Abstract
The acuABC genes of Bacillus subtilis comprise a putative posttranslational modification system. The AcuA protein is a member of the Gcn5-related N-acetyltransferase (GNAT) superfamily, the AcuC protein is a class I histone deacetylase, and the role of the AcuB protein is not known. AcuA controls the activity of acetyl coenzyme A synthetase (AcsA; EC 6.2.1.1) in this bacterium by acetylating residue Lys549. Here we report the kinetic analysis of wild-type and variant AcuA proteins. We contrived a genetic scheme for the identification of AcuA residues critical for activity. Changes at residues H177 and G187 completely inactivated AcuA and led to its rapid turnover. Changes at residues R42 and T169 were less severe. In vitro assay conditions were optimized, and an effective means of inactivating the enzyme was found. The basic kinetic parameters of wild-type and variant AcuA proteins were obtained and compared to those of eukaryotic GNATs. Insights into how the isolated mutations may exert their deleterious effect were investigated by using the crystal structure of an AcuA homolog.
Protein acetylation is a form of posttranslational modification that allows cells to rapidly alter cellular processes in response to a changing environment (14). Acetyltransferase enzymes (referred to as Gcn5-type N-acetyltransferases, or GNATs) in eukaryotic cells responsible for some of these modifications have been extensively studied within the context of the control of gene expression. GNATs modulate the acetylation state of histones facilitating transcription (hyperacetylated histones) or gene silencing (hypoacetylated histones) (4, 11-14, 21). GNATs use acetyl coenzyme A (Ac-CoA) as the donor of acetyl groups to modify ɛ-amino groups of lysyl residues in histone tails. In eukaryotes, GNATs act on different protein substrates and small molecules (1, 7, 10, 15, 16, 24, 25). In contrast, there are only two described examples of prokaryotic metabolic enzymes under GNAT control. The protein acetyltransferase (Pat) enzyme of Salmonella enterica and the AcuA enzyme of Bacillus subtilis (AcuABs) control the activity of the central metabolism enzyme Ac-CoA synthetase (AcsA) (5, 18), and in the case of Pat, the enzyme also controls the activity of propionyl-CoA synthetase (6, 19).
Together with protein deacylases, Pat and AcuA comprise the protein acylation/deacylation system of posttranslational modification that helps the cell maintain CoA homeostasis during growth on acetate or propionate (5, 17, 20) (Fig. 1). To date, the kinetic characterization and mutational analyses of the Pat or AcuA enzyme have not been reported.
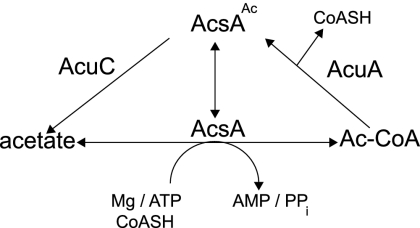
FIG. 1.
Posttranslational control of Ac-CoA synthetase activity by the AcuABs (acetyltransferase) and AcuCBs (deacetylase). AcuA uses Ac-CoA to acetylate (inactivate) AcsA. AcuC deacetylates (activates) acetylated AcsA (AcsAAc), restoring the ability of the Acs enzyme to convert acetate to Ac-CoA. CoASH, free CoA.
To advance our understanding of the function of the bacterial AcuA enzyme, we isolated variants of AcuA that completely or partially lacked activity, quantified the effect of the mutations using an in vitro activity assay, and compared the kinetic parameters of the variant proteins to those of the wild-type protein. The functionality of AcuA variants was also assessed in vivo using strains of S. enterica carrying a null allele of the pat gene in the chromosome.
Residues critical to AcuABs function.
We took a genetic approach to the identification of residues that were critical to AcuABs function. For this purpose, we took advantage of the fact that the growth of an S. enterica pat strain on low acetate (10 mM) is arrested when the AcuA protein is overproduced (5). We introduced single-base mutations into acuA+ (carried by plasmid pACUA1) using the XL1-Red Escherichia coli mutator strain (Stratagene). The strain carrying plasmid pACUA1 was grown as per the manufacturer's instructions. Plasmid DNA was isolated from the mutator strain twice at 24-h intervals, and DNA was transformed into S. enterica strain JE6861 [metE205 ara-9 pat1::Tn10d(tet+)] and plated onto no-carbon E (NCE) medium containing acetate (10 mM) and arabinose (500 μM to induce acuA expression), with selection for growth. Cells from colonies growing on the selective medium were patched on LB agar plates containing ampicillin (Amp) and were replica printed onto plates with NCE plus acetate plus arabinose plus Amp to assess their ability to grow on acetate. Under these conditions, cell growth implied either the lack of acuA expression or the synthesis of AcuA variant proteins with various levels of enzymatic activity. Cells that grew on acetate were inoculated into 2 ml of LB plus Amp and grown overnight. Plasmid DNA was recovered from LB agar- and Amp-grown cells, the acuA allele was cut from the plasmid using the restriction enzymes EcoRI and HindIII, and the fragment was recloned into plasmid pBAD30 cut with the same enzymes. Reconstructed plasmids were electroporated into strain JE6861 [metE205 ara-9 pat1::Tn10d(tet+)], with selection for Ampr cells, and growth on NCE medium containing acetate and arabinose was reassessed. Plasmid DNA was isolated from Ampr cells that grew on acetate in the presence of arabinose, and the acuA alleles from these plasmids were sequenced.
