Abstract
A Lactobacillus reuteri strain isolated from sourdough is known to produce the vitamin cobalamin. The organism requires this for glycerol cofermentation by a cobalamin-dependent enzyme, usually termed glycerol dehydratase, in the synthesis of the antimicrobial substance reuterin. We show that the cobalamin-synthesizing capacity of another L. reuteri strain (20016, the type strain, isolated from the human gut and recently sequenced as F275) is genetically and phenotypically linked, as in the Enterobacteriaceae, to the production of a cobalamin-dependent enzyme which is associated with a bacterial microcompartment (metabolosome) and known as diol dehydratase. We show that this enzyme allows L. reuteri to carry out a disproportionation reaction converting 1,2-propanediol to propionate and propanol. The wide distribution of this operon suggests that it is adapted to horizontal transmission between bacteria. However, there are significant genetic and phenotypic differences between the Lactobacillus background and the Enterobacteriaceae. Electron microscopy reveals that the bacterial microcompartment in L. reuteri occupies a smaller percentage of the cytoplasm than in gram-negative bacteria. DNA sequence data show evidence of a regulatory control mechanism different from that in gram-negative bacteria, with the presence of a catabolite-responsive element (CRE) sequence immediately upstream of the pdu operon encoding diol dehydratase and metabolosome structural genes in L. reuteri. The metabolosome-associated diol dehydratase we describe is the only candidate glycerol dehydratase present on inspection of the L. reuteri F275 genome sequence.
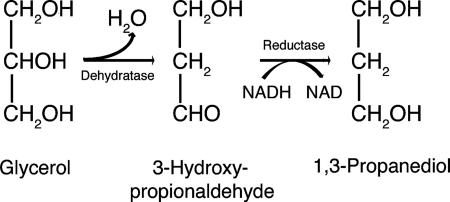
Lactobacillus reuteri is a probiotic bacterium able to colonize the gastrointestinal tract of a wide variety of mammals and birds (12). It produces an antimicrobial agent (45) (reuterin) by fermentation of glycerol. Lactobacillus spp. (including L. reuteri) cannot grow on glycerol as a sole carbon source, but L. reuteri can use beta-hydroxypropionaldehyde (3-HPA), which it derives from glycerol, as a hydrogen acceptor in fermentation of other carbohydrates, including glucose and lactose (44). Unlike most lactobacilli, L. reuteri grown in this way on glycerol and another carbohydrate excretes large amounts of reuterin, consisting of an equilibrium mixture of different monomeric and dimeric forms of 3-HPA, which has been shown to correspond to reuterin (46, 56). There is an optimal ratio of glycerol and glucose for maximal 3-HPA production, and if excess glucose is supplied, 3-HPA is further reduced to 1,3-propanediol by a 1,3-propanediol:NAD oxidoreductase (23) (Fig. 1).
FIG. 1.
3-HPA production and metabolism in L. reuteri.
It has been suggested there are two distinct cobalamin-dependent dehydratases in L. reuteri that can produce HPA from glycerol (47). Certainly, in Klebsiella pneumoniae (49) and a variety of other Enterobacteriaceae (13, 51), two different isofunctional cobalamin-dependent enzymes, glycerol dehydratase (EC 4.2.1.30) and diol dehydratase (EC 4.2.1.28), can catalyze the key reaction of glycerol dehydration to 3-HPA (Fig. 1) (13, 52). They can also convert a different substrate, 1,2-propanediol (1,2-PD), to propionaldehyde. Diol dehydratase genes are associated in many Enterobacteriaceae with a functional cobalamin synthesis pathway (21) and the production of a proteinaceous cellular microcompartment localizing the active enzyme, resembling the carboxysome containing ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) in autotrophic bacteria (10). This structure in heterotrophic Enterobacteriaceae has been termed an enterosome (11) or a carboxysome (29) (based on a hypothesis that carbon dioxide fixation may also occur in these heterotrophic bacteria). Generic terms such as bacterial microcompartment (16) and metabolosome (10) have also been proposed for all such structures, and we use the term metabolosome. Phylogenetic analysis suggests that, despite their size and complexity, linked cobalamin synthesis and metabolosome synthesis operons are frequently horizontally transmitted (21).
In Enterobacteriaceae like Salmonella (27) and Klebsiella (50), the metabolosome-associated propanediol utilization operon specifies enzymes for a dismutation that converts 1,2-PD (via propionaldehyde) to approximately equal amounts of n-propanol (reduced) and propionate (oxidized). ATP is produced via substrate-level phosphorylation: 2 (CH3-CH(OH)-CH2OH) + ADP + Pi → CH3-CH2-COOH + CH3-CH2-CH2OH + ATP + 2H2O.
Because unlike most other lactic acid bacteria, L. reuteri CRL1098 (48) (a lactic acid bacterium isolated from sourdough) produces cobalamin due to the presence of a multigene operon resembling that present in Salmonella and Listeria (37), we hypothesized that, as in Enterobacteriaceae and other gram-negative bacteria, this capacity was due to horizontally acquired genes which also specified production of a metabolosome containing a diol dehydratase. No demonstration of 1,2-PD utilization or bacterial microcompartment production in L. reuteri strains has previously been reported. We show that in L. reuteri 20016 (the type strain, originally isolated from human feces), a bacterial microcompartment is present, and inducible 1,2-PD utilization occurs, with disproportionation to propionate and propanol. Cobalamin is also synthesized. Preliminary analysis of genome sequence data shows the presence of linked cobalamin synthesis and propanediol utilization operons as in gram-negative bacteria, with a distinct gram-positive CRE potentially regulating gene transcription in a Lactobacillus background.
MATERIALS AND METHODS
Bacteria and growth conditions.
L. reuteri NCDO 2589 was obtained from the National Collection of Dairy Organisms, Reading, United Kingdom (now NCIMB Ltd, Aberdeen, United Kingdom). This is also known as DSM 20016, the type strain, and F275, which has recently been sequenced, and was originally isolated from human feces. L. reuteri 100-23 was obtained from Gerald Tannock (University of Otago, Dunedin, New Zealand). This strain is also known as DSM 17509 and has been sequenced. It was originally isolated from the digestive tract of a rat (58). L. reuteri 100-23 was employed only as a negative control in the propanediol metabolism assay and growth curves. All other references to L. reuteri here refer to L. reuteri DSM 20016. L. reuteri strains were grown in de Man-Rogosa-Sharpe (MRS) broth overnight (13a) containing 15 mM glucose at 37°C without shaking. To test for reuterin production by acrolein-based quantitation of HPA, MRS broth containing 250 mM glycerol as well as glucose was used with anaerobic incubation for 24 h at 37°C. For the isolation of metabolosomes and for the purification of diol dehydratase, L. reuteri was grown in conical flasks containing MRS broth supplemented with 50 mM 1,2-PD and 15 mM glucose at 37°C for 36 h. For the detection of metabolosomes using transmission electron microscopy, L. reuteri was grown in MRS broth containing 65 mM 1,2-PD with and without 15 mM glucose at 37°C for 18 h. For dismutation of 1,2-PD, L. reuteri was grown in modified MRS (MRS-MOD) medium, pH 5.7 (19), supplemented with 40 mM 1,2-PD without glucose, at 37°C under anaerobic conditions for 8 days. MRS-MOD is a complex medium containing (per liter) 5 g Bacto-peptone, 4 g Lab-Lemco (Oxoid), 2 g yeast extract, 0.5 ml Tween 80, 1.0 g K2HPO4, 3.0 g NaH2PO4·H2O, 0.6 g CH3COONa, 0.3 g MgSO4·7H2O, and 0.04 g MnSO4·H2O.
Isolation of metabolosomes and protein separation.
Protein preparations were initially made by a modification of a published procedure (14). Briefly, L. reuteri grown in MRS broth containing 1,2-PD and glucose was harvested by centrifugation at 4,000 × g for 10 min. The pelleted cells were washed with 300 ml lysozyme buffer (50 mM Tris-Cl, 0.6 M sucrose, 5 mM EDTA, 0.2% 1,2-PD [pH 8.0]), resuspended in 30 ml of the same buffer containing 5 mg/ml lysozyme, and incubated at 37°C for 2 h with occasional agitation. All further steps were carried out at 4°C. Lysozyme-treated cells were pelleted by centrifugation at 7,500 × g for 15 min, washed with lysozyme buffer, and resuspended in sonication buffer (50 mM Tris-Cl, 2 mM EDTA, 0.2% 1,2-PD [pH 8.0]) at approximately 0.1 g wet cell mass per ml. Cells were lysed by sonication, four 120-s bursts with 1-min cooling intervals on ice, using SoniPrep 150 (MSE UK Ltd). The crude cell extract obtained by sonication was mixed with an equal volume of BPER-II (Pierce, Rockford, IL) supplemented with 400 mM NaCl and 20 mM MgCl2 and incubated for 30 min at 4°C with shaking. Unlysed cells were removed by centrifugation at 12,000 × g for 10 min. The resulting supernatant was subjected to ultracentrifugation (Beckman SW-40 Ti rotor) at 49,000 × g for 90 min. The crude protein pellet was resuspended in 5 ml of TEMP buffer (50 mM Tris-Cl, 1 mM EDTA, 10 mM MgCl2, 0.2% 1,2-PD [pH 8.0]) and clarified by centrifugation at 12,000 × g for 10 min. The clarified preparation was layered onto four 11-ml, 35%-to-65% (wt/vol) sucrose density gradients and centrifuged at 30,000 × g for 16 h. Fractions including the pellet were taken and assayed for diol dehydratase activity. Dehydratase-positive fractions were retained in sucrose buffer, and the pellet was resuspended in 1 ml of TEMP buffer and clarified by centrifugation before electron microscopy. Protein preparations for peptide fingerprinting were made by cell sonication as described above (omitting lysozyme and admixture with BPER-II), with subsequent fractionation of the total crude cell lysate by sucrose density gradient centrifugation, selecting diol dehydratase-positive fractions for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation.
Protein separation.
Aliquots (50 μg) of extracted protein were separated by SDS-PAGE using a 12.5% polyacrylamide gel under denaturing conditions (20) in a MiniProtean apparatus (Bio-Rad) and stained with Coomassie brilliant blue R250.
Peptide fingerprinting.
Bands were excised from the polyacrylamide gel and subjected to in-gel tryptic digestion (40). Peptides were analyzed by MALDI-TOF-MS (matrix assisted laser desorption/ionization-time of flight mass spectroscopy) using a 20-mg/ml solution of 1,4-dihydroxybenzoic acid dissolved in 1 part acetonitrile, 2 parts trifluoroacetic acid as the matrix. Mass spectra were collected on a Bruker UltraFlex mass spectrometer (Bruker Daltonics, Bremen, Germany) that had been calibrated with a peptide calibration standard (1,000 to 4,000 Da) from Bruker (part 206195). Peptide masses were determined using Xmass (version 5.1.5; Bruker). Proteins were identified by peptide mass fingerprinting utilizing the Mascot search engine. Positive matches were ranked using the built-in Mowse score system of Mascot (Matrix Science, Boston, MA).
Electron microscopy.
The L. reuteri cell pellet was prefixed in 2.0% (vol/vol) glutaraldehyde, 2.5% paraformaldehyde in 165 mM phosphate buffer, pH 7.0, for 90 min. The prefixed pellet was postfixed in 2.0% (wt/vol) osmium tetroxide in 165 mM phosphate buffer, pH 7.2, for 120 min, followed by dehydration in an ethanol series. Embedment was done in epoxy resin (43a). Ultrathin sections (90 nm) were poststained with 4% (wt/vol) aqueous uranyl acetate and analyzed in zero-loss bright-field mode in an energy-filtered transmission electron microscope (Zeiss CEM 902; Zeiss, Oberkochen, Germany). Isolated polyhedral bodies were fixed in 1% (vol/vol) glutaraldehyde, and after adsorption to Formvar-carbon-coated grids they were negatively stained with 2% (wt/vol) uranylacetate, pH 4.5. Samples were analyzed with an energy-filtered transmission electron microscope, and images were recorded, in general, with a charge-coupled-device camera (Proscan Electronic Systems, Scheuring, Germany).
Purification of diol dehydratase.
The purification procedure for diol dehydratase was carried out as described previously (38a, 39a). L. reuteri cells harvested by centrifugation at 3,000 × g for 10 min were washed twice in K2HPO4 buffer I (10 mM, pH 7.2, 1 mM dithiothreitol and 1 mM phenylmethylsulfonyl fluoride) and then washed in 10 ml of degassed K2HPO4 buffer II (10 mM, pH 7.2 containing 5 mM dithiothreitol). Cell lysis was performed using SoniPrep 150 (MSE UK Ltd) fitted with a 9-mm-diameter disrupter horn and an output of 12 μm. One milligram of DNase I was added to the lysed cells, and the cell debris was removed by centrifugation at two different reactive centrifugal forces (3,000 × g for 10 min and 15,500 × g for 20 min).
The extract was homogenized with 1 volume of ammonium sulfate solution at 456 g/liter to obtain 40% saturation. The homogenate was incubated on ice for 1 h and centrifuged at 15,500 g for 20 min. The pellet containing the enzyme was resuspended in 1 ml of K2HPO4 buffer II, and the active fraction was purified by gel exclusion chromatography. The enzyme preparation was loaded onto a Sephacryl S300H (Sigma) column (30 by 1.5 cm) equilibrated with K2HPO4 buffer II. Chromatography was conducted at a flow rate of 0.35 ml/min. Fractions possessing the highest dehydratase activity were pooled and stored at −70°C until further use.
Diol dehydratase assay.
The activity of diol dehydratase was measured by the 3-methyl-2-benzothiazolinone hydrazone method (53). One unit of diol dehydratase activity is defined as the amount of enzyme that catalyzes the formation of 1 μmol of propionaldehyde per min per mg protein from 0.2 M 1,2-PD (propanediol is used because of rapid inactivation of the enzyme over periods of more than a minute by glycerol [53]). The presence of differential diol dehydratase and glycerol dehydratase activities in organisms grown on different substrates was sought by establishing the ratio of glycerol dehydrating and 1,2-PD-dehydrating activities, measured by duplicate 1-min assays using glycerol and 1,2-PD as substrates [(glycerol/propanediol)1 min], as described by Toraya and Fukui (49).
Acrolein (prop-2-enal) detection.
Acrolein (prop-2-enal) detection as a quantitative assay of reuterin (3-HPA) production was carried out by the method of Smiley and Sobolev (43), as practiced by Rodriguez et al. (32) with modifications: following induction overnight in MRS-MOD broth plus glycerol (20 mM) and/or 1,2-PD (50 mM), cultures were standardized at the same optical density (600 nm) with the addition of MRS-MOD. Supernatants (300 μl) from 1 ml of culture incubated for 1 h in MRS-MOD with glycerol (200 mM) and/or 1,2-PD (50 mM) were mixed with 150 μl of tryptophan solution (3 g/1 in 0.1 mol/l HCl) and 600 μl of 35% HCl. The mixture was heated at 60°C for 5 min. 3-HPA (reuterin) produced by bacterial metabolism was detected by dehydration to acrolein (prop-2-enal), developing a yellow color assayed at 490 nm against an acrolein standard. Bacterium-free culture media were assayed as controls.
Cobalamin production.
Cobalamin production was determined using a bioassay on sonicated cells grown in synthetic vitamin B12 assay broth (Merck, Darmstadt, Germany) at 37°C for 3 days. Bioassay plates were prepared as described previously (31) with two different indicator strains (Salmonella enterica serovar Typhimurium metE cysG, AR3612, and S. enterica serovar Typhimurium cbiB metE, AR2680) (31). AR2680 requires cobinamide or later intermediates for restoration of growth, whereas AR3612 can grow in the presence of the earlier intermediate cobyric acid.
1,2-PD metabolism.
L. reuteri was grown in MRS-MOD medium supplemented with 50 mM 1,2-PD at 37°C under anaerobic conditions for 8 days. Two-milliliter samples were removed at different time points and pelleted. Supernatant was stored at −20°C until the assays were carried out. We used a gas chromatography assay following a published method (3) using a Chrompack CP-Sil 5 CB column, 25 m × 0.25 mm with a 0.4-μm film thickness (stationary phase, 100% dimethylpolysiloxane). 1,2-PD and its metabolites (1-propanol, propionic acid, and propionaldehyde) were measured by using 20 mM 1-butanol as an internal concentration standard. The temperature program was set at 80°C for 2 min followed by a 20°C/min temperature increase to 160°C. The total time for chromatographic separation of each sample was 10 min.
PCR.
The pdu operon from L. reuteri was amplified using primers containing SalI restriction sites (forward primer, 5′-AGATGTCGACTTTCAACGGTGATGAGTGGA-3′, and reverse primer, 5′-AGATGTCGACTTGTGGCCATGATTTAGCAA-3′). Primers were designed with Primer3 (33) based on a region of the genome of L. reuteri DSM 20016T (genome sequence kindly made available by Gerald Tannock) determined by TBLASTX searching to be more than 70% identical to the published Lactobacillus collinoides diol dehydratase pdu operon (39). PCR amplification was carried out with a hot-start enzyme possessing 3′-to-5′ proofreading activity, Platinum HiFi Taq DNA polymerase (Invitrogen), using the following program: initial denaturation at 94°C for 2 min; 30 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 22 min; and a final elongation at 68°C for 25 min. The amplicon was purified with a gel extraction kit (Qiagen) and digested with the restriction enzyme PstI (New England Biolabs).
DNA sequence analysis.
Artemis (34) was used to define open reading frames in a section of the L. reuteri DSM 20016T genome sequence which was identified by BLASTP (2) similarity with cob-pdu operon genes from L. collinoides (39) and GenBank. Similar segments were sought in other Lactobacillus sequences in GenBank. A CRE motif search was carried out using the program DNA-pattern at the regulatory sequences analysis tools website (http://rsat.ulb.ac.be) (54) with the input string WTGNAANCGNWNNCW (25) on the DNA sequence contig from L. reuteri DSM 20016T incorporating the pdu operon. The pocR-pduA intergenic interval in the various identified Lactobacillus spp. was examined with DNA-pattern, MEME (5), and Virtual Footprint (26). Promoter prediction was performed with BPROM (software available from SoftBerry (Mount Kisco, NY).
Primer extension analysis.
Total RNA was isolated using ToTALLY RNA (Ambion) from L. reuteri grown in MRS-MOD medium supplemented with either 50 mM 1,2-PD or 50 mM 1,2-PD plus 100 mM glucose. Primer extension reactions were carried out as described by Ventura et al. (55), with some modifications. Briefly, around 15 to 20 μg of the RNA from the above-described step was mixed with 1 pmol of primer (5′CAGCTTTTACCATTGCATCAGCAGC-3′) labeled with IRD800 (MWG Biotech) and 2 μl of buffer H (2 M NaCl, 50 mM PIPES [pH 6.4]). The mixture was denatured at 80°C for 5 min followed by incubation for 60 min at 45°C. After addition of 18 μl of 5× first standard buffer (supplied with Superscript III reverse transcriptase [Invitrogen]; 10 μl of 0.1 M dithiothreitol, 20 μl of a deoxynucleoside triphosphate mix [2.5 mM each], 1 μl of 200 U/μl Superscript III reverse transcriptase, and 41 μl of double-distilled water), the mixture was incubated at 45°C for 2 h. The product was then precipitated with 250 μl of ethanol/acetone (1:1), and the pellet was washed with 80% ice-cold ethanol and dissolved in 4 μl of distilled water. The cDNA was separated on an 8% polyacrylamide-urea gel along with the mixture from a sequencing reaction (Thermo Sequence fluorescently labeled primer cycle sequencing kit; Amersham) conducted with the same primer that was used for the primer extension reaction and detected with a LiCor sequencer machine.
Nucleotide sequence accession number.
The DNA sequence shown in Fig. 3 has been deposited in GenBank with accession no. EU167935.
FIG. 3.
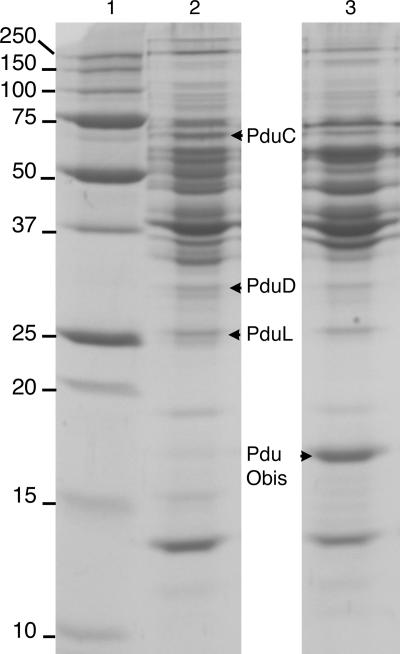
SDS-PAGE separation of L. reuteri total cell protein fractions and MALDI-TOF-identified proteins. Lane 1, protein molecular weight marker. Lane 2, diol dehydratase-positive fraction 1. Lane 3, diol dehydratase-positive fraction 2 (immediately below fraction 1 in sucrose density gradient). L. reuteri was grown in MRS broth supplemented with 15 mM glucose and 50 mM 1,2-PD at 37°C for 36 h.
RESULTS
Electron microscopy.
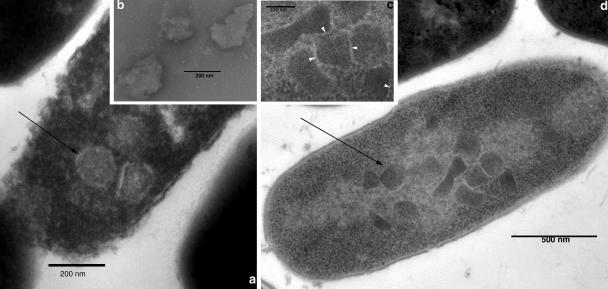
Ultrathin sections of L. reuteri 20016 grown in the presence of 1,2-PD alone or with initial glucose in addition to 1,2-PD showed the presence of polygonal intracellular bodies approximately 150 nm in diameter resembling the metabolosomes described for gram-negative organisms and L. collinoides (38) (Fig. 2). From Fig. 2a and c it is clear that the metabolosome is covered by a single-layer shell. L. reuteri 20016 grown in the absence of 1,2-PD did not show metabolosomes inside the cells (data not shown). Metabolosome extracts showed particles of similar size with evidence of surface layer disruption (Fig. 2b).
FIG. 2.
L. reuteri 20016 produces metabolosomes. (a) Cell section electron micrograph after growth in MRS broth supplemented with 65 mM 1,2-PD and 15 mM glucose at 37°C for 18 h. (b) Extracted metabolosomes from cells grown as for panel a. (c) Enlarged view of metabolosomes shown in panel d. Arrowheads indicate a single-layer shell. (d) L. reuteri grown on MRS broth supplemented with 65 mM 1,2-PD at 37°C for 18 h. Arrows indicate metabolosomes.
Diol dehydratase activity.
L. reuteri 20016 showed maximal diol dehydratase activity when incubated in medium containing 1,2-PD plus glucose (Table 1). It showed minimal diol dehydratase activity when incubated on medium containing glucose only or glucose plus glycerol. L. reuteri 100-23 showed minimal levels of diol dehydratase activity on incubation with glucose-, glycerol-, or 1,2-PD-containing medium. There was no evidence of induction of a distinct glycerol dehydratase with more affinity for glycerol than 1,2-PD by incubation with glycerol in either organism. In K. pneumoniae expressing both glycerol dehydratase and diol dehydratase, the (glycerol/propanediol)1 min dehydratase activity is 2.6 in organisms preincubated with glycerol and 0.7 in those incubated with propanediol (49).
TABLE 1.
L. reuteri diol dehydratase activity after 36 h incubation with different substrates
| Growth substratea | Diol dehydratase activity (U/mg) (measured with 1,2 propanediol substrate)b
|
(Glycerol/ propanediol)1 min dehydratase activityc
|
||
|---|---|---|---|---|
| L. reuteri 20016 | L. reuteri 100-23 | L. reuteri 20016 | L. reuteri 100-23 | |
| Glucose (15 mM) | 0.04 | 0.04 | 0.60 | 0.91 |
| Glucose (15 mM) + 1,2-PD (50 mM) | 0.55 | 0.04 | 0.91 | 0.91 |
| Glucose (15 mM) + glycerol (50 mM) | 0.07 | 0.04 | 0.89 | 0.96 |
Carbon source added to MRS-MOD (L. reuteri 100-23 requires glucose in addition to 1,2-PD to grow in MRS-MOD).
One unit of diol dehydratase activity is defined as the amount of enzyme activity catalyzing the formation of 1 μmol propionaldehyde.
Ratio of dehydrating assay activity detected when glycerol is the assay substrate to that with l,2-propanediol as the substrate, measured by 1-min assays.
SDS-PAGE protein analysis and peptide fingerprinting.
Four predicted proteins from the L. reuteri 275 pdu operon were identified by MALDI-TOF fingerprinting in the diol dehydratase-positive fractions of whole-cell lysate of L. reuteri DSM 20016 grown on MRS medium with glucose and 1,2-PD (Fig. 3; Table 2).
TABLE 2.
Peptide mass fingerprinting of metabolosome components
| Predicted mol wt | Identity assigned in L. reuteri F275 genome (locus tag) | NCBI accession no. | Mascot search resulta
|
||
|---|---|---|---|---|---|
| No. of peptides matched | Mowse score | % Sequence coverage | |||
| 62,566 | Glycerol dehydratase, large subunit PduC (Lreu 1747) | gi 148544953 | 19 | 197 (1.2e-13) | 49 |
| 25,849 | Propanediol dehydratase, medium subunit PduD (Lreu 1746) | gi 148544952 | 8 | 131 (5e-07) | 55 |
| 23,947 | Propanediol utilization protein PduL (Lreu 1740) | gi 148544946 | 11 | 99 (0.00079) | 67 |
| 17,007 | Protein of unknown function DUF336/PduObis (Lreu 1736) | gi 148544942 | 5 | 85 (0.02) | 63 |
Mascot search was used to compare the MALDI-TOF MS data obtained for sample proteins to predicted spectra for proteins present in the National Center for Biotechnology Information (NCBI) database. The Mowse score is the probability that the observed match is a random event. Protein scores greater than 80 are significant (P < 0.05).
Cobalamin and 3-HPA production.
Growth of both Salmonella indicator strains (AR3612 and AR2680) was promoted by cell extracts of L. reuteri DSM 20016, indicating the production of cobinamide or a later intermediate on the route to cobalamin. 3-HPA (reuterin) production from glycerol by L. reuteri DSM 20016 was detected by dehydration to the pigmented aldehyde acrolein (prop-2-enal). Maximal production was associated with overnight induction with both glycerol and 1,2-PD prior to the assay (Table 3). Addition of 1,2-PD to the glycerol substrate for the assay had no inhibitory effect on reuterin production but rather increased it sixfold. L. reuteri 100-23 produced either no detectable reuterin or very small amounts of reuterin at the limits of detection of the assay under all conditions tested.
TABLE 3.
3-HPA (reuterin) production from glycerol and/or propanediol in 1 h by L. reuteri strains induced with glycerol or with glycerol and 1,2-PDa
| MRS-MOD supplement | 3-HPA assay substrate | 3-HPA concn (mM)b
|
|
|---|---|---|---|
| L. reuteri 20016 | L. reuteri 100-23 | ||
| Glycerol | Glycerol | 0.08 | 0.00 |
| Glycerol + 1,2-PD | 0.08 | 0.00 | |
| 1,2-PD | 0.00 | 0.03 | |
| Glycerol + 1,2-PD | Glycerol | 0.46 | 0.03 |
| Glycerol + 1,2-PD | 0.52 | 0.03 | |
| 1,2-PD | 0.08 | 0.03 | |
Overnight induction conditions: 1,2-PD in all cases at 50 mM; glycerol induction, 20 mM; 200 mM glycerol for 3-HPA production assay conditions.
Measured by dehydration to acrolein prop-2-enal.
Growth characteristics and 1,2-PD metabolism.
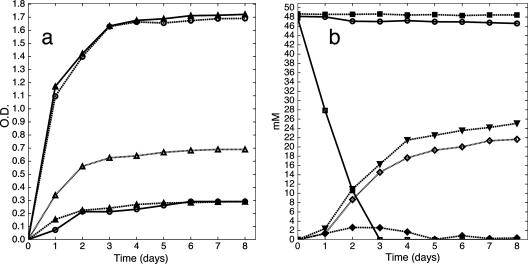
L. reuteri 20016 grew faster to a higher optical density at 600 nm in MRS-MOD medium with the addition of 1,2-PD than in the basal MRS-MOD medium (Fig. 4a) but not as rapidly as when glucose was added. In contrast, L. reuteri 100-23 obtained no growth advantage when 1,2-PD was added to the basal medium but showed growth similar to that of L. reuteri 20016 in glucose-containing medium (Fig. 4a). Approximately equimolar concentrations of 1-propanol (53% of time zero 1,2-PD molar concentration) and propionic acid (45% of time zero 1,2-PD molar concentration) were produced by L. reuteri 20016 (Fig. 4b) from MRS-MOD medium with 1,2-PD, suggesting that a disproportionation reaction was taking place. No decline in propionate concentration was observed in culture supernatant over 8 days, showing that propionate excreted into the culture supernatant was not being taken up and further metabolized. No change in propanediol concentration was observed in a bacterium-free MRS-MOD propanediol medium, and only a 3% decrease in propanediol concentration was seen with incubation of L. reuteri 100-23 in this medium over 8 days (Fig. 4b), showing minimal metabolism. Small amounts of propionaldehyde (an intermediate in the disproportion reaction), a maximum of 2.65 mM, were detected in culture supernatant of L. reuteri 20016 only (Fig. 4b).
FIG. 4.
Growth characteristics of L. reuteri strains and anaerobic propanediol metabolism. (a) Growth curves of L. reuteri 20016 and L. reuteri 100-23 in MRS-MOD at the indicated times postinoculation. O.D., optical density at 600 nm. ▴—▴, L. reuteri 20016 with 50 mM glucose. ▵. . . . .▵, L. reuteri 20016 with 50 mM 1,2-PD. ▴. . . . .▴, L. reuteri 20016, unsupplemented. ○. . . . .○, L. reuteri 100-23 with 50 mM glucose. ○—○, L. reuteri 100-23 with 50 mM 1,2-PD. (b) Propanediol metabolism by L. reuteri 20016 or L. reuteri 100-23 in MRS-MOD with 1,2-PD at the indicated times postinoculation. Metabolite concentrations are shown. ▪. . . . .▪, propanediol concentration in bacterium-free control. ○, L. reuteri 100-23, propanediol concentration. □—□, L. reuteri 20016, propanediol concentration. ▾, L. reuteri 20016, propanol concentration. ◊, L. reuteri 20016, propionate concentration. ⧫, L. reuteri 20016, propionaldehyde concentration.
DNA sequence analysis.
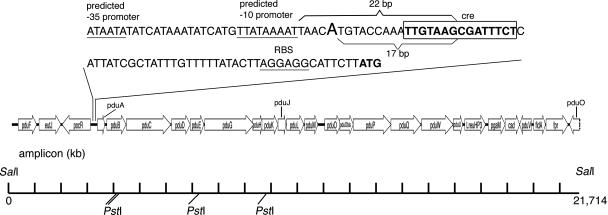
Genes resembling S. enterica serovar Typhimurium linked cob-pdu operons were found in L. reuteri JCM 1112/DSM 20016 (Refseq NZ AAOV00000000) (Fig. 5), L. brevis ATCC 367 (Refseq CP000416) (24), and L. hilgardii (locus AY061969). L. reuteri 100-23 did not contain genes resembling the S. enterica serovar Typhimurium cob-pdu operons. A CRE sequence was detected in the pocR-pduA intergenic interval of L. reuteri DSM 20016 (TTGTAAGCGATTTCT) and L. collinoides (TTGAAAGCGTTTACT). MEME detected the consensus motif GAAAGCGTTT when applied to the data set of the pduA-pocR intergenic sequences of L. reuteri, L. brevis, L. collinoides, and L. hilgardii. This corresponds to part of the CRE consensus sequence. The transcription start site of the L. reuteri pduA gene on induction by 1,2-PD was immediately upstream of the identified CRE sequence (Fig. 5, 6).
FIG. 5.
pdu operon of L. reuteri. Predicted open reading frame gene assignment by comparison with S. enterica serovar Typhimurium (nomenclature of labeled cobalamin synthesis genes follows Salmonella convention). Gene cluster generated with 3BASE sequence file viewer (http://3base.bham.ac.uk/cgi-bin/fileprepare.cgi). The consensus CRE sequence is boxed, predicted −35 and −10 promoter sequences and the ribosomal binding site are underlined, and the start codon of pduA is in bold. The transcriptional start site when induced by propanediol is in larger type. The extent of PCR products and predicted restriction sites are shown below the operon.
FIG. 6.
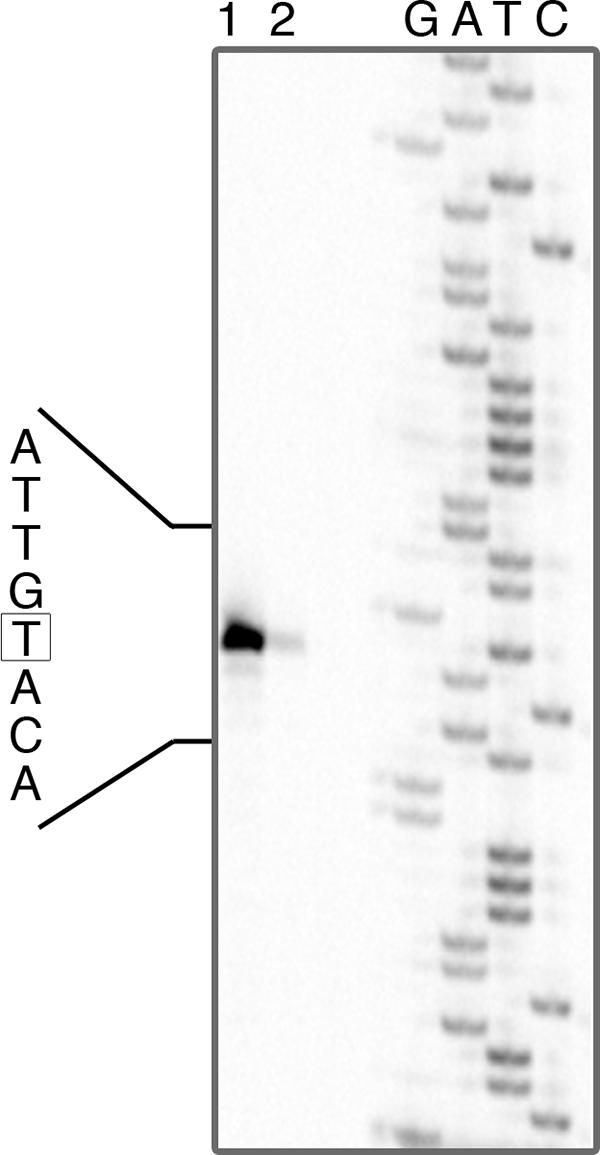
pduA gene transcription start site on propanediol induction. Primer extension products were obtained by using total RNA extracted from L. reuteri grown on MRS-MOD medium supplemented with 50 mM 1,2-PD (lane 1) and 50 mM 1,2-PD plus 100 mM glucose (lane 2). The start point of transcription is boxed.
PCR.
Using genome sequence data from L. reuteri DSM 20016 (also referred to as L. reuteri F275), the putative L. reuteri pdu operon was amplified by PCR from L. reuteri DSM 20016 (NCDO 2589), resulting in an amplicon compatible with the predicted size of 21,714 bp (Fig. 5). In silico digestion of the L. reuteri DSM 20016 pdu locus with the restriction enzyme PstI indicated four restriction sites, which were confirmed by digesting the L. reuteri amplicon with PstI to obtain the predicted size and number of DNA fragments.
DISCUSSION
We present the first demonstration that the antimicrobial agent-producing organism L. reuteri has the capacity to synthesize a bacterial microcompartment (carboxysome or metabolosome). The organism produced a cobalamin-dependent diol dehydratase induced by 1,2-PD, as in gram-negative bacteria containing the pdu operon. Linked cobalamin synthesis and propanediol utilization operons were present in the L. reuteri DSM 20016 genome sequence, and the entire pdu (propanediol utilization) operon was amplified from a laboratory strain of L. reuteri DSM 20016 by PCR, confirming its presence in the propanediol-metabolizing organism. Dismutation of 1,2-PD has been reported from another Lactobacillus sp., Lactobacillus diolivorans, a Lactobacillus buchneri-like organism from maize silage (19). However, no assay of cobalamin production was reported, and metabolosomes were not seen on electron microscopy of L. diolivorans growing on medium incorporating 1,2-PD.
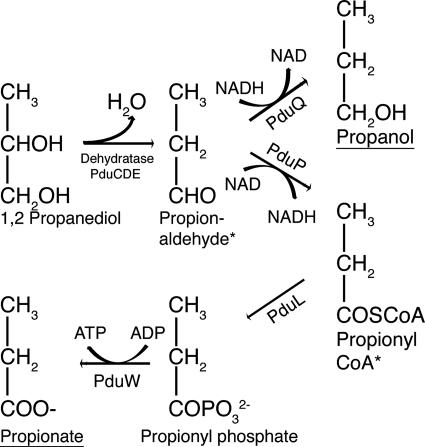
The conversion of 1,2-PD to propanol and propionate with the transient presence of propionaldehyde we have observed (Fig. 4b) suggests a pathway as described for 1,2-PD utilization in Salmonella (8, 22, 36) (Fig. 7). Genes specifying all the enzymes required (Fig. 7) were present in the L. reuteri F275 (DSM 20016) pdu operon (Fig. 5).
FIG. 7.
Proposed pathway of cobalamin-dependent 1,2-PD metabolism in L. reuteri. Metabolic endpoints are underlined; asterisks indicate metabolic intermediates retained within the metabolosome.
However, in Enterobacteriaceae like salmonellae capable of 1,2-PD utilization via a metabolosome-associated diol dehydratase, there are significant further onward metabolic connections for the dismutation products which are not present in lactobacilli. In Salmonella, the propionate product of 1,2-PD utilization can be coupled via the methylcitrate cycle to aerobic respiration (15, 28) or tetrathionate reduction (30), allowing growth on 1,2-PD as a sole carbon and energy source. In the absence of oxygen or tetrathionate, Salmonella spp. can grow only on defined no-carbon media containing added 1,2-PD to which yeast extract has also been added (30). It is proposed that this represents fermentative growth using a carbon source in the yeast extract with energy from propanediol dismutation (30). The pathways by which Salmonella spp. utilize propionate have not been observed in Lactobacillus spp., and no evidence for them is apparent from the L. reuteri genome sequence. We observed a steady increase in propionate levels in culture supernatant from L. reuteri 20016 grown in MRS medium with 1,2-PD over 8 days of continuous culture, suggesting that propionate is excreted and cannot be utilized by the organism. Lower growth rates were seen in MRS-MOD medium when 1,2-PD alone was added to the basal medium than when glucose was also added, but there was an advantage compared with the basal MRS-MOD medium (Fig. 4). It is likely that, as in nonrespiring Salmonella spp., L. reuteri growth on MRS-MOD medium with 1,2-PD is a result of fermentation of other carbon sources such as yeast extract in the complex MRS medium combined with energy from 1,2-PD dismutation (Fig. 7). The control organism L. reuteri 100-23 (in whose genome sequence no pdu operon is apparent) gained no growth advantage from the addition of 1,2-PD to the basal medium and was able to metabolize only a small amount of 1,2-PD over a period of 8 days (Fig. 4).
The enzyme specified by the pdu operon pduCDE genes, diol dehydratase, is responsible for conversion of 1,2-PD to the intermediate propionaldehyde. Interestingly, the enzyme responsible for glycerol conversion to 3-HPA in L. reuteri (Fig. 1) was previously described as a glycerol dehydratase (47) but is capable of acting as a propanediol dehydratase (47). The presence of two isofunctional related enzymes (glycerol and propanediol dehydratase) in L. reuteri, as in K. pneumoniae, was inferred from the existence of two peaks of propanediol dehydratase activity on cell extracts separated by DEAE-cellulose chromatography (47). This left the possibility that reuterin production could be dependent on either one of two isofunctional enzymes. The L. reuteri F275 (DSM 20016) genome sequence has recently been circularized (http://genome.jgi-psf.org/finished_microbes/lacre/lacre.info.html), and BLAST searching does not reveal a distinct glycerol dehydratase in addition to the diol dehydratase linked with cobalamin synthesis. That is, the only candidate enzyme identifiable from the genome sequence for production of 3-HPA from glycerol forming the antimicrobial reuterin (56) is the metabolosome-associated propanediol-induced diol dehydratase we describe. Supporting this, we found no phenotypic evidence of a distinct glycerol-induced dehydratase in L. reuteri 20016 (Table 1), and maximal reuterin production by L. reuteri 20016 was associated with preincubation with 1,2-PD in addition to glycerol (Table 3). Very small amounts of reuterin were produced in the absence of 1,2-PD under preincubation or assay conditions (Table 3). L. reuteri 100-23, lacking the metabolosome-associated diol dehydratase in its unpublished genome sequence, was unable to synthesize more than trace amounts of reuterin (at most, less than 6% of that detected from L. reuteri 20016) (Table 3) and had very low levels of diol dehydratase activity, irrespective of substrate induction (Table 1).
While appearances of individual metabolosomes were consistent with electron microscopy reports on Salmonella (8, 14, 41), fewer metabolosomes were observed in each bacterial cell, and metabolosomes were agglomerated (Fig. 2a, c, and d). Similar electron microscopy appearances have been reported from L. collinoides (38), which also expresses a metabolosome-associated diol dehydratase (39) but does not synthesize cobalamin. Biochemical data supported these qualitative electron microscopy appearances, showing a reduced specific enzyme activity compared with gram-negative organisms: maximal diol dehydratase activity per mg of whole cell extract was comparable with that reported for L. collinoides (39) and approximately a quarter of that reported for Salmonella (14).
Although the pdu operon is substantially similar in gene number and order in Salmonella and L. reuteri, DNA sequence analysis upstream of the pdu operon suggests that it may be regulated differently (Fig. 4). The linked cob and pdu metabolosome operons in a gram-negative background are regulated by Crp and Arc (1). In Lactobacillus spp., as for other gram-positive organisms (35), catabolite repression generally occurs via HPr [HPr(Ser-P)], the small phosphocarrier protein of the phosphoenolpyruvate-sugar phosphotransferase system, and CcpA protein (6, 17), operating via short CREs in the DNA sequence (4, 17, 25). Although 1,2-PD utilization operons have been described for other Lactobacillus species, CREs have not previously been noted in connection with them. We identified a CRE consensus sequence in the L. reuteri pdu operon upstream of pduA, the first gene in the pdu operon. We found complete or partial CRE sequences upstream of pduA in all other available DNA sequences from Lactobacillus spp. containing this operon. In L. reuteri, the center of the CRE is 17 bp downstream of the transcription start site of the initial gene in the pdu operon when induced by 1,2-PD and +22 bp relative to the end of the putative −10 sequence (Fig. 5). In Lactococcus lactis, a CRE in this orientation is associated with strong CcpA-dependent repression (59).
The requirement for a complex 22-gene 1,2-PD utilization operon for this apparently simple process has been attributed to the need to contain the intermediate compound propionaldehyde within a protein compartment or metabolosome, either to reduce toxicity (36) or to prevent its loss as a gas by the cell (29). As reported for metabolosome-containing S. enterica metabolizing 1,2-PD (36), we detected only small amounts of propionaldehyde in culture supernatants of 1,2-PD-metabolizing L. reuteri (Fig. 3b), suggesting retention within the metabolosome. It has been suggested that in the metabolosome associated with the ethanolamine utilization operon in S. enterica, the mechanism of aldehyde retention is based on reduced loss of the aldehyde intermediate (in this case acetaldehyde) by evaporation, possibly by creating a low pH within the compartment, rendering aldehydes more likely to convert to a less volatile acetal (29). However, with regard to the S. enterica 1,2-PD utilization metabolosome, assays of pduA deletion mutants not producing the metabolosome shell but retaining metabolic activity showed that increased propionaldehyde evaporation was not a major factor affecting 1,2-PD metabolism (36). If, as we suggest, the metabolosome-associated diol dehydratase is also responsible for reuterin (3-HPA) production from glycerol, then the fact that this aldehyde is excreted by the organism suggests that either 3-HPA is not produced within the aldehyde-retaining metabolosome (i.e., a significant amount of diol dehydratase is outside the metabolosome in the cytoplasm, unlike the situation in Salmonella [8]) or the NAD-dependent oxidoreductase which removes 3-HPA in L. reuteri by conversion to 1,3-propanediol (Fig. 1) might not be localized in the metabolosome in the same way that PduP coenzyme A-acylating propionaldehyde dehydrogenase is present within the 1,2-PD-metabolizing metabolosome (22) (Fig. 7). That is, effective aldehyde retention by the metabolosome requires the presence of specific aldehyde-metabolizing enzymes within the metabolosome.
Carboxysomes in cyanobacteria affect internal cytoplasmic pH (7) and concentrate protons. There is recent evidence for regulation of the pdu operon by external pH in L. reuteri. During revision of the manuscript it was reported that gene transcription assays using a DNA microarray based on partial genome sequence data from L. reuteri ATCC 55730 showed that 11 genes from the pdu operon were downregulated by dilution and incubation at pH 5.1 versus pH 2.7 (57). Lactobacilli, including L. reuteri, are heterotrophic fermentative organisms that obtain energy by substrate-level phosphorylation and require high levels of different nutrients to maintain a sufficient proton motive force for viability (18). While neutrophilic bacteria like Escherichia coli respond to changes in external pH (pHe) by maintaining a relatively constant internal pH (pHi) at the expense of a large proton gradient across the cell wall, fermentative lactic acid bacteria decrease pHi in response to decreasing pHe, to maintain a constant transmembrane proton gradient (9, 42). Proton concentration within Lactobacillus metabolosomes could potentially raise the pHi of the remaining cytoplasm, compromising efforts to maintain a constant transmembrane proton gradient in acidified growth media.
However, we have shown that the metabolosome-associated propanediol utilization operon is 1,2-PD induced as in gram-negative organisms and functions in a Lactobacillus intracellular background, despite differences in pH homeostasis from the organisms in which it has been mainly studied to date. This finding reinforces the evidence (21) that this very large and complex metabolic operon is nevertheless frequently horizontally transmitted between different bacteria. Further study of the constraints of operating in a fermentative background will shed new light on the electrochemical properties of the metabolosome.
Acknowledgments
This publication emanated from research conducted with the financial support of Science Foundation Ireland to M.B.P. (Research Frontiers Programme 05-RF-GEN053).
Nicolas Sauvageot kindly gave advice on glycerol and propanediol metabolism in L. collinoides and access to thesis data. Dan Walsh and David Cocker helped with propanol and propionate assays. Paddy O'Reilly gave help and advice on Lactobacillus culture media. We thank Mary O'Connell Motherway and Eileen Dilane for assistance with the primer extension protocol. The skillful work of Ingeborg Kristen (HZI, Braunschweig) in electron microscopic sample preparation is gratefully acknowledged The sequence data from L. reuteri 20016T and L. reuteri 100-23 were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) and were generously made available by Gerald Tannock and Jens Walter.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Ailion, M., T. A. Bobik, and J. R. Roth. 1993. Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J. Bacteriol. 1757200-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. C., M. A. Rasmussen, and M. J. Allison. 1993. Metabolism of the plant toxins nitropropionic acid and nitropropanol by ruminal microorganisms. Appl. Environ. Microbiol. 593056-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson, U., D. Molenaar, P. Radstrom, and W. M. de Vos. 2005. Unity in organisation and regulation of catabolic operons in Lactobacillus plantarum, Lactococcus lactis and Listeria monocytogenes. Syst. Appl. Microbiol. 28187-195. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 228-36. [PubMed] [Google Scholar]
- 6.Barrangou, R., M. A. Azcarate-Peril, T. Duong, S. B. Conners, R. M. Kelly, and T. R. Klaenhammer. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. USA 1033816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belkin, S., R. J. Mehlhorn, and L. Packer. 1987. Proton gradients in intact cyanobacteria. Plant Physiol. 8425-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 1815967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booth, I. R. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinsmade, S. R., T. Paldon, and J. C. Escalante-Semerena. 2005. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 1878039-8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannon, G. C., C. E. Bradburne, H. C. Aldrich, S. H. Baker, S. Heinhorst, and J. M. Shively. 2001. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl. Environ Microbiol. 675351-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casas, I., and W. Dobrogosz. 2000. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb. Ecol. Health Dis. 12247-285. [Google Scholar]
- 13.Daniel, R., T. A. Bobik, and G. Gottschalk. 1998. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol. Rev. 22553-566. [DOI] [PubMed] [Google Scholar]
- 13a.de Man, J., M. Ragosa, and M. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23130-135. [Google Scholar]
- 14.Havemann, G. D., and T. A. Bobik. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 1855086-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horswill, A. R., and J. C. Escalante-Semerena. 1999. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J. Bacteriol. 1815615-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerfeld, C. A., M. R. Sawaya, S. Tanaka, C. V. Nguyen, M. Phillips, M. Beeby, and T. O. Yeates. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309936-938. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. H., Y. K. Yang, and G. H. Chambliss. 2005. Evidence that Bacillus catabolite control protein CcpA interacts with RNA polymerase to inhibit transcription. Mol. Microbiol. 56155-162. [DOI] [PubMed] [Google Scholar]
- 18.Konings, W. N. 2002. The cell membrane and the struggle for life of lactic acid bacteria. Antonie Leeuwenhoek 823-27. [PubMed] [Google Scholar]
- 19.Krooneman, J., F. Faber, A. C. Alderkamp, S. Oude Elferink, F. Driehuis, I. Cleenwerck, J. Swings, J. C. Gottschal, and M. Vancanneyt. 2002. Lactobacillus diolivorans sp. nov., a 1,2-propanediol-degrading bacterium isolated from aerobically stable maize silage. Int. J. Syst. Evol. Microbiol. 52639-646. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, J. G., and J. R. Roth. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 14211-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leal, N. A., G. D. Havemann, and T. A. Bobik. 2003. PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch. Microbiol. 180353-361. [DOI] [PubMed] [Google Scholar]
- 23.Luthi-Peng, Q., F. B. Dileme, and Z. Puhan. 2002. Effect of glucose on glycerol bioconversion by Lactobacillus reuteri. Appl. Microbiol. Biotechnol. 59289-296. [DOI] [PubMed] [Google Scholar]
- 24.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 10315611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miwa, Y., A. Nakata, A. Ogiwara, M. Yamamoto, and Y. Fujita. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 281206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munch, R., K. Hiller, A. Grote, M. Scheer, J. Klein, M. Schobert, and D. Jahn. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 214187-4189. [DOI] [PubMed] [Google Scholar]
- 27.Obradors, N., J. Badia, L. Baldoma, and J. Aguilar. 1988. Anaerobic metabolism of the l-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J. Bacteriol. 1702159-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palacios, S., V. J. Starai, and J. C. Escalante-Semerena. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J. Bacteriol. 1852802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penrod, J. T., and J. R. Roth. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 1882865-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 1832463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raux, E., A. Lanois, F. Levillayer, M. J. Warren, E. Brody, A. Rambach, and C. Thermes. 1996. Salmonella typhimurium cobalamin (vitamin B12) biosynthetic genes: functional studies in S. typhimurium and Escherichia coli. J. Bacteriol. 178753-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez, E., J. L. Arques, R. Rodriguez, M. Nunez, and M. Medina. 2003. Reuterin production by lactobacilli isolated from pig faeces and evaluation of probiotic traits. Lett. Appl. Microbiol. 37259-263. [DOI] [PubMed] [Google Scholar]
- 33.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132365-386. [DOI] [PubMed] [Google Scholar]
- 34.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16944-945. [DOI] [PubMed] [Google Scholar]
- 35.Saier, M. H., Jr., S. Chauvaux, J. Deutscher, J. Reizer, and J. J. Ye. 1995. Protein phosphorylation and regulation of carbon metabolism in gram-negative versus gram-positive bacteria. Trends Biochem. Sci. 20267-271. [DOI] [PubMed] [Google Scholar]
- 36.Sampson, E. M., and T. A. Bobik. 2008. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J. Bacteriol. 1902966-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos, F., J. L. Vera, R. van der Heijden, G. Valdez, W. M. de Vos, F. Sesma, and J. Hugenholtz. 2008. The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098. Microbiology 15481-93. [DOI] [PubMed] [Google Scholar]
- 38.Sauvageot, N. 2002. Ph.D. thesis. Etude du métabolisme du glycerol chez Lactobacillus collinoides: aspects physiologiques, génétiques et biochimiques Université de Caen, Caen, France.
- 38a.Sauvageot, N., V. Pichereau, L. Louarme, A. Hartbe, Y. Auffray, and J.-M. Laplace. 2002. Purification, characterization and subunits identification of the diol dehydratase of Lactobacillus collinoides. Eur. J. Biochem. 2695731-5737. [DOI] [PubMed] [Google Scholar]
- 39.Sauvageot, N., C. Muller, A. Hartke, Y. Auffray, and J. M. Laplace. 2002. Characterisation of the diol dehydratase pdu operon of Lactobacillus collinoides. FEMS Microbiol. Lett. 20966-71. [DOI] [PubMed] [Google Scholar]
- 39a.Schutz, H., and F. Radler. 1984. Anaerobic reduction of glycerol to propanediol-1,3 by Lactobacillus brevis and Lactobacillus luchneri. Syst. Appl. Microbiol. 5169-178. [Google Scholar]
- 40.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 41.Shively, J. M., C. E. Bradburne, H. C. Aldrich, T. A. Bobik, J. L. Mehlman, S. Jin, and S. H. Baker. 1998. Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes-polyhedral bodies. Can. J. Bot. Rev. Can. Bot. 76906-916. [Google Scholar]
- 42.Siegumfeldt, H., K. Bjorn Rechinger, and M. Jakobsen. 2000. Dynamic changes of intracellular pH in individual lactic acid bacterium cells in response to a rapid drop in extracellular pH. Appl. Environ. Microbiol. 662330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smiley, K. L., and M. Sobolov. 1962. A cobamide-requiring glycerol dehydrase from an acrolein-forming Lactobacillus. Arch. Biochem. Biophys. 97538-543. [DOI] [PubMed] [Google Scholar]
- 43a.Spurr, A. R. 1969. A low viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 2631-43. [DOI] [PubMed] [Google Scholar]
- 44.Talarico, T. L., L. T. Axelsson, J. Novotny, M. Fiuzat, and W. J. Dobrogosz. 1990. Utilization of glycerol as a hydrogen acceptor by Lactobacillus reuteri: purification of 1,3-propanediol:NAD oxidoreductase. Appl. Environ. Microbiol. 56943-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talarico, T. L., I. A. Casas, T. C. Chung, and W. J. Dobrogosz. 1988. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 321854-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talarico, T. L., and W. J. Dobrogosz. 1989. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 33674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talarico, T. L., and W. J. Dobrogosz. 1990. Purification and characterization of glycerol dehydratase from Lactobacillus reuteri. Appl. Environ. Microbiol. 561195-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taranto, M. P., J. L. Vera, J. Hugenholtz, G. F. De Valdez, and F. Sesma. 2003. Lactobacillus reuteri CRL1098 produces cobalamin. J. Bacteriol. 1855643-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toraya, T., and S. Fukui. 1977. Immunochemical evidence for the difference between coenzyme-B12-dependent diol dehydratase and glycerol dehydratase. Eur. J. Biochem. 76285-289. [DOI] [PubMed] [Google Scholar]
- 50.Toraya, T., S. Honda, and S. Fukui. 1979. Fermentation of 1,2-propanediol with 1,2-ethanediol by some genera of Enterobacteriaceae, involving coenzyme B12-dependent diol dehydratase. J. Bacteriol. 13939-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toraya, T., S. Kuno, and S. Fukui. 1980. Distribution of coenzyme B12-dependent diol dehydratase and glycerol dehydratase in selected genera of Enterobacteriaceae and Propionibacteriaceae. J. Bacteriol. 1411439-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toraya, T., T. Shirakashi, T. Kosuga, and S. Fukui. 1976. Substrate specificity of coenzyme B12-dependent diol dehydrase: glycerol as both a good substrate and a potent inactivator. Biochem. Biophys. Res. Commun. 69475-480. [DOI] [PubMed] [Google Scholar]
- 53.Toraya, T., K. Ushio, S. Fukui, and P. C. Hogenkamp. 1977. Studies on the mechanism of the adenosylcobalamin-dependent diol dehydrase reaction by the use of analogs of the coenzyme. J. Biol. Chem. 252963-970. [PubMed] [Google Scholar]
- 54.van Helden, J. 2003. Regulatory sequence analysis tools. Nucleic Acids Res. 313593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ventura, M., C. Canchaya, R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES Loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 706197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vollenweider, S., and C. Lacroix. 2004. 3-Hydroxypropionaldehyde: applications and perspectives of biotechnological production. Appl. Microbiol. Biotechnol. 6416-27. [DOI] [PubMed] [Google Scholar]
- 57.Wall, T., K. Bath, R. A. Britton, H. Jonsson, J. Versalovic, and S. Roos. 2007. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 733924-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wesney, E., and G. Tannock. 1979. Association of rat, pig and fowl biotypes of lactobacilli with the stomach of gnotobiotic mice. Microb. Ecol. 535-42. [DOI] [PubMed] [Google Scholar]
- 59.Zomer, A. L., G. Buist, R. Larsen, J. Kok, and O. P. Kuipers. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 18916. [DOI] [PMC free article] [PubMed] [Google Scholar]