonucleotide reductase (NrdEFI)-deficient B. subtilis cells require deoxyribonucleosides for growth (13). Our findings with TrxA-deficient cells suggest that TrxA is required also as an electron donor to APS/PAPS reductase and is therefore important for sulfate assimilation.
B. subtilis ResA and StoA are membrane-anchored thiol-disulfide oxidoreductases with a role in cytochrome c and endospore cortex maturation, respectively (7, 8). The catalytic domain of each of these proteins is located on the outer side of the membrane and has a thioredoxin-like fold with a WCE/PPCKKE active-site sequence motif. CcdA in the cytoplasmic membrane of B. subtilis cells is suggested to reduce ResA and StoA (26). We demonstrate here that TrxA-deficient cells are defective in cytochrome c and endospore maturation and that this phenotype is suppressed by BdbD deficiency. This strongly suggests that TrxA is the cytoplasmic electron donor to CcdA. The situation is then similar to that in the gram-negative bacterium E. coli, where TrxA is the electron donor to DsbD in the inner membrane (29).
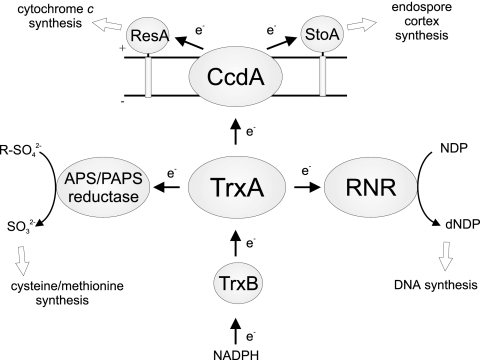
Figure 4 presents a summary of the central functions of TrxA in B. subtilis, consistent with the properties of the TrxA-deficient mutants we have analyzed in this work. It should be noted that TrxA has more functions, for example, to reduce arsenate reductase (24) and to play a role against various types of environmental stress (23, 33).
FIG. 4.
Functions of TrxA in B. subtilis as deduced from the results of this work. APS/PAPS reductase is encoded by the cysH gene, and ribonucleotide reductase (RNR) is encoded by the nrdEFI genes. TrxB is thioredoxin reductase. The arrows and e− indicate the direction of flow of the reducing equivalents. + and − indicate the outer and cytoplasmic sides of the membrane, respectively.
Acknowledgments
This work was supported by grants 621-2004-2762 and 621-2007-6094 from the Swedish Research Council and a grant from the Swedish National Research School in Genomics and Bioinformatics.
We thank Ingrid Stål and Hadija Trojer for technical assistance, Claes von Wachenfeldt for strain LUW327, and Liselotte Andersson for doing the mass spectrometry analysis.
Footnotes
Published ahead of print on 2 May 2008.
REFERENCES
- 1.Backman, K., M. Ptashne, and W. Gilbert. 1976. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 734174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengtsson, J., C. von Wachenfeldt, L. Winstedt, P. Nygaard, and L. Hederstedt. 2004. CtaG is required for formation of active cytochrome c oxidase in Bacillus subtilis. Microbiology 150415-425. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, M. S., Z. Guan, M. Laurberg, and X. D. Su. 2001. Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 9813577-13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berndt, C., C. H. Lillig, M. Wollenberg, E. Bill, M. C. Mansilla, D. de Mendoza, A. Seidler, and J. D. Schwenn. 2004. Characterization and reconstitution of a 4Fe-4S adenylyl sulfate/phosphoadenylyl sulfate reductase from Bacillus subtilis. J. Biol. Chem. 2797850-7855. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho, A. P., P. A. Fernandes, and M. J. Ramos. 2006. Similarities and differences in the thioredoxin superfamily. Prog. Biophys. Mol. Biol. 91229-248. [DOI] [PubMed] [Google Scholar]
- 6.Erlendsson, L. S., and L. Hederstedt. 2002. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J. Bacteriol. 1841423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erlendsson, L. S., R. M. Acheson, L. Hederstedt, and N. E. Le Brun. 2003. Bacillus subtilis ResA is a thiol-disulfide oxidoreductase involved in cytochrome c synthesis. J. Biol. Chem. 27817852-17858. [DOI] [PubMed] [Google Scholar]
- 8.Erlendsson, L. S., M. Möller, and L. Hederstedt. 2004. Bacillus subtilis StoA is a thiol-disulfide oxidoreductase important for spore cortex synthesis. J. Bacteriol. 1866230-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortnagel, P., and E. Freese. 1968. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J. Bacteriol. 951431-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167335-336. [DOI] [PubMed] [Google Scholar]
- 11.Hambraeus, G., C. von Wachenfeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269706-714. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 20463-113. [DOI] [PubMed] [Google Scholar]
