Abstract
To gain a better understanding of epidemiology of resistance in Staphylococcus aureus, we describe the molecular epidemiology of methicillin-resistant Staphylococcus aureus bloodstream isolates in urban Detroit. Bloodstream isolates from July 2005 to February 2007 were characterized. Two hundred ten bloodstream isolates from 201 patients were evaluated. Patient characteristics were as follows: median age, 54 years; 56% male; and 71% African-American. Seventy-six percent of infections were health care associated, with 55% being community-onset infections and 21% hospital acquired, and 24% were community associated. The most common sources were skin/wound (25%), central venous catheters (24%), unknown source (20%), and endocarditis (9%). Ninety percent and 5% of isolates had a MIC of vancomycin of ≤1.0 mg/liter, using automated dilution testing and E-test, respectively. Six percent of isolates showed heteroresistance to vancomycin, all occurring with isolates having a vancomycin E-test MIC of ≥1.5 mg/liter. Results of pulsed-field gel electrophoresis showed 17 strain types. The predominant strains were USA100 (104 isolates) and USA300 (74 isolates). Forty-nine percent of the isolates had staphylococcal cassette chromosome mec II, and 56% had agr II. All USA300 isolates were positive for the Panton-Valentine leukocidin toxin genes and agr I. Forty-seven percent of USA300 bloodstream infections were health care associated (35% community onset and 12% hospital onset). USA300 strains were more common in injection drug users with skin/wound as the predominant source of infection. Thirty percent of the USA100 strains were closely related to vancomycin-resistant Staphylococcus aureus isolates. The results of this study show that vancomycin MICs using automated dilution testing with Vitek-2 and E-test were highly discordant. Most methicillin-resistant S. aureus strains causing bacteremia are health care associated, commonly have MICs of vancomycin that are high within the susceptible range are not detected by routine automated dilution testing, and have significant diversity of molecular characteristics. USA100 strains that are closely related to vancomycin-resistant S. aureus (VRSA) isolates and USA300 strains are common as causes of both hospital and community-onset infection. Infection control measures should focus not only on prevention of the spread of community strains in the hospital but also prevention of the spread of hospital strains associated with VRSA into the community.
Staphylococcus aureus is one of the most common etiologies of bloodstream infections (BSI) (30, 50) and has emerged as the leading cause of infective endocarditis in developed countries (10). Methicillin-resistant Staphylococcus aureus (MRSA) accounts for more than 60% of these isolates in the intensive care unit setting (27). The Centers for Disease Control and Prevention (CDC) estimated more than 30,000 hospitalizations for MRSA septicemia in the United States in the year 2000 (19). In a more recent study published by Klevens et al. (18), those authors estimated an occurrence of more than 90,000 invasive MRSA infections in 2005, with bacteremias accounting for 75% of these infections. Hospitalizations due to MRSA bacteremia have increased more than 80% from 1999 to 2005 (16). Bacteremia due to MRSA has been associated with a significantly higher mortality (6, 37), greater length of hospitalization, and higher hospital charges (5, 22) than is the case with methicillin-susceptible strains (6).
MRSA infections have traditionally been thought to be acquired through health care contact (1); however, community-acquired MRSA infections have been increasing in frequency since this type of infection was first described in 1980s (35). The strain type currently associated with community-acquired MRSA infections has been designated CDC strain USA300 (24). This strain is thought to have unique epidemiologic and clinical characteristics. It has been associated predominantly with skin and soft-tissue infections (7, 15) but recently has also been described as a significant cause of health care-associated and nosocomial infections (17). BSIs have been less commonly reported (11, 12, 36). There is little information describing the molecular, clinical epidemiology, and clinical characteristics of BSIs due to the USA300 strain of MRSA.
It is clear that the epidemiology of MRSA is continuously evolving, with USA300 MRSA increasingly reported as causing hospital-associated infections and the USA100 strain typically associated with nosocomial acquisition seen to arise from the community (9, 17). To gain a better understanding of the epidemiology of resistance in S. aureus, in this article we therefore describe the molecular epidemiology of MRSA bloodstream isolates in urban Detroit, a geographic area where MRSA has been common since the 1980s and where the first reported cases of vancomycin-intermediate S. aureus (VISA) in North America (2) and vancomycin-resistant S. aureus (VRSA) in the world (3) occurred. Since seven out of the nine VRSA confirmed by the CDC in the United States have been isolated in Detroit (38, 53; also unpublished observation), information is urgently needed to evaluate the epidemiology of MRSA to look at possible trends associated with emergence of strains in order to develop rational control measures.
MATERIALS AND METHODS
Setting.
Henry Ford Hospital is a 900-bed tertiary-care, acute-care teaching facility with level 1 trauma and multiorgan transplantation located in urban Detroit.
Patients and isolates.
Consecutive MRSA isolates were collected from July 2005 to February 2007. All the isolates included in this study were from patients who had at least one blood culture positive for oxacillin-resistant S. aureus as identified by the clinical microbiology laboratory at Henry Ford Hospital. Only the first positive blood culture was included for analysis. Subsequent positive bloodstream isolates from patients were included if the blood culture was collected at least 28 days from the first episode and the patient had completed treatment for the previous episode. Demographic data including age, sex, race, and comorbid conditions were collected. The infections were further identified as community associated or health care associated, subdivided between community onset and hospital onset as previously done by Klevens et al. (18). The source of the bacteremia was identified by chart review using CDC definitions of nosocomial infection (14). Isolates were stored at −70°C and were plated on trypticase-soy agar supplemented with 5% sheep blood for subsequent testing.
