Abstract
Cellulose synthase and cellulose synthase-like proteins, responsible for synthesizing β-glucan-containing polysaccharides, play a fundamental role in cellular architectures, such as plant cell and tissue morphogenesis, bacterial biofilm formation, and fruiting-body development. However, the roles of the proteins involved in the developmental process are not well understood. Here, we report that a cellulose synthase-like protein (CslASc) in Streptomyces has a function in hyphal tip growth and morphological differentiation. The cslASc replacement mutant showed pleiotropic defects, including the severe delay of aerial-hyphal formation and altered cell wall morphology. Calcofluor white fluorescence analysis demonstrated that polysaccharide synthesis at hyphal tips was dependent on CslASc. cslASc was constitutively transcribed, and an enhanced green fluorescent protein-CslASc fusion protein was mostly located at the hyphal tips. An extract enriched in morphogenetic chaplin proteins promoted formation of aerial hyphae by the mutant. Furthermore, a two-hybrid experiment indicated that the glycosyltransferase domain of CslASc interacted with the tropomyosin-like polarity-determining DivIVA protein, suggesting that the tip-located DivIVA governed tip recruitment of the CslASc membrane protein. These results imply that the cellulose synthase-like protein couples extracellular and cytoskeletal components functioning in tip growth and cell development.
Cellulose is considered to be the most abundant macromolecule on earth, and cellulose synthase proteins exist in many bacterial species, most groups of algae, the slime mold Dictyostelium, plants, and tunicates. In plants, cellulose synthases and cellulose synthase-like proteins are responsible for cell wall synthesis and are essential for growth and tissue development. In Arabidopsis thaliana, the 10-member cellulose synthase A (CesA) gene family (CesA1 to CesA10) is involved in primary and secondary cell wall synthesis (28). Furthermore, there are another six families of cellulose synthase-like genes (Csl) in Arabidopsis, CslA, CslB, CslC, CslD, CslE, and CslG (28), whose functions are still unknown. It is thought that they may be involved in tissue-specific expression and/or response to environmental stresses. For instance, the Csl gene KOJAK/AtCSLD3 is expressed in hair cells of the epidermis and is involved in the biosynthesis of β-glucan-containing polysaccharides that are required during root hair elongation (14).
The gram-negative bacterium Gluconacetobacter xylinus (formerly Acetobacter xylinum) has long been the model organism for the study of bacterial cellulose biosynthesis (30). Cellulose in bacteria often acts as the extracellular polysaccharide matrix and is associated with the formation of cell aggregates. Cellulose fibrils are also important during biofilm formation as a component of the extracellular matrices of many bacterial species, including Salmonella enterica serovar Typhimurium and Escherichia coli (41). In G. xylinus and enterobacteria, genes determining cellulose biosynthesis are organized in an operon (bcs, for bacterial cellulose synthesis). The cellulose synthase encoded by bcsA catalyzes the polymerization of UDP-glucose by forming β-1,4 glucosidic bonds. The activity of BcsA is allosterically regulated by a cyclic di-GMP binding protein encoded by bcsB. bcsZ encodes a cellulase (family 8 glucosidase), which is required for cellulose synthesis. The bcsC product is an unknown protein, but it is needed for cellulose production in vivo (40, 41). The cellulose-producing bacteria mentioned above all contain genes homologous to the enterobacterial bcsABZC operon on their chromosomes. Similar gene clusters are also present in other bacterial genomes (for instance, those of Burkholderia, Ralstonia, and Aquifex) (29).
Members of the genus Streptomyces are gram-positive soil-dwelling filamentous bacteria that undergo an ordered, complicated colony differentiation process. After spore germination, vegetative hyphae grow from the germ tubes by tip elongation and branching (7, 12, 18). On solid media, vegetative or substrate hyphae intrude into the agar to absorb nutrients for growth. Subsequently, a fluffy aerial mycelium emerges from the colony-air interface (5). In the model species, Streptomyces coelicolor, this transition requires the bld genes and two classes of surface-active molecules, SapB and the chaplins (5, 6, 8, 13, 38, 39). The aerial hyphae finally form long chains of spores. In shaking liquid culture, many Streptomyces species form pellets of tightly tangled hyphae. In standing liquid media, S. coelicolor cultures can show a biofilm-like growth (36).
The S. coelicolor genome has one gene (SCO2836) encoding a cellulose synthase-like protein (1). In this study, we show that this cellulose synthase-like protein is located at hyphal tips, apparently via an interaction with the polarity-determining protein DivIVA, and is involved in the deposition of β-linked glucan at the tips. Elimination of SCO2836 has marked effects on growth and development.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1. S. coelicolor strains were cultivated at 30°C on solid R2YE or MS agar or in TSB or YEME liquid medium, as described previously (21). E. coli DH5α and ET12567 (dam dcm hsdS) were grown and transformed by standard methods (31). ET12567 was used to propagate unmethylated DNA for introduction into S. coelicolor by transformation or conjugation (25).
TABLE 1.
Bacterial strains and oligonucleotides
| Strain or oligonucleotide | Characteristics | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 lacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 31 |
| ET12567/pUZ8002 | dam dcm hsdS cat tet tra neo RP4 | 21 |
| S. coelicolor | ||
| M145 | SCP1− SCP2− | 1 |
| XE | M145ΔcslASc::Hygr cassette | This work |
| Oligonucleotide primers for PCR amplification | ||
| pcslAsc5 | 5′ AATGGATCCTCACACTCCCGGTCGGCAGG3′ | |
| pcslAsc6 | 5′ TCTAGATTACATATGTCATTCCCCCCACACGCGGG3′ | |
| pcslAsc7 | 5′ ATTAGATCTGGTTCTGGTGGTTCTGGTATGACGTCGACGCCGACGGG3′ | |
| pcslAsc8 | 5′ TTATGCGGCCGCTTATCATTCCTTACGTCCCCCAAG3′ | |
| pramS1 | 5′ AGTAGATCTGCCGGTCGTGTCGTAGGTG3′ | |
| pramS2 | 5′ TCTAGATTACATATGTCCGAGGTGCGGAACGCAC3′ | |
| pdivIVA1 | 5′ AGTGGATCCGAGGACGTGCGGAACAAGCAG3′ | |
| pdivIVA2 | 5′ AGTGAATTCCTCGTCGATCAGGAACCCGCG3′ | |
| pcslAsc3 | 5′ AGTGAATTCTTCCTCACCTCCTTCGTGCCC3′ | |
| pcslAsc4 | 5′ AGTAGATCTCGAGCCGCCGATCTGCTTGAG3′ |
DNA manipulations.
Streptomyces genomic DNA was isolated according to a protocol described previously (21). Plasmid preparation, digestion, and ligation followed standard methods (31). For Southern analysis, digoxigenin labeling of DNA probes, hybridization, and detection were performed according to the protocols provided by the manufacturer (Roche). Restriction enzymes and other molecular biology reagents were from commercial sources.
Plasmid constructs for gene replacement, gene complementation, gene fusions, and two-hybrid analysis.
The cslASc gene replacement plasmid pHL151 was constructed as follows. A 3.8-kb BglII-BglII fragment from S. coelicolor cosmid SCE20 containing cslASc was amplified and subcloned into pOJIJ1 between the BglII and BamHI sites (21), yielding pBBE20 (data not published). pBBE20 was digested with BamHI and ligated with the BamHI hyg cassette from pHP45 Ω hyg (3), giving pHL151, in which a 756-bp internal BamHI fragment encoding amino acids (aa) 189 to 439 of the CslASc protein was replaced by the hyg cassette encoding hygromycin resistance. The 2.9-kb PvuII-BglII fragment from pBBE20 was also inserted between the BamHI and EcoRV sites of pSET152 (21) to yield pHL155 for complementation of the cslASc mutant. pHL155 has the ΦC31 phage integration function region and can therefore integrate into the S. coelicolor chromosome.
To translationally fuse egfp to the 5′ end of cslASc, the 551-bp promoter region of cslASc was amplified from genome DNA using the primers pcslAsc5 and pcslAsc6 and then digested with BamHI and NdeI and cloned between the BglII and NdeI sites of pHL117, which has the ΦC31 phage integration function region and can integrate into the S. coelicolor chromosome (23), generating pHL177. Then, the NdeI-NotI egfp gene fragment excised from pHL167 (data not published) was ligated between the NdeI and NotI sites of pHL177, generating pHL178. The whole cslASc gene was amplified from genome DNA using the primers pcslAsc7 and pcslAsc8, digested with BglII and NotI, and cloned between the BglII and NotI sites of pHL178, generating pHL179, in which the enhanced green fluorescence protein (EGFP) was translationally fused to the N terminus of CslAsc in frame and a linker (GSGGSG) was added between EGFP and the CslASc protein.
To construct a transcriptional fusion of the ram gene cluster promoter to egfp, the PCR product generated using the primers pramS1 and pramS2 was digested with BglII and NdeI and cloned between the BglII and NdeI sites of pHL117 (23). This generated pHL171, which contains the major promoter of the ram gene cluster, Piram (20).
pKF59 (a gift from Klas Flardh) is an integrative plasmid containing a divIVA-egfp translational fusion gene (15). It was digested with StuI and cloned at the SmaI site of pOJ260 (2). This generated pHL175, which has an RP4-derived origin of transfer (oriT) region to facilitate E. coli-Streptomyces conjugation.
Plasmids for bacterial two-hybrid analysis were constructed as follows. The divIVA gene PCR product generated using the primers pdivIVA1 and pdivIVA2 was digested with BamHI and EcoRI and cloned between the BamHI and EcoRI sites of the “target” plasmid, pTRG, generating plasmid pHL172. The glycosyltransferase-encoding fragment of cslASc was amplified by PCR using the primers pcslAsc3 and pcslAsc4. The product was digested with BamHI and EcoRI and cloned between the same sites of the “bait” plasmid, pBT, giving pHL173.
cslASc gene replacement and genetic complementation.
pHL151 was introduced into M145 from ET12567/pUZ8002 by E. coli-Streptomyces intergeneric conjugation as described previously (21). Aprr Hygr exconjugants were transferred to selective agar medium containing nalidixic acid and hygromycin, allowed to sporulate, and then transferred to MS agar without antibiotic selection. Spores from the MS agar were then plated on MS agar containing hygromycin to obtain single colonies, which were replicated on agar media containing only hygromycin and both apramycin and hygromycin. Five Aprs Hygr colonies were obtained, all with the same phenotype on MS, MM, and R2YE agars. To confirm gene replacement in these isolates, Southern blots were performed with a digoxigenin-labeled 3.8-kb BglII/PvuII fragment containing cslASc as a probe. Complementation of the mutant was achieved by introducing pHL155 by conjugation. The vector pSET152 (21) was also introduced into the mutant and M145 to provide negative controls.
Microscopy.
Live hyphae were examined by light microscopy as described previously (17, 21). For fluorescence microscopy, the samples were prepared as described above and the hyphae were mounted in 50% glycerol before observation. For staining hyphae or DNA, samples of cells were fixed three times with methanol and then washed with water and covered with poly-l-lysine solution (21). The cells were stained with propidium iodide (PI) or calcofluor work solution for about 10 or 5 min, respectively, and then washed with water and mounted with 50% glycerol before observation under the Olympus BX51 fluorescence microscope (camera, Pixera penguin 150CL).
For transmission electron microscopy (TEM) analysis, the mycelium and spores were harvested from R2YE agar. The cells were fixed with freshly made 2% glutaraldehyde in phosphate buffer for 2 hours and washed with PBS three times; then, the samples were fixed with 1% osmium tetroxide and washed with PBS three times. The cells were embedded in Spuurr resin after dehydration with ethanol. Sections (ca. 50 to 60 nm thick) were examined with an H7650/Hitachi-H-7000 FA transmission electron microscope (21).
For scanning electron microscopy (SEM), about 5- by 5- or 10- by 10-mm pieces made from coverslips were laid flat on R2YE agar before the agar solidified. A little medium was added to the edges of the coverslips, and spores were inoculated at the edges. After sporulation, the samples were fixed with 1% osmium tetroxide for 3 hours. The samples were then examined with the JSM-6390 scanning electron microscope after being sputter coated with gold.
Two-hybrid bacterial analysis.
The BacterioMatch II two-hybrid system (Stratagene) was used to detect protein-protein interactions. The plasmid pairs were used to cotransform the XL1-Blue MRF′ reporter strain on M9 salt agar without 3-amino-1,2,4-triazole (3-AT). Colonies were then restreaked on M9 salt agar containing 5 mM 3-AT at 37°C for 24 h for the first detection of interactions. For confirmation, the colonies were cultured on dual-selective medium containing 5 mM 3-AT and streptomycin as described in the manual.
Purification of chaplin and rodlin proteins.
For purification of chaplin and rodlin proteins, the wild-type strain M145 was grown on cellophane discs on the surface of MS agar medium. After 4 days of growth, the hyphae were harvested and extracted using trifluoroacetic acid as described previously (8, 13).
RESULTS
Bioinformatic analysis of the “cellulose synthase-like” (cslASc) gene in S. coelicolor.
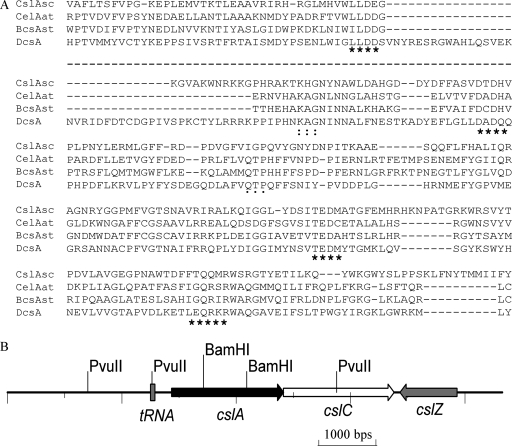
The S. coelicolor genome contains only one gene (SCO2836) encoding a cellulose synthase-like protein. This protein has weak similarity to the catalytic subunits of cellulose synthases from bacteria and plants. We therefore named SCO2836 cslASc (cellulose synthase-like gene of S. coelicolor). Orthologs of cslASc were also found in Streptomyces avermitilis MA-4680 (19) and Streptomyces nanchangensis (35). Structure prediction suggested that the protein has seven transmembrane domains, with the N terminus being cytoplasmic and the C terminus outside the membrane. The large cytoplasmic domain in the middle region contains domains that match entries pfam03552 (cellulose synthase) (E value, 2e−9), from aa 338 to 451, and PF00535 (glycosyltransferase family 2) (E value, 1e−7), from aa 166 to 342 (26). The central active site of cellulose synthases is thought to comprise four particularly conserved subregions (U1 through U4) that contain the conserved Asp (D) residues (in U1 to U3) and the motif QXXRW (in U4) (32). These are considered essential for substrate binding and catalysis (32), and they are present in CslASc (Fig. 1A). The A residue of a normally conserved KAG motif is replaced by H and a QTP motif is not conserved in CslASc, but the functions of these two motifs in cellulose synthase are not yet established.
FIG. 1.
The cslASc gene and comparison of its product with related proteins. (A) Alignment of the highly conserved U1, U2, U3, and U4 regions in the central cytoplasmic loop of the predicted S. coelicolor cellulose synthase-like protein with cellulose synthase protein sequences from Agrobacterium tumefaciens (CelAAt; NP_533806), S. enterica serovar Typhimurium (BcsA; CAC86199), and Dictyostelium discoideum (DcsA; AAF00200). The D, D, and D35QXXRW motifs; KAG motif; and QTP motif are indicated by asterisks, colons, and dots, respectively. The dashed line between the first and second blocks indicates that some regions that do not contain conserved motifs are omitted. (B) The csl genes in S. coelicolor. Compared with the operon of S. enterica serovar Typhimurium/E. coli, Streptomyces does not have the bcsB gene. The csl genes also exist in S. avermitilis.
In addition, cslASc and its downstream genes are organized in a way similar, but not identical, to the bcsABZC cellulose biosynthesis operons in enteric bacteria (41), which encode a cellulose synthase catalytic subunit (bcsA), a cellulose synthase regulatory subunit (bcsB), an endo-1,4-β-glucanase (bcsZ), and an oxidoreductase (bcsC) (41). In S. coelicolor, the immediately downstream gene SCO2837 encodes a secreted copper metalloenzyme that in vitro oxidizes simple alcohols to aldehydes and reduces dioxygen to hydrogen peroxide (37), while the next gene (SCO2838, which converges on SCO2836 and SCO2837) encodes a putative endo-1,4-β-glucanase (Fig. 1B). There is no homolog of bcsB, which encodes a regulatory subunit proposed to stimulate/activate BcsA upon binding of cyclic di-GMP (30).
Inactivation of cslASc affects mycelial differentiation.
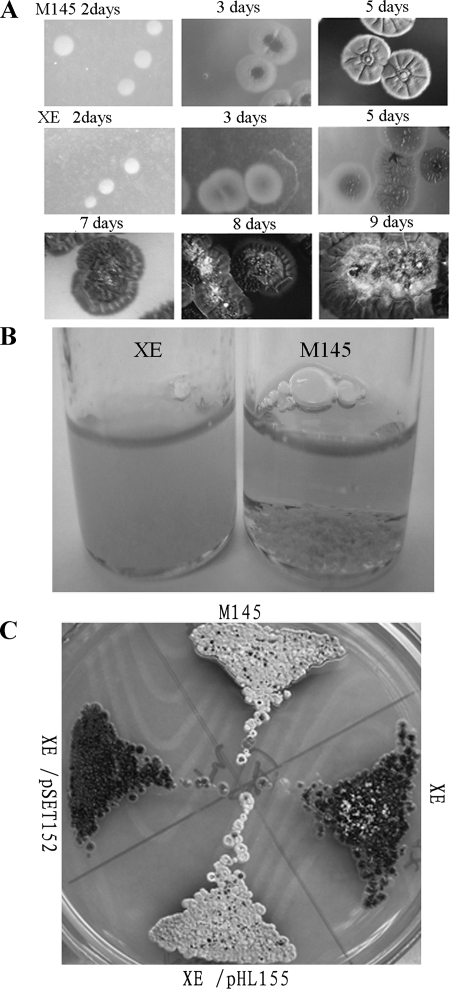
To address the function of cslASc, a mutant was constructed by replacing a BamHI fragment internal to the gene with a hygromycin resistance cassette. The mutant (named XE) produced a very sparse aerial mycelium only after 8 days on R2YE medium, while the wild type produced abundant aerial mycelium after 4 days (Fig. 2A). However, unlike many aerial-mycelium-deficient (bld) mutants, XE retained the ability to produce pigmented antibiotics under the conditions tested. In liquid culture, such as TSB or YEME, the mutant did not form the large aggregates (pellets) typical of the wild type (Fig. 2B), though phase-contrast microscopy confirmed that apparently normal branching mycelial material was abundant.
FIG. 2.
Disrupting cslASc in S. coelicolor. (A) Aerial-mycelium formation by the mutant was severely delayed on solid R2YE medium compared with that of the wild-type M145. (B) Clumping, and resulting sedimentation, of vegetative hyphae in liquid culture (TSB; 24 h) was less pronounced in the cslA mutant than in the wild-type M145. Mycelial clumps of M145 sedimented to the bottom quickly when the bottle was allowed to stand (right). (C) Genetic complementation of the cslA mutant. The mutant containing pHL155 formed normal aerial hyphae on solid R2YE medium, but the mutant containing the empty plasmid pSET152 did not. The wild-type M145, XE, and XE/pSET152 were used as controls.
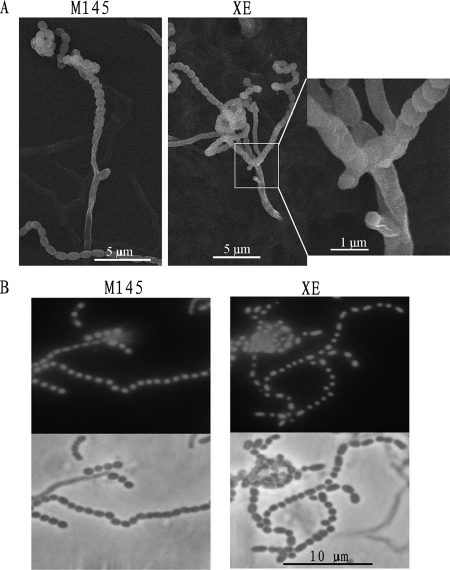
Light microscopy and SEM showed that sporulating hyphae branched from the supporting hyphae at unusually close intervals compared with the wild-type M145 (Fig. 3A). Fluorescence microscopy of samples stained with PI showed that the spores of the mutant all contained normal amounts of DNA (Fig. 3B). This indicated that DNA was replicated and segregated normally, although multiple sporulating hyphae developed.
FIG. 3.
The cslASc gene affected aerial-hyphal development. (A) Multiple closely spaced sporulating hyphae of the mutant often emerged from one supporting hypha. Usually, the sporulating hyphae of wild-type M145 were well separated. (B) Spores of the mutant and wild-type M145 stained with PI. Spores of sporulating hyphae of the mutant contained DNA with the same appearance as that of sporulating wild-type M145. DNA-free spores of the mutant were rare (R2YE solid medium; analysis of the mutant was delayed compared to the wild type until spore chains could be detected).
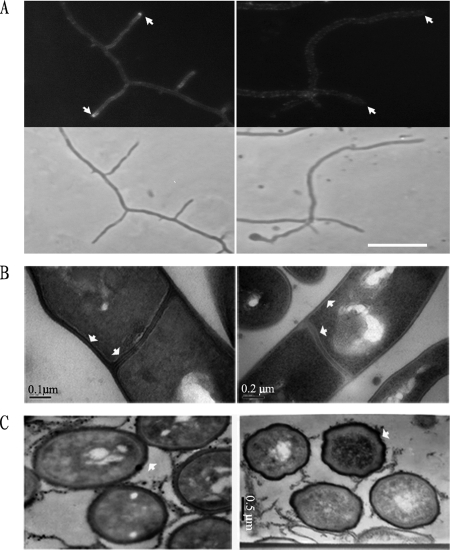
Further analysis by TEM revealed that the mutant had subtle changes in its cell wall and septum structure and spore shape (Fig. 4). The laminated appearance of wild-type septa and lateral walls was not seen in the mutant (Fig. 4B), and the walls of XE spores showed many small deformations compared with the smooth oval contour of the wild-type cell wall (Fig. 4C). Thus, the cslASc gene indeed contributes, directly or indirectly, to normal cell wall structure.
FIG. 4.
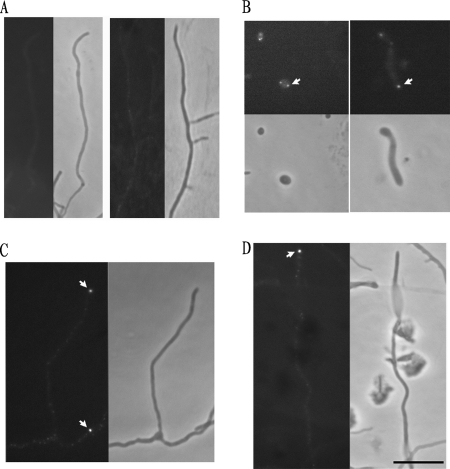
Calcofluor white staining and TEM analysis. (A) Calcofluor white staining revealed that β-1,4-linked polysaccharides accumulated at the tips of vegetative hyphae in the wild type (top left, arrows) but not in the XE mutant (top right). The lower panels show the equivalent phase-contrast images. The strains were cultured on MS solid medium. (B and C) Comparison by TEM of ultrathin sections of wild-type M145 and mutant XE (R2YE solid medium). Wild-type M145 showed normal and classic vegetative hyphal cell wall and septum appearance, with an electron-dense inner layer, while the mutant lacked such layers in its cell wall and septa (B, arrows). The mutant showed abnormal wrinkled spore walls (C, arrows).
To complement the mutant, we constructed pHL155, in which a 2.9-kb fragment containing the intact cslASc open reading frame, along with 0.5-kb upstream and 0.2-kb downstream sequences, was cloned into pSET152. The introduction of pHL155 into XE restored a wild-type appearance to colonies on solid media (Fig. 2C) and pellet formation in liquid culture, confirming that the mutant phenotype was the direct result of cslASc inactivation and ruling out phenotypically significant polarity effects of the disruption cassette on the expression of the possibly cotranscribed downstream gene SCO2837.
cslASc is involved in the accumulation of β-glucan at mycelial tips.
Group 2 glycosyltransferase and cellulose synthase-like proteins catalyze the biosynthesis of various β-linked polysaccharides, such as cellulose, chitin, and curdlan (4, 10). To examine the distribution of β-glucan in S. coelicolor and the XE mutant, we used calcofluor white, a fluorescent dye that specifically binds β-1,4 polysaccharides, such as chitin and cellulose. The vegetative hyphae were stained with calcofluor white after growth on MS solid medium for 16 h. The mycelium of the wild type was weakly fluorescently stained by the dye, but nearly all hyphal tips showed much more intense fluorescence (Fig. 4A). In the XE mutant, the tip-located brighter loci were abolished (Fig. 4A), although some small fluorescent foci were observed along the hyphae. These results showed that cslASc is involved in the preferential biosynthesis of certain β-1,4-linked polysaccharides at hyphal tips.
Fluorescently tagged CslASc protein accumulates at hyphal tips.
To examine the temporal and spatial pattern of CslASc abundance, the EGFP gene was fused in frame to the 5′ end of cslASc. A linker coding sequence was added between EGFP and cslASc, and the promoter region of the cslASc gene was cloned before the egfp gene. The generated reporter plasmid, pHL179, integrated into the chromosome by site-specific recombination at the bacteriophage ΦC31 attachment attB site, did not complement the XE mutant completely, indicating that the fusion protein had partial function. The plasmid pHL179 was introduced into wild-type M145, and the strain M145/pHL179 grew normally compared with M145. M145/pHL179 was grown on MS agar medium and observed at 4, 24, 40, and 60 h (Fig. 5). When spores began to germinate, they became large and round. At this stage, fluorescent foci became visible (Fig. 5B). After germination, fluorescent signals were seen at the tips of hyphae at different stages: germ tubes emerging from spores and vegetative and aerial hyphae (Fig. 5B, C, and D). The result showed that the CslASc protein mainly accumulated at hyphal tips. The C-terminal EGFP fusion was also tested, and the result was the same, with the caveat that the C-terminal domain was likely to be on the outside of the cell membrane, so the C-terminal EGFP fusion could have disturbed both the distribution of the CslASc protein and the activity of EGFP. Aerial hyphae of wild-type M145 were used as a control and showed no fluorescence (Fig. 5A).
FIG. 5.
Localization of CslASc-EGFP fusion protein. The CslASc-EGFP fusion protein was localized at hyphal tips. (A) Controls; aerial hypha (left) and vegetative hyphae (right) of wild-type M145 without an egfp fusion. (B, C, and D) M145/pHL179. (B) Left, spore germination; right, germ tube elongation. (C) Vegetative hyphae. (D) Aerial hyphae. (A, C, and D) Left, fluorescent images; right, phase-contrast images. (B) Top, fluorescent images; bottom, phase-contrast images. The arrows indicate the fluorescent foci at tips. The strains were grown on MS medium. Bar, 10 μm.
Protein-protein interaction between DivIVA and CslASc.
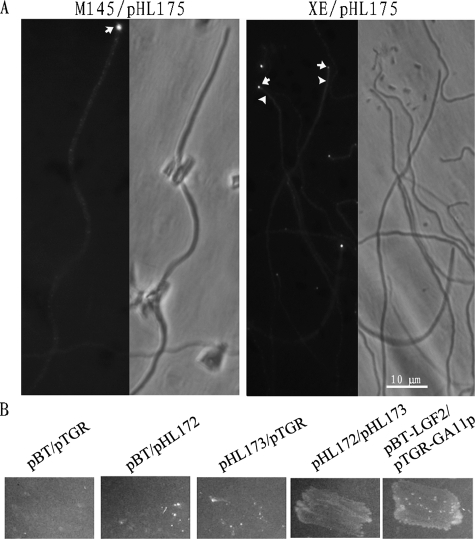
The tip growth and polarity of S. coelicolor are dependent on the tropomyosin-like protein DivIVA, which is principally localized at hyphal tips (15). In view of the apparent tip location of CslASc and the CslASc-dependent β-glucan, the cell wall and aerial growth abnormalities of the XE mutant, and the prediction that CslASc has a substantial cytoplasmic domain, we examined the possibility of a physical interaction between DivIVA and CslASc. First, we examined the location of the DivIVA protein in the mutant strain by fluorescence microscopy using a DivIVA-EGFP fusion protein. In young vegetative hyphae and most aerial hyphae, the distribution of fluorescence intensity at the tips was the same in both XE/pHL175 and M145/pHL175 (Fig. 6A), matching the previous observations (15). Intriguingly, though, in some presumptive young aerial hyphae of the XE mutant, some of the fluorescence was dispersed in the cell (Fig. 6A), suggesting that the CslASc protein might play a role in the location of the DivIVA protein at the time that aerial hyphae start to form. We therefore used the BacterioMatch II two-hybrid system (Stratagene) to investigate possible protein-protein interactions of DivIVA and CslASc. Although we were unable to successfully introduce the entire coding region of the CslASc protein into the bait vector, pBT, we succeeded in cloning the segment predicted to encode the cytoplasmic glycosyltransferase domain, yielding pHL173. The divIVA gene was amplified by PCR and fused to the target plasmid, pTRG, to yield pHL172. As shown in Fig. 6B, the strain with pHL173 and pHL172 (XL1-Blue MRF′/pHL173/pHL172) and the positive control strain (XL1-Blue MRF′/pBT-LGF2/pRGT-GAL11P) grew on the double-selective indicator plate containing 5 mM 3-AT and streptomycin, while the negative control strains (XL1-Blue MRF′/pBT/pRGT, XL1-Blue MRF′/pHL173/pTRG, and XL1-Blue MRF′/pBT/pHL172) did not grow, suggesting that the glycosyltransferase domain of the CslASc protein interacted with the DivIVA protein directly.
FIG. 6.
Interaction between DivIVASc and CslASc. (A) Localization of DivIVASc-EGFP protein in wild-type M145 and XE. The DivIVASc-EGFP protein was mostly at hyphal tips, with some smaller foci along the hyphae. In the XE mutant, the foci were also mostly at the tips (arrows), but in some aerial hyphae, the fluorescence was dispersed through the cell (arrowheads). (B) A bacterial two-hybrid experiment showed that DivIVA interacted with the glycosyltransferase domain of the CslASc protein. The reporter strain XL1-Blue MRF′ with different plasmid pairs was grown on double-selective indicator plates containing 3-AT and streptomycin. Experimental construct, pHL172/pHL173; positive control, pBT-LGF2/pRGT-GAL11P; negative controls, pBT/pRGT, pHL173/pTRG, and pBT/pHL172.
Examination of the expression of amphipathic aerial-hyphal surface proteins in the mutant.
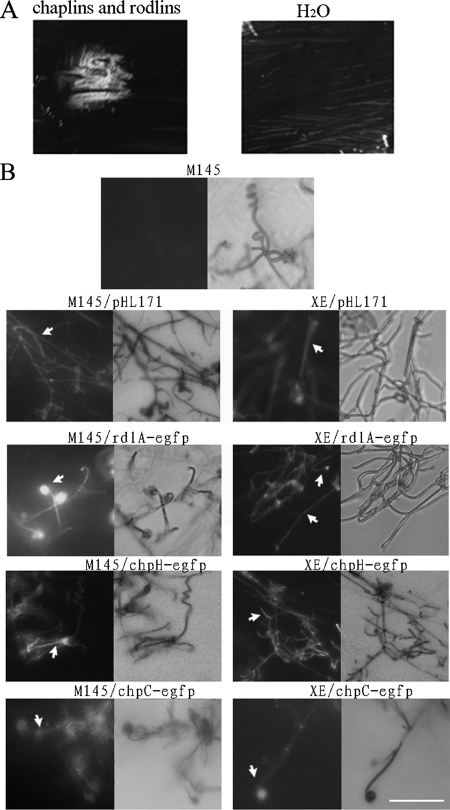
The aerial hyphae of S. coelicolor are covered by different surface-active proteins and peptides (chaplins, rodlins, and SapB) that facilitate aerial growth (8, 13, 38). The addition of such molecules to many kinds of bld mutants can result in partial suppression of the morphological defect. We determined whether the XE mutant could be phenotypically suppressed in this way. We first extracted the chaplin and rodlin proteins from wild-type M145 cultured on MS medium. The extract was expected to include both chaplin and rodlin proteins (8, 9, 13), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis verified that proteins of the expected electrophoretic mobility were present. The XE mutant was cultured for a day on R2YE solid medium, and then the extract was added. After 2 days, the mycelium formed aerial hyphae (Fig. 7A), suggesting that the chaplin and/or rodlin protein could restore aerial growth to the XE mutant. We then investigated the expression of genes for chaplins, rodlins, and SapB production in the mutant, using transcriptional fusions of the egfp reporter gene to the structural genes chpC, chpH, rdlA (8), and P1ram (pHL171). The P1 promoter is the principal promoter of the ramCSAB operon in Streptomyces lividans (20), and ramS encodes the precursor of SapB (22). Fluorescence microscopy revealed that chpC, chpH, rdlA, and P1ram were comparably expressed in the aerial hyphae of both strains (Fig. 7B). The fluorescence of chpH and rdlA was strong, and that of chpC was weak, which matched the previous observations (8, 9). These data implied that replacement of the cslASc gene did not interfere with the aerial-mycelium-specific expression of these genes.
FIG. 7.
Extracellular complementation of the XE mutant by a chaplin/rodlin extract and analysis of the expression of chaplin and rodlin hydrophobins in XE by EGFP fusions. (A) Extracellular chaplins and rodlins promoted the XE mutant to form aerial mycelium. The extracted chaplin and rodlin material or control material (water) was spotted on the lawns after 1 day of mycelial growth on R2YE solid medium. The extract was seen to promote the mutant to form aerial hyphae 1.5 days after addition. Water had no effect. The photographs were captured with a Fuji FinePix S602 2 days after addition. (B) EGFP fluorescence expressed from pHL171 derivatives carrying the P1ram-egfp, rdlA-egfp, chpH-egfp, and chpC-egfp fusions in aerial hyphae of both the wild type and the XE mutant. EGFP expression was observed in aerial hyphae of both strains containing the P1ram-egfp, rdlA-egfp, chpH-egfp, and chpC-egfp reporters. The spores were inoculated on solid R2YE medium, and aerial hyphae attached to the surfaces of coverslips were analyzed 2 days after inoculation in the cases of M145 and its derivatives and after 7 days in the cases of XE and its derivatives. Wild-type M145 without EGFP was set up as the control. Bars, 10 μm.
DISCUSSION
The growth of Streptomyces aerial hyphae from the vegetative mycelium requires the solution of several problems, two of which have been particularly studied: growth away from water and the maintenance of apical dominance to permit rapid and efficient growth into the air. The first gene shown to be very important for tip growth, divIVA (15), encodes a tropomyosin-like protein that can form an alpha-helical coiled-coil structure (34). divIVA could not be deleted unless another copy of the gene was present, and ectopic overexpression of DivIVA caused hyphal tips to appear swollen (15). The targeting of DivIVA is a typical feature of alpha-helical coiled-coil proteins (11). This suggests that DivIVA can act as a scaffold for the recruiting of related proteins to target sites, for example, to the growing tips or division sites (15, 16). Protein-protein interaction between CslASc and DivIVA suggested that the tip location of CslASc might be governed by the tip-located DivIVA protein. This might be particularly important during the emergence of aerial hyphae. Given the diverse organization of cellulose synthase complexes (“rosette terminal complexes”) in different organisms and developmental contexts (33), it will be of interest to investigate the assembly of such complexes during vegetative and aerial growth of S. coelicolor and their effects on the pattern of assembly of β-glucan fibrils. The hyphal tip location of CslASc and the β-glucan accumulated under its direction, the association of CslASc with DivIVA, and the severe delay of aerial-hyphal formation in the cslASc mutant all indicate a role for CslASc in hyphal tip growth. As a working model, we propose that the multiple transmembrane regions of CslASc enable its integration into the cell membrane and that the interaction of its cytoplasmic segment with DivIVA precisely locates it in the tip position for the correctly positioned synthesis of β-glucan-containing polysaccharide. Why does Streptomyces need β-glucan-containing polysaccharides, which are not a component of the gram-positive bacterial cell wall? Perhaps the β-glucan fibers act as a “bandage” to wrap the hyphal tips, which are vulnerable to loss of integrity because of the continuous remodeling of the growing cell wall. The proposed bandage might help to maintain cell integrity, flexibility, and rigidity under different circumstances, notably including growth into the air. During aerial hyphal development, the hyphae must emerge from an aqueous environment and achieve cell wall growth in the air, using chaplins, rodlins, and SapB as hydrophobins (8, 9, 13, 39). The delayed aerial-hyphal formation of the XE mutant was phenotypically suppressed by adding an extract highly enriched for chaplins and rodlins, yet genes for each class of molecule showed the normal pattern of spatially specific expression in aerial hyphae. This suggests that the defect in aerial growth of the mutant is at least partly a quantitative deficiency in the hydrophobin-like molecules at the time of initial emergence. The hydrophobic β-glucan fibers may help to catalyze the correct assembly of the aerial-hyphal surface components in an (indirectly) self-inducing loop, since it appears that emergence into the air is a signal for increased chaplin and rodlin gene expression. In plants, cellulose synthase and cellulose synthase-like proteins cooperate with the cytoskeleton to control cell shape and to shape growth (24). Movement of the cellulose synthase complex is also coincident with the cortical microtubules (24, 27). The colocation of CslASc and DivIVA, and the protein-protein interaction between them, is to some extent a prokaryotic equivalent of the plant cellulose synthase-cytoskeleton correlation, maintaining apical dominance and guiding β-glucan synthesis and deposition.
In addition to its slow aerial-mycelium development, the cslASc mutant appeared to have abnormalities in the lateral and cross walls of vegetative hyphae and in its spore walls. These abnormalities may largely be manifestations of partially uncoordinated synthesis and maturation of the cell wall at tips lacking the β-glucan bandage. The structure of the lateral walls may provide a template for the precise structure of cross walls, accounting for the abnormal cross walls of the mutant.
Finally, the more dispersed growth of the mutant in liquid media may indicate that the β-glucan fibers play a role in the aggregation of clumps. This could be of interest in industrial fermentations of Streptomyces species for antibiotics and other valuable secondary metabolites, in which the degree of clumping has profound rheological effects.
Acknowledgments
We thank Jiaxin Liu, Jianbo Cao, and Lihong Qin for helping to perform the TEM and SEM work; Klas Flardh for gifts of FtsZ and DivIVA-EGFP constructs; and Dennis Claessen and Lubbert Dijkhuizen for the rdl and chp-egfp reporters.
This work was supported by grants from the National Natural Science Foundation of China (NSFC no. 30570030), from the Youth Chenguang Project of Science and Technology of Wuhan City of China to M. Tao, and by a Joint Project grant from the Royal Society and NSFC to K. Chater and Z. Deng.
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 2.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. Nagaraja Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 11643-49. [DOI] [PubMed] [Google Scholar]
- 3.Blondelet-Rouault, M.-H., J. Weiser, A. Lebrihi, P. Branny, and J.-L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the [pi] interposon for use in E. coli and Streptomyces. Gene 190315-317. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47685-713. [DOI] [PubMed] [Google Scholar]
- 6.Chater, K. F. 1989. Multilevel regulation of Streptomyces differentiation. Trends Genet. 5372-377. [DOI] [PubMed] [Google Scholar]
- 7.Chater, K. F., and R. Losick. 1997. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives, p. 149-182. In J. A. Shapiro and M. Dworkin (ed.), Bacteria as multicellular organisms. Oxford University Press, New York, NY.
- 8.Claessen, D., R. Rink, W. de Jong, J. Siebring, P. de Vreugd, F. G. Boersma, L. Dijkhuizen, and H. A. Wosten. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 171714-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claessen, D., H. A. B. Wosten, G. van Keulen, O. G. Faber, A. M. C. R. Alves, W. G. Meijer, and L. Dijkhuizen. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 441483-1492. [DOI] [PubMed] [Google Scholar]
- 10.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328307-317. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, D. H., H. B. Thomaides, and J. Errington. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 192719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliot, M. A., M. J. Buttner, and J. R. Nodwell. 2007. Multicellular development in Streptomyces, p. 419-438. In D. E. Whitworth (ed.), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC.
- 13.Elliot, M. A., N. Karoonuthaisiri, J. Huang, M. J. Bibb, S. N. Cohen, C. M. Kao, and M. J. Buttner. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 171727-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favery, B., E. Ryan, J. Foreman, P. Linstead, K. Boudonck, M. Steer, P. Shaw, and L. Dolan. 2001. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 1579-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flardh, K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 491523-1536. [DOI] [PubMed] [Google Scholar]
- 16.Flardh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6564-571. [DOI] [PubMed] [Google Scholar]
- 17.Flardh, K., K. C. Findlay, and K. F. Chater. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 1452229-2243. [DOI] [PubMed] [Google Scholar]
- 18.Hopwood, D. A. 1999. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 1452183-2202. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21526-531. [DOI] [PubMed] [Google Scholar]
- 20.Keijser, B. J. F., G. P. van Wezel, G. W. Canters, and E. Vijgenboom. 2002. Developmental regulation of the Streptomyces lividans ram genes: involvement of RamR in regulation of the ramCSAB operon. J. Bacteriol. 1844420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 22.Kodani, S., M. E. Hudson, M. C. Durrant, M. J. Buttner, J. R. Nodwell, and J. M. Willey. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 10111448-11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, W., X. Ying, Y. Guo, Z. Yu, X. Zhou, Z. Deng, H. Kieser, K. F. Chater, and M. Tao. 2006. Identification of a gene negatively affecting antibiotic production and morphological differentiation in Streptomyces coelicolor A3(2). J. Bacteriol. 1888368-8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lloyd, C., and J. Chan. 2004. Microtubules and the shape of plants to come. Nat. Rev. Mol. Cell Biol. 513-22. [DOI] [PubMed] [Google Scholar]
- 25.MacNeil, D. J., J. L. Occi, K. M. Gewain, T. MacNeil, P. H. Gibbons, C. L. Ruby, and S. J. Danis. 1992. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115119-125. [DOI] [PubMed] [Google Scholar]
- 26.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paredez, A. R., C. R. Somerville, and D. W. Ehrhardt. 2006. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 3121491-1495. [DOI] [PubMed] [Google Scholar]
- 28.Richmond, T. A., and C. R. Somerville. 2000. The cellulose synthase superfamily. Plant Physiol. 124495-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153205-212. [DOI] [PubMed] [Google Scholar]
- 30.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325279-281. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 32.Saxena, I. M., R. M. Brown, Jr., M. Fevre, R. A. Geremia, and B. Henrissat. 1995. Multidomain architecture of beta-glycosyltransferases: implications for mechanism of action. J. Bacteriol. 1771419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxena, I. M., and R. M. Brown, Jr. 2005. Cellulose biosynthesis: current views and evolving concepts. Ann. Bot. 969-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahlberg, H., E. Kutejova, K. Muchova, M. Gregorini, A. Lustig, S. A. Muller, V. Olivieri, A. Engel, A. J. Wilkinson, and I. Barak. 2004. Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol. Microbiol. 521281-1290. [DOI] [PubMed] [Google Scholar]
- 35.Sun, Y., X. Zhou, H. Dong, G. Tu, M. Wang, B. Wang, and Z. Deng. 2003. A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem. Biol. 10431-441. [DOI] [PubMed] [Google Scholar]
- 36.van Keulen, G., H. M. Jonkers, D. Claessen, L. Dijkhuizen, and H. A. B. Wosten. 2003. Differentiation and anaerobiosis in standing liquid cultures of Streptomyces coelicolor. J. Bacteriol. 1851455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittaker, M. M., and J. W. Whittaker. 2006. Streptomyces coelicolor oxidase (SCO2837p): a new free radical metalloenzyme secreted by Streptomyces coelicolor A3(2). Arch. Biochem. Biophys. 452108-118. [DOI] [PubMed] [Google Scholar]
- 38.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7895-903. [DOI] [PubMed] [Google Scholar]
- 39.Willey, J. M., A. Willems, S. Kodani, and J. R. Nodwell. 2006. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol. Microbiol. 59731-742. [DOI] [PubMed] [Google Scholar]
- 40.Wong, H. C., A. L. Fear, R. D. Calhoon, G. H. Eichinger, R. Mayer, D. Amikam, M. Benziman, D. H. Gelfand, J. H. Meade, A. W. Emerick, et al. 1990. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. USA 878130-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 391452-1463. [DOI] [PubMed] [Google Scholar]