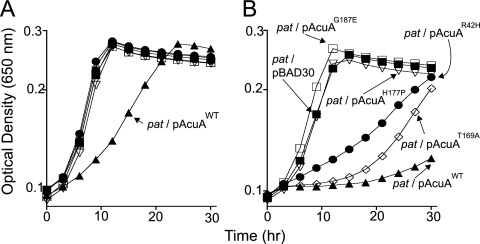
The growth behavior of strain JE6861 carrying mutant alleles of acuA is shown in Fig. 2. Overproduction of the variant proteins AcuAG187E and AcuAH177P did not affect the growth of the strain on acetate, suggesting that changes at residues G187 and H177 completely eliminated AcuA activity or resulted in a level of activity that was too low to affect growth on acetate. Two other AcuA variant proteins with changes at position R42 (AcuAR42H) or T169 (AcuAT169A) retained some activity, as shown by the intermediate growth phenotype (Fig. 2). The two mutant acuA alleles encoding the AcuAR42H and AcuAT169A proteins were cloned into protein overexpression vector pTYB12, AcuA proteins were overproduced and isolated, and the wild-type AcuA (AcuAWT) enzyme was isolated as a control using the same procedure. Gel filtration analysis of the AcuAWT protein was consistent with the enzyme being a monomer (see Fig. S4A in the supplemental material).
FIG. 2.
In vivo assessment of AcuAWT and variant AcuA proteins. All growth experiments were performed at least three times, and each time they were performed in triplicate. In all experiments, the standard deviation was ≤0.5. The size of the symbols obscures the error bars, but they are included in the figure. NCE minimal medium (2) containing acetate (10 mM) as a carbon and energy source was used for these experiments. (A) Behavior of the strains in the absence of l-(+)-arabinose, the inducer used to trigger the synthesis of AcuA proteins. (B) Behavior of the same strains in the presence of 500 μM l-(+)-arabinose in the medium. Filled squares, pat/pBAD30 strain (empty vector control); filled triangles, pat/pAcuAWT strain; open squares, pat/pAcuAG187E strain; open inverted triangles, pat/pAcuAH177Pstrain; circles, pat/pAcuAR42H strain; diamonds, pat/pAcuAT169A strain. For a complete description of the strain and plasmid genotypes, see Table S2 in the supplemental material. Details of the construction of plasmids and strains, the growth conditions, and medium supplements are described in the supplemental material.
Optimization of reaction conditions.
We determined optimal conditions for AcuABs activity using as the substrate a synthetic peptide of AcsA from B. subtilis and Ac-CoA. The pH optimum was found to be 7.5, and the temperature optimum was 37°C (see Fig. S4B and C in the supplemental material). We could not test AcuA activity at temperatures above 37°C because the rate of spontaneous Ac-CoA cleavage was higher than the rate of consumption of Ac-CoA by AcuA. In addition, the spontaneous cleavage of Ac-CoA at temperatures above 37°C decreased the known concentration, and therefore, precise rate calculations were not possible. Increasing the ionic strength of the reaction mixture decreased the activity of AcuA (see Fig. S4D in the supplemental material).
Strong ionic interactions maintain AcuABs in its active conformation.
In previous work, we noted the unusual heat stability of AcuABs (5). This feature of the enzyme was not understood, but it presented us with the challenge of finding a way of inactivating the enzyme. During the optimization of the assay conditions, we learned that the AcuABs activity was sensitive to salt concentration; however, a high concentration of KCl (500 mM) was needed to completely inactivate the enzyme (see Fig. S4D in the supplemental material). The predicted isoelectric point for AcuABs is 5.3, and it therefore harbors a net negative charge at pH 7.5, where activity was highest. The addition of KCl screens away ionic interactions of AcuABs, decreasing its activity. This finding strongly suggests that the biologically active form of AcuABs is maintained by strong ionic interactions. This new knowledge provides the means to efficiently terminate AcuABs reactions.
Kinetic analysis of AcuAWT and variant AcuA enzymes.
The rates of the reactions catalyzed by AcuAWT and variant AcuA proteins as a function of substrate concentration were determined by using the graphing and statistical software package Prism v4 (GraphPad) (see Fig. S5 in the supplemental material). Under optimal assay conditions, the AcuAR42H and AcuAT169A variants had the same kinetic parameters but differed from the wild-type protein in their reduced Km for Ac-CoA (two- to threefold lower than for AcuAWT) and their reduced turnover number (threefold slower) (Table 1). Although the catalytic efficiency of the AcuAR42H and AcuAT169A variants was the same as that of the AcuAWT protein, those proteins did not inhibit growth when overproduced, suggesting that they were unstable in vivo.
TABLE 1.
Kinetic parameters of AcuAWT and variant AcuABs enzymes
| Protein | Peptide substrate
|
Ac-CoA substrate
|
||||||
|---|---|---|---|---|---|---|---|---|
| Km (mM) | Vmax (mM min−1 mg−1) | kcat (s−1) | kcat/Km (s−1 M−1) | Km (mM) | Vmax (mM min−1 mg−1) | kcat (s−1) | kcat/Km (s−1 M−1) | |
| AcuAWT | 20 ± 2 | 0.9 ± 0.02 | 0.3 | 1.5 × 104 | 22 ± 2 | 0.8 ± 0.03 | 0.3 | 1.4 × 104 |
| AcuAR42Ha | 37 ± 5 | 0.3 ± 0.01 | 0.1 | 2.7 × 103 | 8 ± 1 | 0.2 ± 0.01 | 0.1 | 1.3 × 104 |
| AcuAT169Aa | 37 ± 5 | 0.3 ± 0.01 | 0.1 | 2.7 × 103 | 9 ± 1 | 0.2 ± 0.01 | 0.1 | 1.1 × 104 |
| yGCN5b | 490 ± 80 | 1.7 ± 0.1 | 3.5 × l03 | 2.5 ± 1.4 | ||||
| P/CAFc | 532 ± 81 | 2.3 ± 0.07 | 4.3 × 103 | 0.98 ± 0.31 | ||||
We probed whether or not the R42H and T169A mutations had a structural effect by assessing changes in the known heat stability of the AcuA enzyme (5). After heating AcuAWT and variant AcuA proteins to 100°C for 30 min, the acetyltransferase activity of the enzyme was assayed in vitro. The AcuAR42H protein retained 54% ± 11% of the activity of the wild-type enzyme treated under the same conditions, whereas the AcuAT169A protein retained 60% ± 13% of the activity (data not shown).
The AcuABs protein is less responsive to low levels of Ac-CoA but more responsive to its peptide substrate than eukaryotic GNATs.
AcuABs requires more Ac-CoA substrate to reach saturation than yeast GCN5 (yGCN5) and P/CAF (p300/CREB-binding-protein-associated factor) (22, 23). AcuABs was substantially less responsive to low levels of Ac-CoA, as shown by a Km for Ac-CoA that is 1 order of magnitude higher than that of yGCN5 and P/CAF (Table 1). In contrast, the Km of AcuABs for its peptide substrate was 25-fold lower than that of yGCN5, suggesting that either B. subtilis synthesizes less AcsA or the activity of AcsA in this bacterium is very high and needs to be tightly controlled. The latter would be a plausible explanation given the deleterious effect that high levels of Acs have on cell growth (9; J. Garrity, S. R. Brinsmade, and J. Escalante-Semerena, unpublished results).
The explanation for the difference in Km values between AcuABs and yGCN5 may lie in the intracellular concentration of the CoA pool in B. subtilis, which was estimated at 71 pmol/mg (dry weight) (3).
The sixfold-slower turnover of AcuABs relative to those of yGCN5 and P/CAF may be explained by the absence of physiological pressures for the evolution of a faster AcuABs enzyme in B. subtilis. It would be of interest to isolate faster variants of AcuABs to determine what effect such variants would have on B. subtilis physiology.
The decreased catalytic efficiency of the AcuAR42H and AcuAT169A enzymes explains the growth phenotype shown in Fig. 2B. That is, since the AcuAR42H and AcuAT169A proteins retained some enzymatic activity (∼25%) (Table 1), acetylation (i.e., inactivation) of the entire pool of Ac-CoA synthetase (AcsA) was not achieved, thus resulting only in impaired growth on low acetate, the conditions where AcsA was required.
On the other hand, the G187E mutation had such a severe effect on the activity of the enzyme that the S. enterica overproduction variant AcuAG187E grew as well as the control strain carrying an empty vector, suggesting that the G187E mutation either completely inactivated or completely destabilized AcuA. The H177P mutation resulted in a growth phenotype similar to the one caused by the G187E mutation.
Structural modeling using the Exiguobacterium sibiricum AcuA crystal structure.
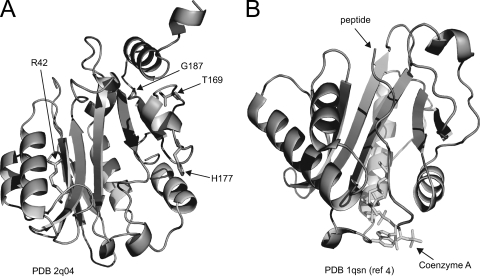
The crystal structure of AcuA from Exiguobacterium sibiricum (AcuAEs) was recently solved (Fig. 3) (Protein Data Bank accession no. 2q04). AcuABs and AcuAEs share substantial end-to-end identity (62%) and similarity (76%) (see Fig. S6 in the supplemental material). Residues R42, T169, G187, and H177 are conserved in both proteins. We used the structure of the AcuAEs protein to visualize where the AcuABs residues changed by mutation would lie in three dimensions so that we could try to explain the changes in enzymatic activity.
FIG. 3.
Modeling AcuABs mutations into the structures of its homologs. (A) Reported structure of the bacterial apo-AcuAEs. (B) Structure of Tetrahymena Gcn5 with substrates in the active site. This structure provides a point of reference for the location of the active site in AcuAEs. PDB 2q04 and PDB 1qsn, Protein Data Bank accession numbers; ref, reference.
There are four characteristic motifs in GNATs, motifs A, B, C, and D (see Fig. S6 in the supplemental material). Residue R42 is part of motif C and is located before the α2 helix of the protein. We used the PyMOL software program (W. L. DeLano, the PyMOL molecular graphics system version 0.98) to introduce mutations into the AcuAEs structure, measured distances to neighboring side chain functions, and compared the distances to those found in the wild-type AcuAEs protein (see Fig. S7 in the supplemental material). These measurements are hypothetical and assume that no significant conformational change of the protein is caused by the mutation. In wild-type AcuAEs, the imine N of the guanidinium group of R42 forms a salt bridge with the hydroxyl of the γ-carboxylate function of Q47, and these functions are 2.6 Å apart. We hypothesize that in the AcuAR42H variant protein, this interaction is likely absent. Residue T169 is conserved in both proteins but is not part of any consensus GNAT motif; T169 is located after the β8 sheet of the AcuAEs protein. In the AcuAEs structure, the side chain hydroxyl group of T169 is 3.3 Å away from the peptide bond N of D171. This salt bridge is likely absent in the AcuAT169A variant protein. Residue H177 is located immediately after the α10 helix of the protein. The carbonyl of H177 forms a salt bridge with the δN of N180 (2.9 Å). This ionic interaction may be present in the AcuAH177P protein, but the negative effect of the mutation may arise from the proximity of the resulting vicinal prolyl side chains (P177 and P178). We suspect that such a change results in a loss of flexibility that could restrict conformational changes necessary for catalysis.
Residue G187 is part of hydrogen-bonded turn 12 of the protein after sheet β9 and before helix α11. In the AcuAG187E protein, interactions between the γ-carboxylate group of Glu at position 187 with the peptide bond N atoms of residues H127 and M125 may form and restrict the flexibility of the enzyme to the point of rendering it completely inactive.
Variant proteins AcuAG187E and AcuAH177P could not be overproduced, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown), suggesting that these variants are rapidly turned over by the cell.
GNAT motifs, structural considerations, and AcuABs activity.
As shown in Fig. S6 in the supplemental material, AcuABs contains four consensus motifs found in GNATs. Briefly, motif A contains the β sheet that is the core of the GNAT domain, specifically β6, which is involved in binding both substrates; motif B contains a portion of the β9 sheet, which also is involved in binding Ac-CoA; motif C is the least conserved of the GNAT motifs, and it is involved in orienting the protein substrate; and motif D forms part of the β sheet core and, together with motif A, makes up half of the catalytic site (4, 12, 13).
Noteworthy is the observation that two of the mutated residues (T169 and H177) map outside motifs A, B, C, and D. The other two mutations that we isolated, namely, the R42H and G187 mutations, were found in motifs C and B, respectively (see Fig. S6 in the supplemental material). We hypothesize that the G187 and H177P changes greatly disrupt the structure of AcuABs. Using heat stability as a parameter of structural integrity, we assessed the activities of the AcuAR42H and AcuAT169A variant proteins and found that both of these variants lost about 50% of the wild-type activity. Based on these results, we suggest that these residues have a structural role but that they may also be involved in substrate binding, an idea supported by our kinetic data (Table 1).
A potential role for the AcuA enzyme in B. subtilis physiology.
Recently, temporal gene expression during spore germination and outgrowth was examined (8). In this study, the acuA gene was upregulated during the 80 to 100 min of spore germination and into the outgrowth phase. Detailed genetic analyses of acuA expression and the identification of additional protein substrates in Bacillus subtilis are needed to gain insights into the scope of the AcuA-mediated posttranslational regulation in this bacterium.
Supplementary Material
Acknowledgments
This work was supported by PHS grant R01-GM622023 to J.C.E.-S.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 81243-1254. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, D., J. M. Hushon, H. J. Whitfield, Jr., J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chohnan, S., H. Furukawa, T. Fujio, H. Nishihara, and Y. Takamura. 1997. Changes in the size and composition of intracellular pools of nonesterified coenzyme A and coenzyme A thioesters in aerobic and facultatively anaerobic bacteria. Appl. Environ. Microbiol. 63553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dyda, F., D. C. Klein, and A. B. Hickman. 2000. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 2981-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner, J. G., F. J. Grundy, T. M. Henkin, and J. C. Escalante-Semerena. 2006. Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD+ involvement in Bacillus subtilis. J. Bacteriol. 1885460-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrity, J., J. G. Gardner, W. Hawse, C. Wolberger, and J. C. Escalante-Semerena. 2007. N-Lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 28230239-30245. [DOI] [PubMed] [Google Scholar]
- 7.Hallows, W. C., S. Lee, and J. M. Denu. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA 10310230-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keijser, B. J. F., A. Ter Beek, H. Rauwerda, F. Schuren, R. Montijn, H. van der Spek, and S. Brul. 2007. Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. J. Bacteriol. 1893624-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumari, S., R. Tishel, M. Eisenbach, and A. J. Wolfe. 1995. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 1772878-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeDizet, M., and G. Piperno. 1987. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc. Natl. Acad. Sci. USA 845720-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marmorstein, R. 2001. Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell Biol. 2422-432. [DOI] [PubMed] [Google Scholar]
- 12.Marmorstein, R. 2001. Structure of histone acetyltransferases. J. Mol. Biol. 311433-444. [DOI] [PubMed] [Google Scholar]
- 13.Marmorstein, R. 2001. Structure of histone deacetylases: insights into substrate recognition and catalysis. Structure 91127-1233. [DOI] [PubMed] [Google Scholar]
- 14.Marmorstein, R., and S. Y. Roth. 2001. Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 111555-1561. [DOI] [PubMed] [Google Scholar]
- 15.Polevoda, B., and F. Sherman. 2002. The diversity of acetylated proteins. Genome Biol. 3reviews0006.1-reviews0006.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwer, B., J. Bunkenborg, R. O. Verdin, J. S. Andersen, and E. Verdin. 2006. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA 10310224-10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 2982390-2392. [DOI] [PubMed] [Google Scholar]
- 18.Starai, V. J., and J. C. Escalante-Semerena. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 612020-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starai, V. J., and J. C. Escalante-Semerena. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 3401005-1012. [DOI] [PubMed] [Google Scholar]
- 20.Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2003. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sternglanz, R., and H. Schindelin. 1999. Structure and mechanism of action of the histone acetyltransferase Gcn5 and similarity to other N-acetyltransferases. Proc. Natl. Acad. Sci. USA 968807-8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner, K. G., M. R. Langer, and J. M. Denu. 2000. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 3911961-11969. [DOI] [PubMed] [Google Scholar]
- 23.Tanner, K. G., M. R. Langer, Y. Kim, and J. M. Denu. 2000. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J. Biol. Chem. 27522048-22055. [DOI] [PubMed] [Google Scholar]
- 24.Vetting, M. W., L. P. S. de Carvalho, M. Yu, S. S. Hegde, S. Magnet, S. L. Roderick, and J. S. Blanchard. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433212-226. [DOI] [PubMed] [Google Scholar]
- 25.Wardleworth, B. N., R. J. M. Russell, S. D. Bell, G. L. Taylor, and M. F. White. 2002. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 214654-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.