Antimicrobial susceptibility testing.
In vitro susceptibilities were determined following Clinical and Laboratory Standards Institute (CLSI) standards. In vitro susceptibility to vancomycin was initially determined by the clinical microbiology laboratory using automated microdilution with Vitek-2 (bioMerieux, Durham, NC). Inducible clindamycin resistance was determined using the D-zone test as described by the CLSI (4). In vitro susceptibilities to vancomycin and daptomycin were determined using the E-test, following the instructions provided by the manufacturer (AB-Biodisk, Solna, Sweden). Isolates were screened for heteroresistance to vancomycin using the macrodilution method E-test (AB-Biodisk, Solna, Sweden) as previously described (51). The following Staphylococcus aureus strains were used in performance of the tests: ATCC 29213, Mu3, and Mu50.
PFGE.
Genomic DNA was prepared and digested with SmaI (New England BioLabs, Beverly, MA) using a previously described method (24). SmaI fragments were separated using a CHEF-DR III apparatus (Bio-Rad, Hercules, CA) using the following settings: initial switch time, 5 s; final switch time, 35 s; voltage, 6 V/cm; run time, 21 h; temperature, 14°C. The reference standard that was used was NCTC 8325, which was run in lanes 1, 8, and 15 of a 15-well gel. Pulsed-field gel electrophoresis (PFGE) patterns were compared using the BioNumerics software program (Applied Maths, Belgium). All of the MRSA BSI isolates in this study were compared to MRSA strains USA100 to -1100 (Network on Antimicrobial Resistance in Staphylococcus aureus, Herndon, VA) as described by the CDC (24). The first seven VRSA isolates in the United States (NARSA, Herndon, VA) were also included as controls (53). Isolates were determined to be of the same PFGE strain group if their SmaI restriction patterns were ≥80% similar using the Dice coefficient (39, 45).
PCR.
Multiplex PCR was performed to determine the staphylococcal cassette chromosome mec (SCCmec) types I, II, III, and IV (28). If an SCCmec type could not be identified using this method, an additional PCR assay used to type SCCmec I to V was done (52). Subtyping of SCCmec IV was also done (52). agr typing was performed for all isolates (42), as was detection of the Panton-Valentine leukocidin (PVL) toxin genes, lukS-PV and lukF-PV (20).
Statistical analysis.
Continuous variables were compared utilizing the t test. Dichotomous data were compared using either a χ2 test or Fisher's exact test where appropriate. A P value of less than 0.05 was considered statistically significant. Spearman's correlation was used to examine the relationship between vancomycin and daptomycin E-test MICs.
RESULTS
MRSA caused 63.8% of our Staphylococcus aureus BSIs in 2006. There were 345 consecutive episodes of MRSA bacteremia in 314 patients from July 2005 to February 2007. Of these, 210 isolates from 201 patients were available for evaluation. Demographic characteristics for these patients are listed in Table 1. The median age was 54 years, with 56% males and 71% African Americans. Seventy-six percent of infections were health care associated, with 55% community-onset and 21% hospital-acquired infections, and 24% were community associated. A trend toward younger age in the community-associated MRSA bacteremia patients compared with patients with health care-associated infection was found, whereas older patients were more common in the health care-associated groups (Table 1). Comorbidities, such as chronic kidney disease, hemodialysis, cardiovascular disease, neurologic disease, and malignancy, were significantly more common in patients with health care-associated MRSA bacteremia, while intravenous drug users commonly had community-associated BSIs (Table 1). Ten percent of patients had no identifiable comorbidities, with the majority found in the community-associated group.
TABLE 1.
Demographic characteristics of patients with MRSA bacteremiaa
| Characteristic | Value for case groupb
|
P value | |||
|---|---|---|---|---|---|
| All cases | Community associated | Health care associated, community onset | Health care associated, hospital onset | ||
| Total subjects | 201 | 50 | 110 | 41 | |
| Sex | |||||
| Male | 112 (56) | 29 (58) | 63 (57) | 20 (49) | NS |
| Female | 89 (44) | 21 (42) | 47 (43) | 21 (51) | NS |
| Age (yr) | |||||
| <18 | 2 (1) | 2 (4) | 0 (0) | 0 (0) | NS |
| 18-34 | 21 (10) | 9 (18) | 8 (7) | 4 (10) | NS |
| 35-49 | 48 (24) | 18 (36) | 22 (20) | 8 (19) | 0.062 |
| 50-64 | 66 (33) | 15 (30) | 36 (33) | 15 (37) | NS |
| ≥65 | 64 (32) | 6 (12) | 44 (40) | 14 (34) | 0.003 |
| Race | |||||
| Black | 143 (71) | 32 (64) | 84 (76) | 27 (66) | NS |
| White | 54 (27) | 16 (32) | 24 (22) | 14 (34) | NS |
| Other | 4 (2) | 2 (4) | 2 (2) | 0 (0) | NS |
| Comorbidities | |||||
| None | 20 (10) | 14 (28) | 3 (3) | 3 (7) | <0.001 |
| HIV | 8 (4) | 2 (4) | 6 (5) | 0 (0) | NS |
| Diabetes mellitus | 79 (39) | 16 (32) | 47 (43) | 16 (39) | NS |
| Chronic kidney disease | 72 (36) | 2 (4) | 54 (49) | 16 (39) | <0.001 |
| Hemodialysis | 49 (24) | 0 (0) | 37 (34) | 12 (29) | <0.001 |
| COPD | 23 (11) | 3 (6) | 14 (13) | 6 (15) | NS |
| Cardiovascular disease | 85 (42) | 4 (8) | 56 (51) | 25 (23) | <0.001 |
| Neurologic disease | 50 (25) | 4 (8) | 32 (29) | 14 (34) | 0.005 |
| Liver disease | 52 (26) | 19 (38) | 23 (21) | 10 (24) | 0.071 |
| Malignancy | 30 (15) | 0 (0) | 24 (22) | 6 (15) | 0.002 |
| IVDU | 45 (22) | 28 (56) | 11 (10) | 6 (15) | <0.001 |
Statistical comparisons were made between the three epidemiological groups (community associated, health care associated—community onset, and health care associated—hospital onset). NS, not significant; HIV, human immunodeficiency virus; COPD, chronic obstructive pulmonary disease.
Values are no. (%) of patients unless otherwise noted.
The most commonly identified source of bacteremia (Table 2) was skin/wound (25%), followed by central venous catheters (24%), respiratory tract (10%), and endocarditis (9%). A source could not be identified in 20% of bacteremias. Other identified sources are listed in Table 2. The most common source for community-associated bacteremia was skin/wound (42%), with 24% due to infective endocarditis. For health care-associated infections, central venous catheters were most commonly implicated in 30% of community-onset infections and 36% of hospital-onset infections. Skin/wound as a source of infections was also common (24%) for patients who acquired their infections in the community but had health care risk factors. For hospital-onset infection, a central venous catheter source was most common (36%). Health care-related infections were associated with SCCmec II, agr II, and PVL toxin gene-negative isolates, while community-associated infections were associated with SCCmec type IV, agr I, and PVL toxin gene-positive isolates, driven primarily by the distribution of USA100 and USA300 strains.
TABLE 2.
Source of infection; PFGE and PCR results versus epidemiologic sourcea
| Characteristic | No. (%) of cases
|
P value | |||
|---|---|---|---|---|---|
| Total | Community associated | Health care associated, community onset | Health care associated, hospital onset | ||
| All cases | 210 | 50 | 115 | 45 | |
| Source | |||||
| CVC | 50 (24) | 0 (0) | 34 (30) | 16 (36) | <0.001 |
| Graft infection | 8 (4) | 0 (0) | 6 (5) | 2 (4) | NS |
| Infective endocarditis | 19 (9) | 12 (24) | 6 (5) | 1 (2) | <0.001 |
| Skin/wound | 52 (25) | 21 (42) | 28 (24) | 3 (7) | <0.001 |
| Respiratory tract | 22 (10) | 5 (10) | 10 (9) | 7 (16) | NS |
| Genitourinary tract | 9 (4) | 2 (4) | 6 (5) | 1 (2) | NS |
| Intra-abdominal | 3 (1) | 0 (0) | 3 (3) | 0 (0) | NS |
| Multiple | 5 (2) | 2 (4) | 1 (1) | 2 (4) | NS |
| Unknown | 42 (20) | 8 (16) | 21 (18) | 13 (29) | NS |
| PFGE type | |||||
| USA100 | 104 (50) | 7 (14) | 70 (61) | 27 (60) | <0.001 |
| USA300 | 74 (35) | 39 (78) | 26 (23) | 9 (20) | <0.001 |
| USA400 | 2 (1) | 1 (2) | 1 (1) | 0 (0) | NS |
| USA500 | 2 (1) | 1 (2) | 1 (1) | 0 (0) | NS |
| USA600 | 8 (4) | 0 (0) | 7 (6) | 1 (2) | NS |
| USA700 | 1 (0) | 0 (0) | 1 (1) | 0 (0) | NS |
| USA900 | 1 (0) | 0 (0) | 1 (1) | 0 (0) | NS |
| Unique | 18 (9) | 2 (4) | 8 (7) | 8 (18) | 0.037 |
| SCCmec type | |||||
| II | 103 (49) | 6 (12) | 71 (62) | 26 (58) | <0.001 |
| IV | 101 (48) | 42 (84) | 43 (37) | 16 (36) | <0.001 |
| Other | 6 (3) | 2 (4) | 1 (1) | 3 (7) | NS |
| agr group | |||||
| I | 90 (43) | 40 (80) | 36 (31) | 14 (31) | <0.001 |
| II | 118 (56) | 9 (18) | 78 (68) | 31 (69) | <0.001 |
| III | 2 (1) | 1 (2) | 1 (1) | 0 (0) | NS |
| PVL toxin gene | 76 (36) | 40 (80) | 27 (23) | 9 (20) | <0.001 |
Statistical comparisons were made between the three epidemiological groups (community associated, health care associated—community onset, and health care associated—hospital onset. NS, not significant; CVC, central venous catheter.
Ninety percent of the isolates had a Vitek MIC of ≤1.0 mg/liter; however, only 5% of the isolates had an E-test MIC of ≤1.0 mg/liter. Of the 190 isolates with a vancomycin Vitek MIC of ≤1.0 mg/liter, 6%, 78%, and 16% had an E-test MIC of 1.0, 1.5, and 2.0 mg/liter, respectively. All of the isolates with a vancomycin Vitek MIC of 2.0 mg/liter had an E-test MIC of either 1.5 (30%) or 2.0 mg/liter (70%). Thirteen of the 210 isolates (6.2%) were heteroresistant to vancomycin according to the macrodilution method E-test, with increasing distribution across higher vancomycin MICs; 5 out of 154 isolates (3.2%) with a vancomycin E-test MIC of 1.5 mg/liter were heteroresistant to vancomycin, while 8 out of 45 isolates (18%) with a MIC of 2.0 mg/liter were heteroresistant (P = 0.002). Twelve of the thirteen heteroresistant isolates were in patients with health care risk factors, with eight cases of community onset.
All isolates were susceptible to daptomycin, with 3%, 52%, and 45% having MICs of 0.25, 0.5, and 1.0 mg/liter, respectively. Using Spearman's correlation to examine the relationship between vancomycin E-test and daptomycin E-test MICs, a significant (P < 0.001) correlation of ρ = 0.286 was found. Daptomycin MICs were 0.25, 0.5, and 1.0 mg/liter in 9%, 55%, and 36%, respectively, of the 11 isolates with a vancomycin MIC of 1.0 mg/liter. Daptomycin MICs were 0.25, 0.5, and 1.0 mg/liter in 4%, 59%, and 37%, respectively, of the 154 isolates with a vancomycin MIC of 1.5 mg/liter and 0%, 27% and 73%, respectively, of 45 isolates with a vancomycin MIC of 2.0 mg/liter.
Results of PFGE showed 17 strain types. Of the 210 isolates, 104 (49.5%) were CDC strain USA100 and 74 (35%) were USA300. Comparison of the USA100 and USA300 cases showed significant differences in the comorbid conditions, epidemiologic source, and source of bacteremia (Table 3). Chronic kidney disease, hemodialysis, cardiovascular disease, neurologic disease, and malignancy were significantly more common with USA100 infections, while liver disease and the presence of no comorbidities were associated with USA300 infections. Intravenous drug use (IVDU) was more common in the USA300 group.
TABLE 3.
Comparison of patients infected with CDC strain USA100 with those infected with USA300a
| Patient characteristic | Value for group infected withb:
|
P value | |
|---|---|---|---|
| USA100 | USA300 | ||
| Total patients | 104 | 74 | |
| Median age (yr) | 59 | 47 | <0.001 |
| Male sex | 52 (50) | 46 (62) | NS |
| Race | |||
| Black | 75 (72) | 51 (69) | NS |
| White | 28 (27) | 21 (28) | NS |
| Other | 1 (1) | 2 (3) | NS |
| Comorbidity | |||
| None | 4 (4) | 14 (19) | 0.001 |
| HIV | 2 (2) | 4 (5) | NS |
| Diabetes mellitus | 43 (41) | 24 (32) | NS |
| Chronic kidney disease | 45 (43) | 14 (19) | <0.001 |
| Hemodialysis | 31 (30) | 8 (11) | 0.003 |
| COPD | 11 (11) | 6 (8) | NS |
| Cardiovascular disease | 56 (54) | 13 (18) | <0.001 |
| Neurologic disease | 30 (29) | 10 (14) | 0.016 |
| Liver disease | 16 (15) | 30 (41) | <0.001 |
| Malignancy | 24 (23) | 8 (11) | 0.036 |
| IVDU | 4 (4) | 37 (50) | <0.001 |
| Epidemiologic source | |||
| Community associated | 7 (7) | 39 (53) | <0.001 |
| Health care associated, community onset | 70 (67) | 26 (35) | <0.001 |
| Health care associated, hospital onset | 27 (26) | 9 (12) | 0.024 |
| Source | |||
| CVC | 36 (35) | 9 (12) | 0.001 |
| Graft infection | 6 (6) | 1 (1) | NS |
| Infective endocarditis | 4 (4) | 14 (19) | 0.001 |
| Skin/wound | 19 (18) | 24 (32) | 0.030 |
| Respiratory tract | 9 (9) | 6 (8) | NS |
| Genitourinary tract | 8 (8) | 1 (1) | 0.054 |
| Intra-abdominal | 3 (3) | 0 (0) | NS |
| Multiple | 2 (2) | 3 (4) | NS |
| Unknown | 17 (16) | 16 (22) | NS |
| Vancomycin E-test MIC | |||
| 1.0 | 4 (4) | 6 (8) | NS |
| 1.5 | 71 (68) | 62 (84) | 0.019 |
| 2.0 | 29 (28) | 6 (8) | 0.001 |
| Daptomycin MIC | |||
| 0.25 | 6 (6) | 0 (0) | 0.037 |
| 0.5 | 60 (58) | 38 (51) | NS |
| 1.0 | 38 (37) | 36 (49) | NS |
| Vancomycin heteroresistance | 7 (7) | 1 (1) | 0.086 |
NS, not significant; COPD, chronic obstructive pulmonary disease.
Values are no. (%) of patients unless otherwise noted.
Eighty-five percent of the USA100 isolates had SCCmec type II, while the remaining was type IV. All of these were agr group II isolates. All USA300 isolates except one had SCCmec type IV (only two isolates were not subtypes a to d), all with agr group I. There were eight USA600 isolates, all with SCCmec II and agr I. Nine percent of the isolates could not be classified into any previously published CDC strain type. Seven, seven, and one isolate had SCCmec type I, II, and V, respectively, while the remaining three isolates did not have types I to V. Twelve isolates belonged to agr group II and the rest to agr group I. All of the USA300 and USA400 isolates were positive for the PVL toxin genes. PVL was not present in other PFGE groups. Ninety-nine percent of the USA300 isolates were susceptible to trimethoprim-sulfamethoxazole, while 97% were susceptible to clindamycin, with only one isolate having inducible clindamycin resistance as determined by use of the D-zone test. Results of vancomycin in vitro susceptibility testing in relation to PFGE strain type are shown in Table 3. A vancomycin E-test result of 1.5 was more common for USA300, whereas an E-test result of 2.0 was more common for USA100. The PFGE distribution of the vancomycin-heteroresistant isolates was as follows: seven USA100 isolates, four USA600 isolates, one USA300 isolate, and one unique PFGE strain type. Seven of these isolates had agr group II, and the remaining had agr group I.
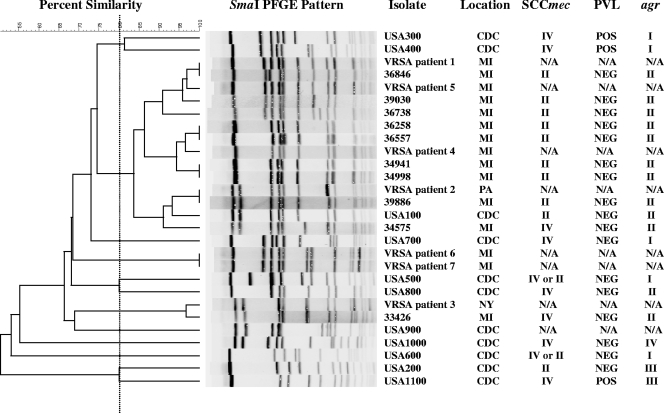
A comparison of USA100 strains showed that strains varied from zero to three bands. When USA100 MRSA strains were compared with the VRSA isolates, 31 out of the 104 USA100 MRSA isolates were very similar to or differed by only 1 or 2 bands from four of the VRSA isolates in the USA100 lineage (Fig. 1). One MRSA isolate was indistinguishable from the VRSA strain isolated in Pennsylvania. One unique MRSA strain was also very similar to the VRSA isolate from New York.
FIG. 1.
Dendrogram of selected clinical MRSA bacteremia isolates, CDC USA100 to -1100, and the first seven VRSA isolates in the United States obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus showing PFGE results, location of isolation, SCCmec types, PVL toxin gene presence, and agr group. Abbreviations: N/A, not available; CDC, Centers for Disease Control and Prevention; MI, Michigan; PA, Pennsylvania; NY, New York; POS, positive; NEG, negative.
DISCUSSION
Antimicrobial resistance in S. aureus is an issue of increasing clinical importance. Vancomycin MICs have been reported to have increased yet have remained within the susceptible range over time (41, 48). Our investigation supports these observations and gives the highest rate reported to date, with 73% and 21% of MRSA isolated from blood overall during the period 2005 to 2007 having MICs of 1.5 and 2.0 mg/liter, respectively. Earlier studies have reported bacteremia isolates with a MIC of >1.0 mg/liter, representing 8.6 to 74% of isolates. (13, 25, 40).
We found there to be very poor correlation between the vancomycin MIC determined by automated dilution testing and the E-test MIC, with discordant results for 85% of 210 isolates tested. These findings have important implications, including the need to consider routine use of more-reliable testing methods, such as the E-test, to verify the MIC for serious infections. This may have the most important implications with serious infections such as bacteremia, since many reports now describe poor outcomes when the vancomycin MIC is >1.0 mg/liter (13, 23, 26, 33, 40). We also found there to be an association with higher vancomycin and daptomycin MICs; the clinical significance of this observation is unsettled. Previous investigators have reported a reduced susceptibility of S. aureus to daptomycin for isolates with a reduced susceptibility to vancomycin (4 to 16 mg/liter) (8, 29). More recently, a shift toward higher daptomycin MICs was noted for patients with previous exposure to vancomycin (25). Our study supported these observations and noted this shift even without specifically selecting for patients with prior vancomycin use.
Our study shows that there is significant heterogeneity of strain types causing bloodstream infections, with 17 strain types observed, including 10 strain types not previously reported using the CDC typing scheme. The predominant strain type for patients with health care risk factors for bacteremia remains USA100, and that for those without such risk factors is USA300; however, it is clear that these strains are not restricted to these clinical settings. This supports the findings of other investigators (9, 17, 36), indicating that clinical epidemiologic characteristics are increasingly less useful in stratifying patients by molecular strain type (11). As with the previous study done by Seybold et al. (36), strain USA300 accounted for more than a third of our bloodstream infections, 47% of which occurred in patients with health care risk factors and 12% of which were of hospital onset. IVDU, along with IVDU-related viral hepatitis, is typically more common with community-associated USA300 infections (12). The predominant source for infection with the USA300 strain is skin and soft-tissue infections, but a significant proportion of patients also have infective endocarditis, which underscores the increasing spectrum of illness due to this strain type. Central venous catheters remain the primary source for USA100 BSI but are also seen in USA300 infections. These data emphasize the emergence of USA300 strains as an important cause of health care-associated and nosocomial MRSA BSI. Importantly, the results of this study show that even with the introduction of strain USA300 into the hospital environment, USA300 isolates are still consistently susceptible in vitro to trimethoprim-sulfamethoxazole and clindamycin, with inducible clindamycin resistance as a very rare event in this cohort, consistent with the first reports of the susceptibility of this strain (45).
USA100 isolates have traditionally been described as predominant in the hospital setting. We found these strains to be present in 14% of our patients without any known health care risk factors. As expected, comorbidities including chronic kidney disease, hemodialysis, cardiovascular disease, neurologic disease, and malignancy are significantly more common with this strain. All of the USA100 bloodstream isolates in this study had agr group II, which has been associated with poorer clinical outcomes in previous studies (26).
Southeastern Michigan is unique in that the first patient with a vancomycin-intermediate S. aureus strain in North America was identified in that location (2), as were seven of the nine known VRSA cases in the United States (38, 53; also unpublished observation). The reason for these findings is undetermined. In a recent paper from Zhu et al. (53), those authors identified an Inc18-like, vanA-encoding conjugative plasmid in vancomycin-resistant Enterococcus as a putative likely factor in the emergence of VRSA. Other clinical or molecular characteristics of S. aureus also need to be considered, such as recognized reservoirs for infection or a unique strain of MRSA that is hypersusceptible to gene transfer (43). Microbial biofilms (49) and skin/wound infections allowing cell-to-cell contact and facilitating gene transfer also have to be considered. USA100 is known to be the predominant hospital-related strain type worldwide; however, the amount of molecular diversity within this strain type has not been previously evaluated. In the present study, USA100 was found to be the most common cause of MRSA bacteremia and was present as a cause of infection both in and out of the hospital setting. We found that USA100 strains varied by up to three bands by PFGE and yet remained within the same group. Almost 30% of our USA100 isolates, however, were closely related to VRSA isolates. Given that the USA100 lineage is the most commonly isolated, that these infections frequently occur in patients with indwelling medical devices, which are notorious for biofilm formation, and that in the present study we found skin/wound infections implicated as a source in 18% of USA100 infections, along with its emergence in the community, MRSA strains that are susceptible to gene transfer from enterococci will continue to need to be carefully monitored.
Strains of MRSA having a vancomycin MIC of 2.0 mg/liter were significantly more common with USA100 isolates versus USA300 isolates, but 84% of the USA300 isolates had an E-test MIC of 1.5 mg/liter, which would be clinically reported as 2.0 mg/liter by CLSI standards. This implies that higher MICs that have been associated with a poor outcome are not restricted to strain types associated with health care-associated infections. Heteroresistance to vancomycin was more likely to occur with the USA100 strain than with the USA300 strain, but this did not reach statistical significance (P = 0.086). In earlier studies, VISA and heteroresistant VISA have also been found to be most commonly associated with agr group II (31, 32, 33); however, this was not supported by our investigation. The above findings may be due to the small number of heteroresistant strains in our study. In this study, heteroresistance testing was done with the macrodilution method E-test. Earlier studies have verified good correlation between this test and the gold standard population analysis profile-area under the curve ratio method (47). The results of our study suggest the macrodilution E-strip may have underestimated the rate of vancomycin heteroresistance. Our findings may also have been due to strain variability, since the likelihood of heteroresistance to vancomycin with MICs of 1.5 to 2.0 mg/liter was found to be much higher in some studies (34, 46) but lower in others (21, 44). Heteroresistance to vancomycin was not restricted to USA100 and USA300, since 50% of USA600 strains were heteroresistant to vancomycin, which has not been reported in earlier studies.
Since all data were derived from a single institution, the applicability to other facilities is limited. The population studied was predominantly urban and African-American, so the applicability to other ethnicities is not known. The importance of the findings of this study, however, are that MRSA has been common in Detroit earlier than in other parts of the world and that southeastern Michigan has been the source of the first VISA isolate in North America and has seven out the nine known VRSA isolates in the United States at this time.
Having an understanding of the epidemiology of MRSA in an urban area where MRSA has been common for years has important implications for developing rational measures for control. Infection control measures should focus not only on prevention of the spread of community strains in the hospital but also on prevention of the spread of hospital strains associated with VRSA into the community.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Boyce, J. M., R. L. White, and E. Y. Spruill. 1983. Impact of methicillin-resistant Staphylococcus aureus on the incidence of nosocomial staphylococcal infections. J. Infect. Dis. 148763. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 1997. Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morb. Mortal. Wkly. Rep. 46765-766. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51565-567. [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing: 15th informational supplement. Document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Cosgrove, S. E., Y. Qi, K. S. Kaye, S. Harbarth, A. W. Karchmer, and Y. Carmeli. 2005. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect. Control Hosp. Epidemiol. 26166-174. [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 3653-59. [DOI] [PubMed] [Google Scholar]
- 7.Crum, N. F., R. U. Lee, S. A. Thornton, O. C. Stine, M. R. Wallace, C. Barrozo, A. Keefer-Norris, S. Judd, and K. L. Russell. 2006. Fifteen-year study of the changing epidemiology of methicillin-resistant Staphylococcus aureus. Am. J. Med. 119943-951. [DOI] [PubMed] [Google Scholar]
- 8.Cui, L., E. Tominaga, H. M. Neoh, and K. Hiramatsu. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 501079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, S. L., M. B. Perri, S. M. Donabedian, C. Manierski, A. Singh, D. Vager, N. Z. Haque, K. Speirs, R. R. Muder, B. Robinson-Dunn, M. K. Hayden, and M. J. Zervos. 2007. Epidemiology and outcomes of community-associated methicillin-resistant Staphylococcus aureus infection. J. Clin. Microbiol. 451705-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowler, V. G., Jr., J. M. Miro, B. Hoen, C. H. Cabell, E. Abrutyn, E. Rubinstein, G. R. Corey, D. Spelman, S. F. Bradley, B. Barsic, P. A. Pappas, K. J. Anstrom, D. Wray, C. Q. Fortes, I. Anguera, E. Athan, P. Jones, J. T. van der Meer, T. S. Elliott, D. P. Levine, A. S. Bayer, and ICE Investigators. 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 2933012-3021. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez, B. E., A. M. Rueda, S. A. Shelburne III, D. M. Musher, R. J. Hamill, and K. G. Hulten. 2006. Community-associated strains of methicillin-resistant Staphylococccus aureus as the cause of healthcare-associated infection. Infect. Control Hosp. Epidemiol. 271051-1056. [DOI] [PubMed] [Google Scholar]
- 12.Haque, N. Z., S. L. Davis, C. L. Manierski, D. Vager, S. M. Donabedian, M. B. Perri, R. Sabbagh, F. Cheema, and M. J. Zervos. 2007. Infective endocarditis caused by USA300 methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 3072-77. [DOI] [PubMed] [Google Scholar]
- 13.Hidayat, L. K., D. I. Hsu, R. Quist, K. A. Shriner, and A. Wong-Beringer. 2006. High-dose vancomycin therapy for methicillin-resistant Staphylococcus aureus infections: efficacy and toxicity. Arch. Intern. Med. 1662138-2144. [DOI] [PubMed] [Google Scholar]
- 14.Horan, T. C., and R. P. Gaynes. 2004. Surveillance of nosocomial infections, p. 1672-1689. In C. G. Mayhall (ed.), Hospital epidemiology and infection control, 3rd ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 15.Johnson, J. K., T. Khoie, S. Shurland, K. Kreisel, O. C. Stine, and M. C. Roghmann. 2007. Skin and soft tissue infections caused by methicillin-resistant Staphylococcus aureus USA300 clone. Emerg. Infect. Dis. 131195-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein, E., D. L. Smith, and R. Laxminarayan. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg. Infect. Dis. 131840-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klevens, R. M., M. A. Morrison, S. K. Fridkin, A. Reingold, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, G. Fosheim, L. K. McDougal, F. C. Tenover, and Active Bacterial Core Surveillance of the Emerging Infections Program Network. 2006. Community-associated methicillin-resistant Staphylococcus aureus and healthcare risk factors. Emerg. Infect. Dis. 121991-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, S. K. Fridkin, and Active Bacterial Core Surveillance (ABCs) MRSA Investigators. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 19.Kuehnert, M. J., H. A. Hill, B. A. Kupronis, J. I. Tokars, S. L. Solomon, and D. B. Jernigan. 2005. Methicillin-resistant Staphylococcus aureus hospitalizations, United States. Emerg. Infect. Dis. 11868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 291128-1132. [DOI] [PubMed] [Google Scholar]
- 21.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 473040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodise, T. P., and P. S. McKinnon. 2005. Clinical and economic impact of methicillin resistance in patients with Staphylococcus aureus bacteremia. Diagn. Microbiol. Infect. Dis. 52113-122. [DOI] [PubMed] [Google Scholar]
- 23.Maclayton, D. O., K. J. Suda, K. A. Coval, C. B. York, and K. W. Garey. 2006. Case-control study of the relationship between MRSA bacteremia with a vancomycin MIC of 2 microg/ml and risk factors, costs, and outcomes in inpatients undergoing hemodialysis. Clin. Ther. 281208-1216. [DOI] [PubMed] [Google Scholar]
- 24.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 415113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moise, P. A., D. S. Smyth, N. El-Fawal, D. A. Robinson, P. N. Holden, A. Forrest, and G. Sakoulas. 2008. Microbiological effects of prior vancomycin use in patients with methicillin-resistant Staphylococcus aureus bacteraemia. J. Antimicrob. Chemother. 6185-90. [DOI] [PubMed] [Google Scholar]
- 26.Moise-Broder, P. A., G. Sakoulas, G. M. Eliopoulos, J. J. Schentag, A. Forrest, and R. C. Moellering, Jr. 2004. Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. 381700-1705. [DOI] [PubMed] [Google Scholar]
- 27.National Nosocomial Infections Surveillance System. 2004. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control. 32470-485. [DOI] [PubMed] [Google Scholar]
- 28.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 404289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel, J. B., L. A. Jevitt, J. Hageman, L. C. McDonald, and F. C. Tenover. 2006. An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin. Infect. Dis. 421652-1653. [DOI] [PubMed] [Google Scholar]
- 30.Pfaller, M. A., R. N. Jones, G. V. Doern, and K. Kugler. 1998. Bacterial pathogens isolated from patients with bloodstream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program (United States and Canada, 1997). Antimicrob. Agents Chemother. 421762-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., R. P. Novick, L. Venkataraman, C. Wennersten, P. C. DeGirolami, M. J. Schwaber, and H. S. Gold. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187929-938. [DOI] [PubMed] [Google Scholar]
- 32.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 461492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakoulas, G., P. A. Moise-Broder, J. Schentag, A. Forrest, R. C. Moellering, Jr., and G. M. Eliopoulos. 2004. Relationship of MIC and bactericidal activity to efficacy of vancomycin for treatment of methicillin-resistant Staphylococcus aureus bacteremia. J. Clin. Microbiol. 422398-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sancak, B., S. Ercis, D. Menemenlioglu, S. Colakoglu, and G. Hasçelik. 2005. Methicillin-resistant Staphylococcus aureus heterogeneously resistant to vancomycin in a Turkish university hospital. J. Antimicrob. Chemother. 56519-523. [DOI] [PubMed] [Google Scholar]
- 35.Saravolatz, L. D., N. Markowitz, L. Arking, D. Pohlod, and E. Fisher. 1982. Methicillin-resistant Staphylococcus aureus. Epidemiologic observations during a community-acquired outbreak. Ann. Intern. Med. 9611. [DOI] [PubMed] [Google Scholar]
- 36.Seybold, U., E. V. Kourbatova, J. G. Johnson, S. J. Halvosa, Y. F. Wang, M. D. King, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin. Infect. Dis. 42647-656. [DOI] [PubMed] [Google Scholar]
- 37.Shurland, S., M. Zhan, D. D. Bradham, and M. C. Roghmann. 2007. Comparison of mortality risk associated with bacteremia due to methicillin-resistant and methicillin-susceptible Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 28273-279. [DOI] [PubMed] [Google Scholar]
- 38.Sievert, D., J. T. Rudrik, J. B. Patel, L. C. McDonald, M. J. Wilkins, and J. C. Hageman. 2008. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin. Infect. Dis. 46668-674. [DOI] [PubMed] [Google Scholar]
- 39.Singh, A., R. V. Goering, S. Simjee, S. L. Foley, and M. J. Zervos. 2006. Application of molecular techniques to the study of hospital infection. Clin. Microbiol. Rev. 19512-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soriano, A., F. Marco, J. A. Martínez, E. Pisos, M. Almela, V. P. Dimova, D. Alamo, M. Ortega, J. Lopez, and J. Mensa. 2008. Influence of vancomycin minimum inhibitory concentration on the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 46193-200. [DOI] [PubMed] [Google Scholar]
- 41.Steinkraus, G., R. White, and L. Friedrich. 2007. Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001-05. J. Antimicrob. Chemother. 60788-794. [DOI] [PubMed] [Google Scholar]
- 42.Strommenger, B., C. Cuny, G. Werner, and W. Witte. 2004. Obvious lack of association between dynamics of epidemic methicillin-resistant Staphylococcus aureus in central Europe and agr specificity groups. Eur. J. Clin. Microbiol. Infect. Dis. 2315-19. [DOI] [PubMed] [Google Scholar]
- 43.Sung, J. M., and J. A. Lindsay. 2007. Staphylococcus aureus strains that are hypersusceptible to resistance gene transfer from enterococci. Antimicrob. Agents Chemother. 512189-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tallent, S. M., T. Bischoff, M. Climo, B. Ostrowsky, R. P. Wenzel, and M. B. Edmond. 2002. Vancomycin susceptibility of oxacillin-resistant Staphylococcus aureus isolates causing nosocomial bloodstream infections. J. Clin. Microbiol. 402249-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenover, F. C., and R. C. Moellering, Jr. 2007. The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin. Infect. Dis. 441208-1215. [DOI] [PubMed] [Google Scholar]
- 47.Walsh, T. R., A. Bolmström, A. Qwärnström, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 392439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, G., J. F. Hindler, K. W. Ward, and D. A. Bruckner. 2006. Increased vancomycin MICs for Staphylococcus aureus clinical isolates from a university hospital during a 5-year period. J. Clin. Microbiol. 443883-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weigel, L. M., R. M. Donlan, D. H. Shin, B. Jensen, N. C. Clark, L. K. McDougal, W. Zhu, K. A. Musser, J. Thompson, D. Kohlerschmidt, N. Dumas, R. J. Limberger, and J. B. Patel. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob. Agents Chemother. 51231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39309-317. [DOI] [PubMed] [Google Scholar]
- 51.Wootton, M., A. P. MacGowan, T. R. Walsh, and R. A. Howe. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 45329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 435026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, W., N. C. Clark, L. K. McDougal, J. Hageman, L. C. McDonald, and J. B. Patel. 2008. Vancomycin-resistant Staphylococcus aureus associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]