- 13.Härtig, E., A. Hartmann, M. Schatzle, A. M. Albertini, and D. Jahn. 2006. The Bacillus subtilis nrdEF genes, encoding a class Ib ribonucleotide reductase, are essential for aerobic and anaerobic growth. Appl. Environ. Microbiol. 725260-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204305-320. [DOI] [PubMed] [Google Scholar]
- 15.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54237-271. [DOI] [PubMed] [Google Scholar]
- 16.Holmgren, A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 2549627-9632. [PubMed] [Google Scholar]
- 17.Hullo, M.-F., S. Auger, O. Soutourina, O. Barzu, M. Yvon, A. Danchin, and I. Martin-Verstraete. 2007. Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J. Bacteriol. 189187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 1004678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar, J. K., S. Tabor, and C. C. Richardson. 2004. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 1013759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390249-256. [DOI] [PubMed] [Google Scholar]
- 21.Larsson, J. T., A. Rogstam, and C. von Wachenfeldt. 2007. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol. Microbiol. 66669-684. [DOI] [PubMed] [Google Scholar]
- 22.Laurent, T. C., E. C. Moore, and P. Reichard. 1964. Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 2393436-3444. [PubMed] [Google Scholar]
- 23.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 1851967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y., Y. Hu, X. Zhang, H. Xu, E. Lescop, B. Xia, and C. Jin. 2007. Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis. J. Biol. Chem. 28211078-11083. [DOI] [PubMed] [Google Scholar]
- 25.Meima, R., C. Eschevins, S. Fillinger, A. Bolhuis, L. W. Hamoen, R. Dorenbos, W. J. Quax, J. M. van Dijl, R. Provvedi, I. Chen, D. Dubnau, and S. Bron. 2002. The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J. Biol. Chem. 2776994-7001. [DOI] [PubMed] [Google Scholar]
- 26.Möller, M., and L. Hederstedt. 2006. Role of membrane-bound thiol-disulfide oxidoreductases in endospore-forming bacteria. Antioxid. Redox Signal. 8823-833. [DOI] [PubMed] [Google Scholar]
- 27.Prinz, W. A., F. Åslund, A. Holmgren, and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 27215661-15667. [DOI] [PubMed] [Google Scholar]
- 28.Raina, S., and D. Missiakas. 1997. Making and breaking disulfide bonds. Annu. Rev. Microbiol. 51179-202. [DOI] [PubMed] [Google Scholar]
- 29.Ritz, D., and J. Beckwith. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 5521-48. [DOI] [PubMed] [Google Scholar]
- 30.Roos, G., A. Garcia-Pino, K. Van belle, E. Brosens, K. Wahni, G. Vandenbussche, L. Wyns, R. Loris, and J. Messens. 2007. The conserved active site proline determines the reducing power of Staphylococcus aureus thioredoxin. J. Mol. Biol. 368800-811. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 33.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Völker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 1801869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiött, T., and L. Hederstedt. 2000. Efficient spore synthesis in Bacillus subtilis depends on the CcdA protein. J. Bacteriol. 1822845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiött, T., C. von Wachenfeldt, and L. Hederstedt. 1997. Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis. J. Bacteriol. 1791962-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulz, H., E. C. Pellicioli, H. Hennecke, and L. Thöny-Meyer. 2000. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc. Natl. Acad. Sci. USA 966462-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smits, W. K., J. Y. Dubois, S. Bron, J. M. van Dijl, and O. P. Kuipers. 2005. Tricksy business: transcriptome analysis reveals the involvement of thioredoxin A in redox homeostasis, oxidative stress, sulfur metabolism, and cellular differentiation in Bacillus subtilis. J. Bacteriol. 1873921-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by ribonucleate. Proc. Natl. Acad. Sci. USA 441072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsang, M. L. 1981. Assimilatory sulfate reduction in Escherichia coli: identification of the alternate cofactor for adenosine 3′-phosphate 5′-phosphosulfate reductase as glutaredoxin. J. Bacteriol. 1461059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Wachenfeldt, C., and L. Hederstedt. 2001. Respiratory cytochromes, other heme-proteins, and heme biosynthesis, p. 163-179. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC.