Abstract
Francisella tularensis is a highly infectious, facultative intracellular bacterial pathogen that is the causative agent of tularemia. Nearly a century ago, researchers observed that tularemia was often fatal in North America but almost never fatal in Europe and Asia. The chromosomes of F. tularensis strains carry two identical copies of the Francisella pathogenicity island (FPI), and the FPIs of North America-specific biotypes contain two genes, anmK and pdpD, that are not found in biotypes that are distributed over the entire Northern Hemisphere. In this work, we studied the contribution of anmK and pdpD to virulence by using F. novicida, which is very closely related to F. tularensis but which carries only one copy of the FPI. We showed that anmK and pdpD are necessary for full virulence but not for intracellular growth. This is in sharp contrast to most other FPI genes that have been studied to date, which are required for intracellular growth. We also showed that PdpD is localized to the outer membrane. Further, overexpression of PdpD affects the cellular distribution of FPI-encoded proteins IglA, IglB, and IglC. Finally, deletions of FPI genes encoding proteins that are homologues of known components of type VI secretion systems abolished the altered distribution of IglC and the outer membrane localization of PdpD.
Francisella tularensis, the causative agent of the zoonotic disease tularemia, is a gram-negative, facultative intracellular bacterial pathogen (10). F. tularensis is remarkable in that it is both highly infectious and capable of infecting a very broad array of animal species. During a short period of time in the early 20th century, F. tularensis was independently found to be the cause of zoonotic diseases in Europe, Asia, and North America. It was also observed that the clinical outcome of tularemia was most severe in North America, where fatalities occurred at a rate 10- to 100-fold higher than the rate found in Europe or Asia. Exchange of F. tularensis strains among Russian, Japanese, and American scientists led to the discovery that two major biotypes existed, a pan-Northern Hemisphere F. tularensis subsp. holarctica (or type B) biotype and a North America-specific biotype, F. tularensis subsp. tularensis (or type A). Both American and Russian scientists used rabbit models of infection to discriminate the highly virulent from less virulent forms (4, 30). Cumulatively, the human clinical disease pattern and the experimental rabbit infection results led to the widely held belief that the F. tularensis type A strains are much more virulent in humans than their type B counterparts. However, a review of the literature shows that there is no solid experimental or clinical basis for this conventional belief. Indeed, the notion has been challenged recently by a retrospective study, conducted by Centers for Disease Control and Prevention (CDC) scientists, of fatal and nonfatal cases of tularemia in the United States (37). Although that study was limited by the strong bias generated by the strains sent to the CDC, it does highlight the possibility that a subset of type A strains (A.II) (17), found mostly in the western part of the United States, are less virulent than the A.I subset, found mostly in the eastern part of the United States.
The recent availability of genomic information for F. tularensis has enabled comparison of type A and type B genomes, as well as those of European and North American type B strains, down to the nucleotide level (3, 20, 34). While there are many genomic rearrangements and single-nucleotide-polymorphism differences among strains, there are very few cases of genes being absent from one biotype and present in another. Of those overt differences, only one operon, anmK-pdpD, is clearly associated with a cluster of known virulence genes, namely, those found in the Francisella pathogenicity island (FPI) (27, 28).
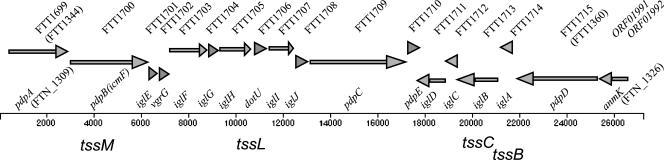
The recently identified FPI is an ∼30-kb genetic element with an average G+C content that differs from the core genome by 2.2% for the anmK-iglD operon and by 6.6% for the larger pdpA-pdpE region (Fig. 1) (27, 28). Aberrant G+C contents are an important signature of DNA introduced into a chromosome by ancient horizontal DNA transfer. All of the type A and type B biovars have two identical copies of the FPI, while the F. novicida biotype contains a single copy of the FPI. In every instance to date, inactivation of FPI genes has led to decreased intramacrophage growth and decreased virulence (8, 13, 14, 35, 40, 43). Although the functions of the proteins encoded by the FPI have not been determined, at least four of the genes in the FPI encode proteins that are homologues of proteins that are part of a type VI secretion system (T6SS) found in other pathogens. The T6SS is thought to mediate the export of virulence effector proteins in a sec-independent manner in a variety of animal (44) and plant (6) pathogens or symbionts, including Vibrio cholerae (33) and Pseudomonas aeruginosa (25). There is substantial evidence that T6SSs are tightly controlled and up-regulated during an infection (8, 26, 36), and these properties may explain why proteins in Francisella that are secreted by its T6SS have not been identified in the past. Recent evidence suggests that the V. cholerae T6SS produces a secreted structure that is predicted to have cell-puncturing properties (32), but the full picture of secretion by T6SS has yet to be elucidated.
FIG. 1.
Diagrammatic representation of the F. novicida form of the FPI with a consensus nomenclature of the FPI genes. The anmK and pdpD genes are the first and second ORFs in an apparent operon that runs from anmK through iglD. This operon has a G+C content of 30.6%, in contrast to the average 33% G+C content for the Francisella chromosome. The pdpA-to-pdpE region, which is the region of the lowest G+C content in the chromosome, has a G+C content of 26.6%. The F. novicida form of the FPI differs from the F. tularensis type B form in having anmK and pdpD. F. tularensis type A biotypes have pdpD but have one of two different-sized anmK forms. The names of the FPI genes are indicated above the line that indicates the base pair numbers starting from the start of pdpA. Below the line are the consensus designations for T6SS genes. For reference, the F. tularensis Schu4 genomic designations for one of the Schu4 FPI copies are included above the arrows that indicate the direction of each gene. For the first and last genes in the FPI, the numbers in parentheses indicate alternative genomic designations for the Schu4 strain (FTT) or the F. novicida species (FTN).
As a gram-negative bacterial pathogen, Francisella is expected to have mechanisms to secrete proteinaceous virulence factors to the surface of the bacterium or into the extracellular milieu. Gil and coworkers (11) found tolC and a tolC-like homologue in Francisella and showed that these genes contribute to resistance to small bactericidal molecules. They also showed that the tolC homologue is required for virulence and suggested that their evidence indicated a role for TolC in a type I secretion system. Bina and colleagues (5) showed that an AcrB RND efflux pump contributes to drug resistance and virulence in the F. tularensis LVS strain. Importantly, Hager et al. (15) showed true secretion of seven proteins in F. novicida that is dependent on genes that are homologous to type IV pilus genes. Surprisingly, inactivation of secretion components or one of the secreted proteins generated strains that had enhanced virulence and enhanced dissemination of F. novicida when introduced via an intradermal injection.
In this work, we examined the roles of anmK and pdpD in Francisella virulence, and in the course of these studies, we discovered an interaction of homologues of T6SS components with PdpD. Thus, here we examine the role in virulence of a protein found in the North America-specific biotype of F. tularensis that is missing in the pan-Northern Hemisphere biotype and provide new knowledge about a poorly understood secretion system.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Detailed descriptions of ΔiglB, ΔpdpB, and ΔdotU strains will be described in later studies, but they were made essentially as described below (see “Mutagenesis and complementation”). F. novicida and F. tularensis LVS were grown using Trypticase soy broth or agar supplemented with 0.1% cysteine (TSB-C or TSA-C). When needed, erythromycin (Em) or kanamycin (Km) was added to a final concentration of 25 μg/ml and 15 μg/ml, respectively. For deletion mutagenesis experiments, filter-sterilized sucrose was added to medium to a final concentration of 10%. Escherichia coli strains were grown using LB broth or agar supplemented with Km (30 μg/ml), Em (100 μg/ml), or ampicillin (100 μg/ml) as needed.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid and/or genotype | Description or relevant characteristic(s)a | Source and/or reference |
|---|---|---|
| Bacterial strains | ||
| F. novicida | ||
| U112 | Wild-type Francisella novicida | 19 |
| JL0 | U112 with a deletion in FTN_1390, which is the site where the integrating pJL-SKX vector inserts; this strain has growth and virulence phenotypes identical to those of U112 | 23 |
| GB2 | U112 with a point mutation in the global virulence regulator, mglA | 2 |
| SC92 | O-antigen mutant of U112 | 7 |
| JL12 | An ermC allelic-exchange mutant of pdpD | 28 |
| ODB2 | JL0 with deletion of iglA | 8 |
| 10b (ΔpdpD) | An in-frame deletion mutant missing codons 513 to 789 of pdpD | This work |
| 20d (ΔpdpD) | An in-frame deletion mutant missing codons 215 to 1138 of pdpD | This work |
| ΔanmK | An in-frame deletion mutant missing codons 18 to 355 of anmK | This work |
| ΔpdpD/pJL-SKX::pdpD | 20d complemented with the integrating pJL-SKX::pdpD construct | This work |
| ΔpdpD/pJL-SKX::anmK-pdpD | 20d complemented with the integrating pJL-SKX::anmK-pdpD construct | This work |
| ΔpdpD/pMP633::KmranmK-pdpD | 20d complemented with the pMP633::KmranmK-pdpD construct | This work |
| ΔanmK/pJL-SKX::anmK | ΔanmK complemented with the integrating pJL-SKX::anmK construct | This work |
| ΔiglAB/pJL-SKX::anmK-pdpD | An ΔiglA mutant complemented with the integrating pJL-SKX::anmK-pdpD construct | This work |
| ΔiglB/pJL-SKX::anmK-pdpD | An ΔiglB mutant complemented with the integrating pJL-SKX::anmK-pdpD construct | This work |
| ΔpdpB/pJL-SKX::anmK-pdpD | A ΔpdpB mutant complemented with the integrating pJL-SKX::anmK-pdpD construct | This work |
| ΔdotU/pJL-SKX::anmK-pdpD | A ΔdotU mutant complemented with the integrating pJL-SKX::anmK-pdpD construct | This work |
| SC92/pJL-SKX::anmK-pdpD | An LPS mutant complemented with the integrating pJL-SKX::anmK-pdpD construct | This work |
| F. tularensis subsp. holarctica | ||
| LVS | Live vaccine strain, type B biotype | American Type Culture Collection (9) |
| LVS(pMP633::KmranmK-pdpD) | The LVS strain complemented with the pMP633::KmranmK-pdpD construct | This work |
| E. coli DH5α | supE44 Δ(lacIZYA-argF)U169 (ϕ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen (16) |
| Plasmids | ||
| pWSK29 | Low-copy-number cloning vector, Apr | 42 |
| pJL-SKX | pWSK29 (with XhoI site removed) with integrating pJL-SKX cassette inserted at the BamHI and KpnI sites, Kmr Apr | 23 |
| pMP633 | Francisella shuttle plasmid, Hygr | 22 |
| pJL-ES-X | An ermC-sacB cassette with flanking XhoI restriction sites | 23 |
| pWSK29::pdpD | A pdpD clone with 2,100 bp of the upstream and downstream flanking regions | This work |
| pWSK29::ΔpdpD10b | A construct containing a 277-codon in-frame pdpD deletion and flanking XhoI restriction sites | This work |
| pWSK29::ΔpdpD20d | A construct containing a 924-codon in-frame pdpD deletion and flanking XhoI restriction sites | This work |
| pJL-SKX::pdpD | An integrating complementation vector carrying the pdpD locus | This work |
| pJL-SKX::anmK | An integrating complementation vector carrying the anmK locus and 397 bp of upstream sequence | This work |
| pJL-SKX::anmK-pdpD | An integrating complementation vector carrying the anmK-pdpD loci and 397 bp upstream of anmK | This work |
| pMP633::KmranmK-pdpD | Francisella shuttle plasmid carrying pJL-SKX::anmK-pdpD, Kmr Hygr; the insert in pMP633 contains the Kmr cassette through the end of pdpD, which was PCR amplified from pJL-SKX::anmK-pdpD and cloned into the EcoRV site of pMP633 | This work |
Ap, ampicillin; Hyg, hygromycin.
Transformation of Francisella.
Genetic constructs were introduced into F. novicida by a previously described chemical transformation protocol (1) with the modification that Chamberlain's broth was replaced with TSB-C supplemented with 0.4% glucose (23).
SDS-PAGE and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to standard techniques (18). To normalize the amount of protein added to each lane, the concentrations of protein samples were determined by use of the bicinchoninic acid assay (Pierce). Separated proteins were transferred onto an Immobilon-FL (Millipore) membrane and blocked with 5% skim milk (Difco) in phosphate-buffered saline (PBS). Rabbit anti-IglA was used at dilutions of 1:4,000, while rabbit anti-PdpD was used at dilutions of 1:1,000. To detect bound antibody, blots were incubated with IRDye800DX-conjugated goat anti-rabbit immunoglobulin G (Rockland, Gilbertsville, PA) and visualized in a LiCor Odyssey imaging system. In some blots, monoclonal anti-IglB, anti-IglC, and anti-PdpB were used, and these were detected using IRDye800-conjugated goat anti-mouse antibody. All of the rabbit antisera and the mouse hybridomas described above have been deposited with the American Type Culture Collection's BEI program. Anti-FopA rabbit serum was kindly supplied by Michael Norgard.
The three different anti-PdpD rabbit antisera used in this study were made by New England Peptide (Gardner, MA) by immunizing New Zealand White rabbits with injections of either a recombinant fragment of PdpD (amino acids 748 to 966) or keyhole limpet hemocyanin-conjugated peptides (amino acids 259 to 272 and 963 to 976). Rabbit anti-VgrG was made by injections with purified recombinant protein.
Fractionation of Francisella.
Approximately 2 × 1011 CFU of two-day-old plate-grown F. novicida were harvested and resuspended in 50 ml of cold PBS supplemented with 35 μl of a bacterial protease inhibitor solution. Agar-plate-grown F. novicida cells were used because they appeared to produce more PdpD than broth-grown cells. Cells were lysed by repeated passage through a French pressure cell (American Instruments Co., Silver Spring, MD) at ∼1,200 lb/in2. Unbroken cells were removed by 20 min of centrifugation at 10,000 × g at 4°C, and a sample was taken as the total protein fraction. The lysate was subjected to ultracentrifugation (Beckman L8-70, rotor type 45 Ti) for 2 h at 100,000 × g at 4°C to pellet the membranes. The supernatant (soluble protein fraction) was removed, while the membrane pellet was resuspended in 2.5 ml of 1% Sarkosyl (Sigma). The Sarkosyl-soluble (inner membrane) and the Sarkosyl-insoluble fractions (outer membrane) were separated by a second ultracentrifugation for 2 h at 100,000 × g at 4°C in a Beckman TLA-100.3 ultramicrocentrifuge. The pelleted membrane fraction was resuspended in SDS-PAGE running buffer, and all samples were separated and blotted using standard techniques.
Biotinylation of Francisella outer membrane proteins.
The biotinylation of potentially surface-exposed proteins was carried out using the EZ-Link sulfo-NHS-LC-LC-biotin (Pierce) labeling agent. Plate-grown F. novicida strains were resuspended in 10 ml of cold PBS, washed three times by pelleting at 10,000 × g, and resuspended in cold PBS. Following the final wash, cells were resuspended in 5 ml of PBS, and a 500-μl aliquot was transferred to a 1.5-ml tube containing 250 μl of a 15-mg/ml solution of sulfo-NHS-LC-LC-biotin. Cells were incubated for 30 min at room temperature, pelleted at 8,000 × g, and washed in 1 ml of biotinylation salt solution (50 mM Tris, 300 mM NaCl, pH 7.5), and two 1-ml washes with cold PBS were performed. Following the final wash, bacteria were resuspended in 50 μl of PBS, lysed by adding 500 μl of B-PERII (Pierce), and centrifuged at 15,200 × g for 1 min. The supernatant was transferred to a new 1.5-ml tube and 200 μl of Ultralink-immobilized NeutrAvidin beads (Pierce) was added. Tubes were incubated with gentle rocking for 30 min at room temperature, followed by five washes in which the mixture was pelleted at 1,000 × g for 1 min and resuspended in 1 ml of Tris-buffered saline (50 mM NaCl, 25 mM Tris, pH 7.5, 0.2% Tween 20). Protein was recovered by resuspending the pelleted NeutrAvidin beads in 40 μl of standard SDS-PAGE sample buffer and boiling them at 95°C for 15 min. The heated mixture was gently pelleted to remove NeutrAvidin beads, and 15 μl of supernatant was separated on a 4 to 10% NuSep gradient gel.
Mutagenesis and complementation of anmK and pdpD.
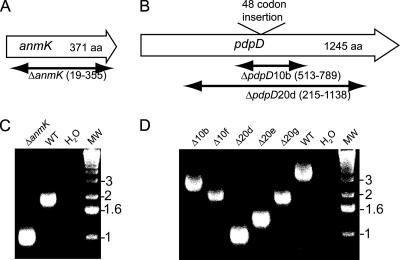
To create deletion mutants, an ∼8-kb clone which encompasses the entire pdpD gene as well as ∼2 kb flanking each side of pdpD was cloned into pWSK29. This recombinant contained a unique PacI site that lay near the middle of pdpD, at position 2021. The clone was linearized by PacI digestion and subjected to exonuclease Bal 31 digestion. Bal 31 reactions were stopped at 5-min intervals for 30 min by heat inactivation of Bal 31, and the resulting digested plasmids were ligated and electroporated into E. coli DH5α. From hundreds of transformants, five clones each from the 10-, 20-, and 30-min Bal 31 reactions were sequenced, and five were found to have in-frame pdpD deletions of various sizes. These deletion constructs were used to create F. novicida deletion mutants as previously described (23). Briefly, the in-frame pdpD deletion mutant clones were digested with XhoI, ligated to an ermC-sacB cassette, and transformed into F. novicida JL0 to form Emr cointegrates. Colonies that resulted from excision of the cointegrate were scored by the loss of a mucoid phenotype in the presence of 10% sucrose and the loss of Emr. Ultimately, these mutagenesis experiments led to the creation of five pdpD mutants (named 10b, 10f, 20d, 20e, and 20g) whose PdpD deletions ranged in size from 462 to 968 amino acid residues, relative to the 1,245 residues observed in the wild-type form.
Complementation of the anmK and pdpD mutations was done by inserting anmK, anmK-pdpD, or pdpD into the integrating vector, pJL-SKX, and introducing linear recombinant DNA into F. novicida JL0. The pJL-SKX vector inserts into the F. novicida chromosome at the FTN_1758 locus, which is located at bp 1887821 in the chromosome (pdpD is at bp 1399803) (34). The FTN_1758 open reading frame (ORF) was deleted in F. novicida JL0. As the FTN_1758 locus is missing from F. tularensis LVS, we were unable to integrate the vector into the LVS chromosome. Instead, we PCR amplified the pJL-SKX::anmK-pdpD recombinant from the promoter that lies in front of the Kmr cassette to the end of pdpD, cloned the resulting amplicon into pMP633 (22), and introduced this recombinant into F. tularensis LVS. The sequence of the primers used for this amplification and all others used in this work will be made available upon request.
Intracellular growth assays.
Bone marrow cells were isolated from femurs of healthy BALB/c male mice and cultivated in 96-well cell culture plates at 4 × 105 cells/well (Costar) for one week in complete Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum, 1% l-glutamine, 1% minimal essential medium nonessential amino acids, 1% HEPES buffer solution, and 10% conditioned L929 supernatant. The resultant bone marrow-derived macrophages (BMDMs) were infected with F. novicida strains at a multiplicity of infection (bacterium to macrophage) of 20:1. Infected monolayers were incubated for 1 h in complete DMEM to allow for phagocytosis to occur, washed five times in Dulbecco's PBS, and incubated at 37°C in 5% CO2. To determine bacterial replication, infected macrophages were lysed in 0.1% deoxycholate at 0, 24, and 48 h postinfection. The lysates were serially diluted in Dulbecco's PBS containing 0.1% gelatin and plated on TSA-C. It has been previously demonstrated that F. tularensis extracellular growth in standard DMEM is not supported, and this fact makes the macrophage infection assay an appropriate determination of intracellular growth. As a negative control, the F. novicida mglA mutant GB2 (2), which does not grow in macrophages, was incorporated into all macrophage growth experiments.
The various cell lines were grown in DMEM essentially as described for BMDMs. The cell lines that were used included the J774A.1 mouse macrophage cell line, the NIH 3T3 mouse embryonic fibroblast cell line, the COS-7 monkey kidney fibroblast cell line, the HeLa human cervical epithelial cell line, the C2C12 mouse muscle fibroblast cell line, the HEK-293 human kidney epithelial cell line, the MDCK dog kidney epithelial cell line, the 4T1 mouse mammary gland epithelial cell line, the CMT-93 mouse rectum epithelial cell line, the C6 rat brain fibroblast cell line, the LLC-PK1 pig kidney epithelial cell line, and the Caco-2 human colon epithelial cell line.
Chicken embryo and mouse infections.
For the in vivo analysis of mutants, F. tularensis strains were grown to the late log phase (optical density at 600 nm, 1.0) and diluted in PBS for injection. The inoculating dose was calculated retrospectively by determining the number of CFU following dilution and plating on TSA-C. Fertilized White Leghorn eggs were obtained from the University of Alberta Poultry Research Station, and chicken embryos were incubated at 37°C with high humidity for seven-days prior to infection. Throughout the experiment the embryos were mechanically tilted to a 45° angle every 40 min. Following the seven-day initial incubation, chicken embryos were injected with various doses of 100 μl of F. novicida diluted in PBS under the chorioallantoic membrane as described previously (29). Chicken embryos were then monitored daily for death for up to 6 days.
For in vivo infections, six- to eight-week-old male specific-pathogen-free BALB/cByJ mice were purchased from the Jackson Laboratory. Animals were housed in sterile microisolator cages in a barrier environment at the Center for Biologics Evaluation and Research. Mice were fed autoclaved food and water ad libitum, and all experiments were performed under Institutional Animal Care and Use Committee guidelines. Mice were given 0.1 ml of appropriately diluted bacteria intradermally at the base of the tail; actual doses of inoculated bacteria were simultaneously determined by plate count. All materials used for animals, including bacteria, were diluted in PBS (BioWhittaker) containing <0.01 ng/ml endotoxin. Graphing and statistical analyses (including standard errors of the means and the P value of an unpaired t test) of experiments were done using GraphPad Prism 4.03 software.
Nucleotide sequence accession numbers.
The sequences of the two mutants described in this work, which represent the smallest and largest deletions, have been deposited in GenBank and have been assigned the accession numbers EU341813 (pdpD from strain 10b) and EU341814 (pdpD from strain 20d). The sequence for the F. tularensis B38 form of the anmK region has been deposited in GenBank and has been assigned the number EU341812.
RESULTS AND DISCUSSION
Variation of the anmK-pdpD region among F. tularensis biotypes.
We previously reported the presence of pdpD in F. novicida and in F. tularensis type A strains and the absence of the pdpD gene in five F. tularensis type B strains (28). The presence of pdpD in type A strains was interesting in that strains of this biotype are considered more virulent than strains of the type B biotype. In our original description of the FPI, we identified the gene upstream of pdpD, anmK, as a gene encoding a putative molecular chaperone (the encoding gene originally called pmcA). Recently, the conserved orthologous group associated with anmK (pmcA), COG2377, has been reannotated, and proteins encoded by members of this group are now recognized as being part of the anhydro-N-acetylmuramic acid kinase family. This enzyme is responsible for the utilization of exogenous or recycled 1,6-anhydro-N-acetylmuramic acid, which is a component of cell wall peptidoglycan. The E. coli anmK homologue (ydhH or b1640) is known not to be essential for viability, and since anmK is missing from many strains of F. tularensis, it is clear that it is not essential for Francisella as well.
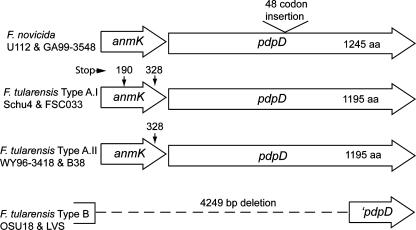
The release of several F. tularensis genomes and our analysis of the anmK region of the type A strain B38 revealed that, in addition to the difference in this region between type A and type B strains, there are also differences in the anmK-pdpD region between the recently identified clades, F. tularensis types A.I and A.II (Fig. 2). In the two representative strains of the type A.I clade, strains Schu4 and FSC033, the anmK region has two premature stop codons, at positions 190 and 328, compared to the anmK form found in F. novicida, which has only one stop codon at position 372. In the A.I form of anmK, the stop codon at position 190 is followed with a start codon at position 194. The deduced AnmK proteins found in most bacteria are about 380 amino acids (371 in F. novicida) in length, and hence, the stop codon at position 190 in the clade A.I form of anmK suggests that AnmK is not functional in this strain. In two representatives of the type A.II clade, WY96-3418 and B38, the anmK gene has a stop codon at position 328 in addition to the stop codon at position 372 found in all forms of anmK. Outside of the internal stop codon regions, the deduced amino acid sequences of the ORF proteins encoded by the anmK genes show 96% identity between the clade A.I and A.II forms and 98% identity between F. novicida and the clade A.I forms.
FIG. 2.
The anmK and pdpD loci vary in F. tularensis subspecies. The F. novicida form of the FPI differs from the type B biotype forms in having anmK and pdpD, while the type A biotypes have pdpD but two distinct, truncated forms of anmK. The F. novicida strains U112 and GA99-3548 encode an intact AnmK protein consisting of 371 amino acid (aa) residues. The anmK genes of F. tularensis type A.I strains Schu4 and FSC033 contain two premature stop codons at positions 190 and 328, while the anmK genes of F. tularensis type A.II strains WY96-3418 and B38 have a single premature stop codon at position 328. The anmK locus is absent from F. tularensis subsp. holarctica (type B) strains OSU18 and LVS.
Homologues of AnmK are very widely found in bacteria, and since the large majority of these bacteria are not pathogens, it seems likely that anmK does not have a role in virulence that involves pathogen-host interactions. However, the presence of anmK could increase the overall fitness of a strain, and we provide evidence below that the loss of anmK has a small effect on F. novicida virulence. Nevertheless, we have reasoned that anmK does not have a specific biological role in the virulence of Francisella, and we have included analysis of anmK mutants only as a necessary component of our study of pdpD.
The F. novicida pdpD gene encodes a 1,245-amino-acid protein (140,663 molecular weight), and the pdpD genes found in both type A.I and type A.II clades of F. tularensis encode proteins of 1,195 amino acids (135,394 molecular weight). The deduced amino acid sequence identity between pdpD gene products found in the two clades is 100 percent. There is no significant identity of PdpD with other proteins as determined by a BLASTP search of the nonredundant protein databases. The F. novicida form of PdpD has 50 more amino acids than PdpD from type A strains of F. tularensis, and 48 of these amino acids constitute a hydrophilic stretch of amino acids near the center of PdpD.
Mutagenesis of the anmK-pdpD region.
The product of the pdpD gene had not been detected in previous studies, including a proteomic analysis of type A F. tularensis (31, 41). Hence, it was important to generate mutants in the pdpD gene in order to help identify the presumptive product of pdpD, in addition to determining whether PdpD plays a role in virulence. Since our previous gene replacement mutation of pdpD had affected the expression of the downstream gene iglA, we thought it was important to construct deletion mutations, which usually have minimal polar effects on transcription/translation coupling. To make deletion mutants, we took advantage of the unique PacI site that lies near the center of pdpD. A recombinant clone containing pdpD and surrounding regions was digested with PacI, and deletion mutants were created by treatment with the processive exonuclease Bal 31 and the subsequent recovery of a number of deletion mutant clones (Fig. 3). Five in-frame deletion mutants were recovered, and the analyses of two of them are presented in this work. A mutant with a deletion of the complete anmK gene was made as well (Fig. 3). Genetic complementation of the ΔanmK or the ΔpdpD mutation was accomplished by introducing anmK or anmK-pdpD into a chromosomal integrating vector, pJL-SKX, which directs insertions into the chromosome 0.48 Mb from pdpD. The region 397 bp upstream of anmK, which is the presumptive promoter region, was included in both complementation constructs.
FIG. 3.
Mutagenesis of pdpD. The extents of the deletions in anmK (A) and pdpD (B) are diagrammed. aa, the number of amino acids encoded by the gene. (C and D) The anmK and five in-frame deletion mutants of pdpD were examined by PCR using primers which flank the anmK and pdpD loci. The DNA sequences of mutated loci for strains used in this work were submitted to GenBank. WT, wild type; MW, molecular weight marker.
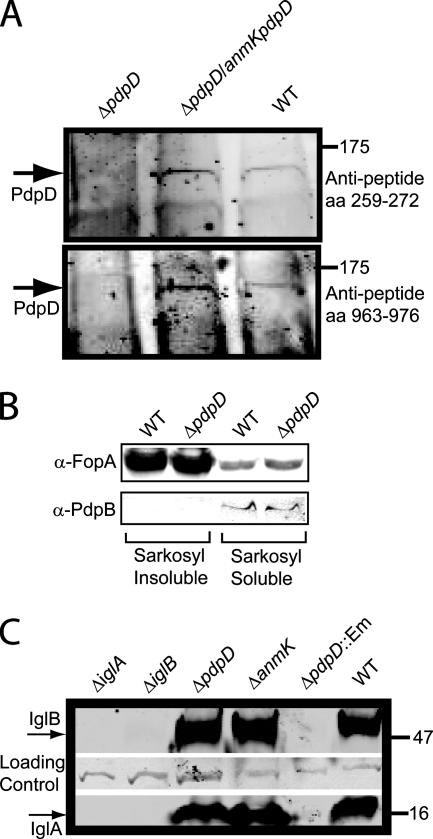
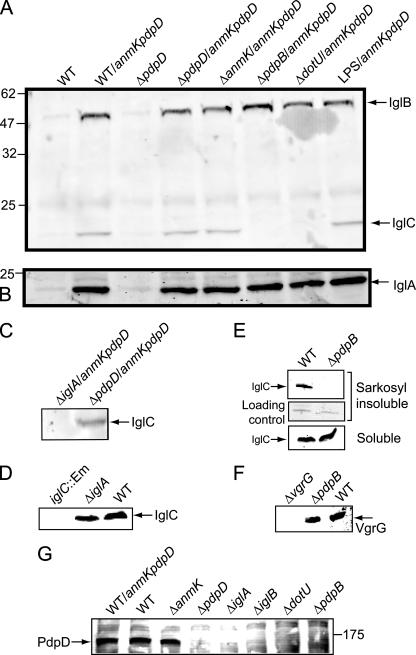
In order to detect PdpD by antibody reactivity, we generated three antisera. Two antisera were raised against peptide sequences found in PdpD at amino acid positions 259 to 272 and 963 to 976, which were predicted to be antigenic by online bioinformatic tools (e.g., http://bio.dfci.harvard.edu/Tools/antigenic.pl). A third antiserum was produced against a recombinant fragment (amino acids 748 to 996) of PdpD. Although the antirecombinant and antipeptide (amino acids 963 to 976) sera reacted well with recombinant protein, they did not reproducibly react with any protein band in whole-cell extracts of wild-type F. novicida that was absent from a pdpD mutant strain. However, preparation of Sarkosyl-insoluble extracts of F. novicida apparently enriched PdpD sufficiently to allow its detection with all three antisera (Fig. 4A and data not shown). As well, overexpression of PdpD occurred in strains carrying the complementation construct, pJL-SKX::anmK-pdpD, and this complementation allowed enhanced detection of PdpD (Fig. 4A). Using Sarkosyl-insoluble fractions from a strain overexpressing PdpD allowed us to visualize a ca. 140,000-relative-molecular-weight protein, by using all three types of antisera, that was absent from the pdpD mutant. The identical patterns of reactivity generated by the three antisera and the absence of the band in the ΔpdpD mutant strains provided confidence that the reactive bands corresponded to PdpD. This is especially important, since detection of PdpD was very difficult, even when the protein was concentrated before Western blotting.
FIG. 4.
Immunoblot analysis of pdpD mutants. (A) Immunoblots developed with anti-PdpD peptide antibodies. Samples were normalized to 15 μg protein per lane prior to separation on a 4 to 12% gradient gel. aa, amino acids. (B) Sarkosyl solubilization separates markers for the inner and outer membranes. Membrane fractions were separated by Sarkosyl solubilization and subjected to Western blotting. All lanes were loaded with 5 μg of protein. α-FopA and α-PdpB, anti-FopA and anti-PdpB antibodies. (C) IglA and IglB expression profiles in F. novicida mutants. In all blots, molecular weight markers (wild type [WT]) are shown on the right.
Sarkosyl was shown to separate inner and outer membrane fractions by testing for the presence of the outer membrane protein FopA in the Sarkosyl-soluble and -insoluble fractions (Fig. 4B). Measurement of the fluorescence intensity of the blots indicates that the outer membrane protein FopA is enriched about 15-fold in the Sarkosyl-insoluble fraction. The inner membrane protein PdpB was detected only in the Sarkosyl-soluble fraction. These results indicate that the Sarkosyl treatment was generating a valid separation of inner and outer membrane proteins.
The deletion mutations in anmK and pdpD did not have a detectable effect on the expression of the downstream genes iglA and iglB (Fig. 4C). The amounts of IglA and IglB expressed in the pdpD deletion strain contrasted sharply with the amounts expressed in the pdpD gene replacement mutant (Fig. 4C). The lack of both IglA and IglB in both the ΔiglA and ΔiglB mutants (first two lanes of Fig. 4C) highlights the previous finding (8) that the lack of expression of one protein leads to the apparent degradation of the other.
Overexpression of PdpD affects the surface localization of IglA, IglB, and IglC and the localization of IglC is dependent on T6SS component homologues.
The enrichment of PdpD in the Sarkosyl-insoluble fraction suggested that it localized to the outer membrane. However, because the antibody-reactive bands were so faint, probably due to the small amounts of PdpD that were made, it was difficult to determine the distribution of PdpD in the bacterial cell. To help ascertain if PdpD localized to the surface of F. novicida, we reacted surface-exposed proteins in the PdpD-overexpressing strain (carrying pJL-SKX::anmK-pdpD) with biotin. After biotin labeling, we separated the biotin-labeled proteins by using streptavidin binding, and analyzed proteins were eluted from the streptavidin by Western blotting. Probing the blots with anti-PdpD antisera failed to yield reactive bands of the appropriate relative molecular weight for PdpD. Since small amounts of IglA were previously shown to be exposed to surface biotinylation (24), we probed our Western blots with anti-IglA; we also reacted the blots with monoclonal antibodies against IglB, IglC, and PdpB, which we expected to serve as negative controls. Although antibody against the inner membrane protein, PdpB, did not detect a protein band on the blot, antibody against IglA, IglB, and IglC did react, and these reacted more strongly in samples overexpressing PdpD (data not shown; see below).
IglA and IglB are homologues of proteins that are part of T6SSs. There are at least two other FPI genes, pdpB and dotU, that encode proteins that are homologous to the IcmF and DotU families of proteins that are parts of both type IV secretion systems and T6SSs. To assess the role of T6SS proteins in the surface localization of IglA, IglB, and IglC, we introduced pJL-SKX::anmK-pdpD into a number of strains and carried out surface biotinylation of these strains (Fig. 5). Our results showed that increased amounts of IglA and IglB were exposed to surface biotinylation when pJL-SKX::anmK-pdpD was present, regardless of the genetic background (Fig. 5A and B). However, the amount of IglC exposed to biotinylation was dramatically affected by the absence of pdpB and dotU (Fig. 5A). The wild-type F. novicida showed a small amount of IglC in a biotinylation-accessible state, but even this small amount failed to be biotinylated in the pdpB and dotU backgrounds, even when PdpD was overexpressed (Fig. 5A). Similarly, in an ΔiglA or ΔiglB background, IglC failed to be biotinylated, even when pJL-SKX::anmK-pdpD was present (Fig. 5C and data not shown), even though IglC was shown to be in the soluble fraction of the wild-type and ΔiglA strains (Fig. 5D). Biotinylation of IglA, IglB, or IglC in an O-antigen mutant strain of F. novicida was identical to that in the wild-type strain (Fig. 5A, far right).
FIG. 5.
Effect of PdpD overexpression on surface biotinylation of IglA, IglB, and IglC. WT, wild type. (A) Reactivity of surface-exposed biotinylated proteins with anti-IglB and anti-IglC monoclonal antibodies. LPS, O-antigen mutant SC92. (B) Reactivity with anti-IglA polyclonal serum. (C) Surface biotinylation of IglC in an ΔiglA strain. (D) Control for the experiment whose blot is shown in panel C. IglC is found in the cytoplasm in the ΔiglA strain. (E) IglC fails to localize to the outer membrane in a ΔpdpB background. (F) The PdpB deletion does not affect expression of the downstream gene vgrG. (G) Presence or absence of PdpD in the outer membrane in the F. novicida ΔanmK, ΔpdpD, ΔiglA, ΔiglB, ΔdotU, and ΔpdpB strains.
Previous work showed that IglC was primarily localized to the cytoplasm (12), with a small amount being found in the outer membrane. Any amount of this protein found outside of the cytoplasm might be attributed to cross-contamination among cell fractions. To discriminate between this possibility and a true physiological localization of IglC to a site accessible to biotinylation, we prepared Sarkosyl-insoluble fractions from wild-type F. novicida and from a ΔpdpB strain (Fig. 5E). We reasoned that a lack of outer membrane localization of IglC in a strain lacking a canonical component of both type IV secretion systems and T6SSs would support the interpretation that this experimental approach provided a meaningful localization profile. In the wild-type strain, but not in the ΔpdpB strain, IglC was detected in the Sarkosyl-insoluble pellet, which represents an enrichment of outer membrane components (Fig. 5E). It should be noted that the amount of total protein loaded for the outer membrane fractions (Fig. 5E, top panel) was about 10-fold greater than the amount analyzed in the soluble portion of the bacterial extract (Fig. 5E, bottom panel). Hence, the results presented here are consistent with previous studies showing the IglC is localized predominantly to the cytoplasm (12). To demonstrate that the ΔpdpB mutation did not disrupt expression of downstream genes which may encode unidentified components of the T6SS protein homologues, we assessed the expression of VgrG, which is encoded by a gene downstream of pdpB. This analysis (Fig. 5F) showed that VgrG was expressed at wild-type levels.
The enhanced localization of IglC to the outer membrane by the overexpression of PdpD and the dependence of this localization on T6SS homologues suggested that PdpD localization to the outer membrane would be dependent on the T6SS. To test this idea, we extracted proteins from the Sarkosyl-insoluble (outer membrane) fraction of wild-type F. novicida and mutant strains with deletions in genes encoding T6SS homologue components and performed Western blotting to detect PdpD (Fig. 5G). Since it was difficult to consistently obtain numerous samples of Sarkosyl-insoluble fractions with clear banding patterns, biotinylation of proteins followed by streptavidin extraction was used to prepare samples of outer membrane proteins. As with IglC, localization of PdpD to the outer membrane required IglA, IglB, DotU, and PdpB but not AnmK. These results are consistent with the growing consensus of the required components of a T6SS. PdpB, which contains an IcmF motif, together with IglAB may form the foundation of a T6SS, as these three proteins make up the IcmF-associated homologous protein group, which was the bioinformatic basis for defining the T6SS. DotU homologues are often associated with IcmF-related proteins.
We interpret the localization data for IglA, IglB, and IglC as indications that these proteins interact with PdpD. The enhanced exposure of IglA, IglB, and IglC to surface biotinylation when PdpD is overexpressed suggests that there is some form of colocalization of the four proteins. The fact that the enhanced localization of IglC is eliminated in strains with ΔiglA, ΔdotU, and ΔpdpB mutations argues that the altered surface exposure of IglC is not simply due to leakage of IglC through a membrane when PdpD is overexpressed. It is noteworthy that the amounts of surface biotinylation of IglC in the ΔiglA, ΔdotU, and ΔpdpB genetic backgrounds appear to be below the level found in the wild-type F. novicida strain and, thus, the effect of these mutations is independent of the expression level of PdpD. Although our biotinylation studies suggest protein interactions or cosecretion, they do not define the nature of the secretion or the final destination of IglA, IglB, IglC, or PdpD. The IglA, IglB, IglC, and PdpD proteins all lack an N-terminal region that corresponds to a signal peptide used by either the Sec or the twin-arginine transport system, and hence, the transport of these FPI-encoded proteins is presumably not mediated by Sec or twin-arginine transport processes.
We previously showed that immunoprecipitations of IglA coprecipitated one protein that was identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry as IglB (8). We also showed that the absence of IglB by mutation led to the loss of IglA, presumably by protease digestion. IglC was made in the absence of IglA or IglB. Hence, there is evidence that IglA and IglB interact, but there is no evidence that IglC interacts with IglA or IglB.
Intracellular growth of PdpD mutants.
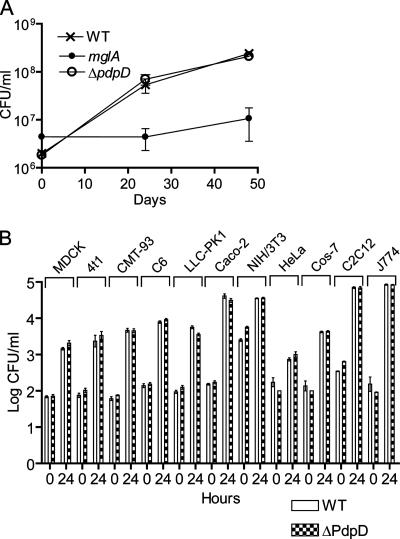
We previously reported that an insertion mutation in pdpD reduced the virulence and intramacrophage growth ability of the resultant strain (28). We noted that the insertion depressed the expression of the downstream-encoded IglA protein (Fig. 4C), which was later shown to be needed for intracellular growth and virulence (8). To more accurately assess the role of pdpD in intracellular growth, we tested one ΔpdpD strain for growth in BMDMs. We found that the ΔpdpD mutant strain 20d (unless otherwise stated, use of the term “the ΔpdpD strain” herein refers to 20d) grew in BMDMs in a manner identical to that of the wild-type strain (Fig. 6A). The ΔanmK strain also showed no defect in intramacrophage growth (data not shown). We also tested the ΔpdpD strain in 11 cell lines of different tissue and species origins (Fig. 6B) and found that in each case, the ΔpdpD strain grew like wild-type F. novicida. Although mouse macrophages, including the mouse macrophage cell line J774, are the most commonly used cell types to study Francisella intracellular growth, our results suggest that other cell lines, such as the human colon epithelial cell line Caco-2, might serve as an equally important host cell for in vitro studies of Francisella intracellular growth.
FIG. 6.
Intracellular growth of ΔpdpD mutant. WT, wild type. (A) Macrophages were infected at a multiplicity of infection of 50:1, and viable F. novicida cells were counted at 0, 24, and 48 h after infection. The mglA mutant strain, which is defective for intracellular growth, served as a negative control. (B) The growth of a ΔpdpD mutant was examined in 11 different cell lines and found to grow as well as wild-type F. novicida. Error bars represent the standard errors of the means.
The contrasting intracellular growth phenotypes for the pdpD gene replacement mutant and the ΔpdpD mutant highlight a phenomenon that we have observed for many mutants with lesions in FPI genes. In several cases, we found that insertion mutations and some small partial in-frame deletion mutations generate strains with phenotypes that are different from complete markerless deletion mutations. This suggests that small perturbations of FPI gene expression interfere with intracellular growth and virulence, and this is to be expected if all of the FPI genes encode virulence-associated proteins.
Virulence phenotype of anmK and pdpD deletion mutants in chicken embryos and in mice.
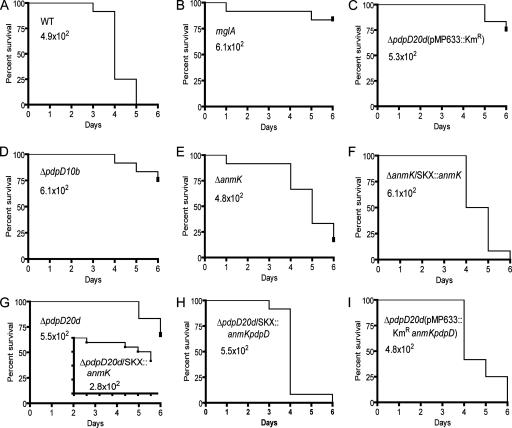
To assess the effects of gene deletions on virulence, we used a combination of infections of chicken embryos and mice. To reduce animal suffering, we performed the bulk of the infections with chicken embryos and compared these results with smaller experiments using mouse infections. Both the large and small deletions in pdpD resulted in strains with attenuated virulence in chicken embryos (Fig. 7D and G). Complementation of the genetic defect by using the integrating recombinant pJL-SKX::anmK-pdpD completely restored virulence (Fig. 7H). Similarly, genetic complementation using a plasmid vector restored virulence (Fig. 7I), but a mock complementation with the plasmid without the anmK-pdpD insert did not (Fig. 7C). Complementation of the ΔpdpD strain with pJL-SKX::anmK alone did not restore virulence (insert in Fig. 7G). Deletion of anmK had a small effect on virulence, and this defect could be reversed by genetic complementation with a wild-type copy of anmK (Fig. 7E and F). A strain that we use for a universal negative control, GB2 (mglA), showed its usual low virulence (Fig. 7B). When pdpD was introduced into the F. tularensis type B strain LVS, the resulting strain always generated more-rapid deaths than the parent strain (data not shown). However, this slight increase in apparent virulence was not statistically significant on a consistent basis.
FIG. 7.
Virulence of ΔanmK and ΔpdpD mutants in chicken embryos. The time-to-death profiles of seven-day-old chicken embryos are shown for the wild-type strain (A) and a standard avirulent strain (B) of F. novicida. The mutant 10b (ΔpdpD), ΔanmK, and 20d (ΔpdpD) strains are shown in panels D, E, and G, respectively. A mock-complemented strain is shown in panel C, and genetically complemented strains are shown in panels F, H, and I. The F. novicida strain and the inoculating dose are shown inside each graph. The P values for the differences between the wild-type strain and all of the mutant strains were less than 0.001. The P values for the differences between the wild type and all of the complemented strains were greater than 0.05, showing that they are not significantly different; however, in some of the repetitions, the ΔpdpD/pJL-SKX::anmK-pdpD complemented strain was more virulent at a statistically significant level. All of the F. novicida experiments were done with 12 chicken embryos and were repeated three times.
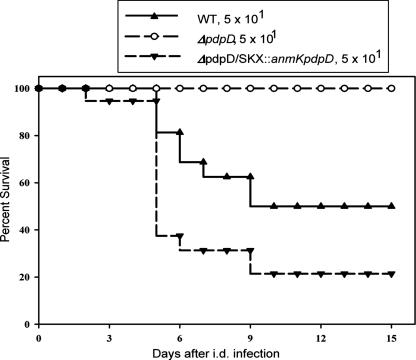
The pattern of the virulence phenotypes for the ΔpdpD 20d strain in mouse experiments was the same as that observed for chicken embryos (Fig. 8). The time to death for the ΔpdpD/pJL-SKX::anmK-pdpD strain was slightly shorter than for the wild-type F. novicida strain, but this difference was statistically significant at a low level of confidence (P = 0.04) by using an unpaired t test and, thus, is of marginal biological significance.
FIG. 8.
Survival patterns of mice infected intradermally (i.d.) with a low dose of F. novicida or pdpD mutants of F. novicida. BALB/cByJ male mice were infected with 5 × 101 CFU of the indicated bacterial strain intradermally and monitored for survival. Results are expressed as percentages of surviving mice within each group. Results through day 15 after infection are shown; mice were further monitored through day 30, and no other deaths occurred. Total group sizes were 16 mice in the group infected with wild-type F. novicida, 17 mice in the group infected with the ΔpdpD mutant, and 14 mice infected with the ΔpdpD/pJL-SKX::anmK-pdpD mutant. The P value for the wild-type (WT) infection survival curve compared to that for the ΔpdpD strain is 0.0001, and that for the wild-type infection survival curve compared to that for the complemented strain is 0.039.
The complementation of the ΔpdpD strain with pJL-SKX::anmK-pdpD restored virulence, but complementation with pJL-SKX::pdpD failed to restore virulence. In the ΔpdpD/pJL-SKX::pdpD strain, both anmK and pdpD were present in wild-type forms but were separated on the chromosome, with a copy of anmK in the FPI and a copy of pdpD inserted into pJL-SKX. Presumably, pdpD requires the promoter region located in front of anmK to express properly.
The biochemical roles of FPI-encoded proteins are unknown, and the biochemical properties of only a few of them have been studied. IglC was the first of the FPI-encoded proteins to be discovered as a protein that is highly induced following F. tularensis infection of macrophages (12). Subsequent work has shown that mutants with a deleted or disrupted iglC gene fail to grow in macrophages (13), are deficient in ability to escape from phagosomes (21), and fail to down-regulate the proinflammatory response in macrophages (39). Although these studies provide insights into the cell biology events surrounding F. tularensis infection, they do not ascribe a biochemical role to IglC. The role of IglC could be direct, or it could be through its interactions with one or several other proteins.
There is some published evidence that IglC primarily localizes to the cytoplasm, with a small proportion of IglC localizing to the outer membrane (12), and this is consistent with the data presented in this work. Recently, the structure of IglC was determined and found to have limited structural similarity to gp5, a component of the hole-poking device of bacteriophage T4 (38). Conceivably, IglC could play a role in a secretion channel in the outer membrane of Francisella, or alternatively, it could play a role in forming a channel in a host cell membrane structure. Whatever the role of IglC, it seems likely that it interacts with PdpD, and this interaction is linked to events carried out by T6SS homologues encoded by the FPI.
In this work, we have shown that the pdpD gene is required for full virulence of F. novicida but not for intracellular growth. We have provided evidence that the PdpD protein localizes to the outer membrane in a fashion dependent on homologues of the T6SS. Overexpression of PdpD increased the amounts of IglA, IglB, and IglC that are exposed to surface biotinylation. Our data suggest that the localization of IglC to a surface biotinylation-susceptible site requires IglA, IglB, PdpB, and DotU, which are all putative components of the FPI-encoded T6SS. These proteins are required to localize PdpD to the outer membrane. Hence, there appears to be some form of interaction of PdpD with IglA, IglB, and IglC, and the processes that affect the secretion of IglC to the outer membrane appear to be the same that affect the secretion of PdpD.
Acknowledgments
This work was supported by grant 5R01 AI056212-02 from the National Institute of Allergy and Infectious Diseases.
We thank K. Klose for suggesting the revised FPI nomenclature and M. Pavelka, D. Monack, T. Zahrt, and A. Sjöstedt for helpful comments on the naming of genes.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Anthony, L. S., M. Z. Gu, S. C. Cowley, W. W. Leung, and F. E. Nano. 1991. Transformation and allelic replacement in Francisella spp. J. Gen. Microbiol. 1372697-2703. [DOI] [PubMed] [Google Scholar]
- 2.Baron, G. S., and F. E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29247-259. [DOI] [PubMed] [Google Scholar]
- 3.Beckstrom-Sternberg, S. M., R. K. Auerbach, S. Godbole, J. V. Pearson, J. S. Beckstrom-Sternberg, Z. Deng, C. Munk, K. Kubota, Y. Zhou, D. Bruce, J. Noronha, R. H. Scheuermann, A. Wang, X. Wei, J. Wang, J. Hao, D. M. Wagner, T. S. Brettin, N. Brown, P. Gilna, and P. S. Keim. 2007. Complete genomic characterization of a pathogenic A.II strain of Francisella tularensis subspecies tularensis. PLoS ONE 2e947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, J. F., C. R. Owen, and C. L. Larson. 1955. Virulence of Bacterium tularense in mice, guinea pigs, and rabbits. J. Infect. Dis. 97162-166. [DOI] [PubMed] [Google Scholar]
- 5.Bina, X. R., C. L. Lavine, M. A. Miller, and J. E. Bina. 2008. The AcrAB RND efflux system from the live vaccine strain of Francisella tularensis is a multiple drug efflux system that is required for virulence in mice. FEMS Microbiol. Lett. 279226-233. [DOI] [PubMed] [Google Scholar]
- 6.Bladergroen, M. R., K. Badelt, and H. P. Spaink. 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 1653-64. [DOI] [PubMed] [Google Scholar]
- 7.Cowley, S. C., C. J. Gray, and F. E. Nano. 2000. Isolation and characterization of Francisella novicida mutants defective in lipopolysaccharide biosynthesis. FEMS Microbiol. Lett. 18263-67. [DOI] [PubMed] [Google Scholar]
- 8.de Bruin, O. M., J. S. Ludu, and F. E. Nano. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eigelsbach, H. T., and C. M. Downs. 1961. Prophylactic effectiveness of live and killed tularemia vaccines. J. Immunol. 87415-425. [PubMed] [Google Scholar]
- 10.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gil, H., G. J. Platz, C. A. Forestal, M. Monfett, C. S. Bakshi, T. J. Sellati, M. B. Furie, J. L. Benach, and D. G. Thanassi. 2006. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc. Natl. Acad. Sci. USA 10312897-12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golovliov, I., M. Ericsson, G. Sandstrom, A. Tarnvik, and A. Sjostedt. 1997. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 652183-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golovliov, I., A. Sjostedt, A. Mokrievich, and V. Pavlov. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222273-280. [DOI] [PubMed] [Google Scholar]
- 14.Gray, C. G., S. C. Cowley, K. K. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 21553-56. [DOI] [PubMed] [Google Scholar]
- 15.Hager, A. J., D. L. Bolton, M. R. Pelletier, M. J. Brittnacher, L. A. Gallagher, R. Kaul, S. J. Skerrett, S. I. Miller, and T. Guina. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62227-237. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 17.Johansson, A., J. Farlow, P. Larsson, M. Dukerich, E. Chambers, M. Bystrom, J. Fox, M. Chu, M. Forsman, A. Sjostedt, and P. Keim. 2004. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 1865808-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 19.Larson, C. L., W. Wicht, and W. L. Jellison. 1955. An organism resembling P. tularensis isolated from water. Public Health Rep. 70253-258. [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37153-159. [DOI] [PubMed] [Google Scholar]
- 21.Lindgren, H., I. Golovliov, V. Baranov, R. K. Ernst, M. Telepnev, and A. Sjostedt. 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53953-958. [DOI] [PubMed] [Google Scholar]
- 22.LoVullo, E. D., L. A. Sherrill, L. L. Perez, and M. S. Pavelka, Jr. 2006. Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology 1523425-3435. [DOI] [PubMed] [Google Scholar]
- 23.Ludu, J. S., E. B. Nix, B. N. Duplantis, O. M. de Bruin, L. A. Gallagher, L. M. Hawley, and F. E. Nano. 2008. Genetic elements for selection, deletion mutagenesis and complementation in Francisella spp. FEMS Microbiol. Lett. 27886-93. [DOI] [PubMed] [Google Scholar]
- 24.Melillo, A., D. D. Sledjeski, S. Lipski, R. M. Wooten, V. Basrur, and E. R. Lafontaine. 2006. Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol. Lett. 263102-108. [DOI] [PubMed] [Google Scholar]
- 25.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 3121526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mougous, J. D., C. A. Gifford, T. L. Ramsdell, and J. J. Mekalanos. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9797-803. [DOI] [PubMed] [Google Scholar]
- 27.Nano, F. E., and C. Schmerk. 2007. The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 1105122-137. [DOI] [PubMed] [Google Scholar]
- 28.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 1866430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nix, E. B., K. K. Cheung, D. Wang, N. Zhang, R. D. Burke, and F. E. Nano. 2006. Virulence of Francisella spp. in chicken embryos. Infect. Immun. 744809-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsufjev, N. G., and O. S. Emelyanova. 1963. Immunological relationships between Old and New World varieties of tularaemic bacteria. J. Hyg. Epidemiol. Microbiol. Immunol. 7178-185. [PubMed] [Google Scholar]
- 31.Pavkova, I., M. Reichelova, P. Larsson, M. Hubalek, J. Vackova, A. Forsberg, and J. Stulik. 2006. Comparative proteome analysis of fractions enriched for membrane-associated proteins from Francisella tularensis subsp. tularensis and F. tularensis subsp. holarctica strains. J. Proteome Res. 53125-3134. [DOI] [PubMed] [Google Scholar]
- 32.Pukatzki, S., A. T. Ma, A. T. Revel, D. Sturtevant, and J. J. Mekalanos. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. USA 10415508-15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 1031528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohmer, L., C. Fong, S. Abmayr, M. Wasnick, T. J. Larson Freeman, M. Radey, T. Guina, K. Svensson, H. S. Hayden, M. Jacobs, L. A. Gallagher, C. Manoil, R. K. Ernst, B. Drees, D. Buckley, E. Haugen, D. Bovee, Y. Zhou, J. Chang, R. Levy, R. Lim, W. Gillett, D. Guenthener, A. Kang, S. A. Shaffer, G. Taylor, J. Chen, B. Gallis, D. A. D'Argenio, M. Forsman, M. V. Olson, D. R. Goodlett, R. Kaul, S. I. Miller, and M. J. Brittnacher. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human-pathogenic strains. Genome Biol. 8R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santic, M., M. Molmeret, J. R. Barker, K. E. Klose, A. Dekanic, M. Doric, and Y. Abu Kwaik. 2007. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell. Microbiol. 92391-2403. [DOI] [PubMed] [Google Scholar]
- 36.Schell, M. A., R. L. Ulrich, W. J. Ribot, E. E. Brueggemann, H. B. Hines, D. Chen, L. Lipscomb, H. S. Kim, J. Mrazek, W. C. Nierman, and D. Deshazer. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 641466-1485. [DOI] [PubMed] [Google Scholar]
- 37.Staples, J. E., K. A. Kubota, L. G. Chalcraft, P. S. Mead, and J. M. Petersen. 2006. Epidemiologic and molecular analysis of human tularemia, United States, 1964-2004. Emerg. Infect. Dis. 121113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, P., B. P. Austin, F. D. Schubot, and D. S. Waugh. 2007. New protein fold revealed by a 1.65 Å resolution crystal structure of Francisella tularensis pathogenicity island protein IglC. Protein Sci. 162560-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Telepnev, M., I. Golovliov, and A. Sjostedt. 2005. Francisella tularensis LVS initially activates but subsequently down-regulates intracellular signaling and cytokine secretion in mouse monocytic and human peripheral blood mononuclear cells. Microb. Pathog. 38239-247. [DOI] [PubMed] [Google Scholar]
- 40.Tempel, R., X. H. Lai, L. Crosa, B. Kozlowicz, and F. Heffron. 2006. Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 745095-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Twine, S. M., N. C. Mykytczuk, M. D. Petit, H. Shen, A. Sjostedt, J. Wayne Conlan, and J. F. Kelly. 2006. In vivo proteomic analysis of the intracellular bacterial pathogen, Francisella tularensis, isolated from mouse spleen. Biochem. Biophys. Res. Commun. 3451621-1633. [DOI] [PubMed] [Google Scholar]
- 42.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
- 43.Weiss, D. S., A. Brotcke, T. Henry, J. J. Margolis, K. Chan, and D. M. Monack. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. USA 1046037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, J., and K. Y. Leung. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 661192-1206. [DOI] [PubMed] [Google Scholar]