Abstract
Glutamine synthetase (GS) plays an important role in nitrogen assimilation. The major GS of Mycobacterium tuberculosis is GlnA1, a type I GS whose activity is controlled by posttranscriptional modification by GlnE. GlnE is an adenylyl transferase comprised of an adenylylating domain and a deadenylylating domain which modulate GS activity. We previously demonstrated that GlnE is essential in M. tuberculosis in normal growth medium. In this study, we further show that GlnE is required under multiple medium conditions, including in nitrogen-limited medium. We demonstrate that adenylylation is the critical activity for M. tuberculosis survival, since we were able to delete the deadenylylation domain with no apparent effect on growth or GS activity. Furthermore, we identified a critical aspartate residue in the proposed nucleotidyltransferase motif. Temperature-sensitive mutants of GlnE were generated and shown to have a defect in growth and GS activity in nitrogen-limited medium. Finally, we were able to generate a GlnE null mutant in the presence of l-methionine sulfoximine, a GS inhibitor, and glutamine supplementation. In the presence of these supplements, the null mutant was able to grow similarly to the wild type. Surprisingly, the GlnE mutant was able to survive and grow for extended periods in liquid medium, but not on solid medium, in the absence of GS inhibition. Thus, we have confirmed that the unusual requirement of M. tuberculosis for GlnE adenylylation activity is linked to the activity of GS in the cell.
A recent report from the WHO noted that approximately 1.7 million people died from tuberculosis and that at least 14.6 million people lived with the disease in 2004. Current antibiotic treatments are unable to address the high prevalence or stem the emergence of multidrug-resistant and extremely drug resistant strains. Therefore, much recent research has focused on the identification and validation of novel drug targets to combat the causative agent of tuberculosis, Mycobacterium tuberculosis.
Glutamine synthetase (GS) catalyzes the ATP-dependent condensation reaction between glutamate and ammonia to produce the amino acid l-glutamine (l-Gln), a precursor for several other amino acids as well as glucosamine-6-phosphate, AMP, and CTP. M. tuberculosis has four GS homologues, GlnA1 to -4 (3). GlnA1, GlnA3, and GlnA4 are involved in the synthesis of l-Gln, whereas GlnA2 is involved in the synthesis of d-Gln and d-iso-Gln (15).
GlnA1 is a dodecamer formed of two hexameric units on overlaying planes, where each subunit contains an active site which binds ATP and glutamate (1). GlnA1 is secreted from M. tuberculosis at high levels, and deletion mutants are both auxotrophic for glutamine and attenuated in the macrophage and guinea pig models of infection (13, 33, 34). GlnA2 is localized at the cell wall, which may allow the incorporation of d-iso-Gln into position 2 of the peptide side chain of the peptidoglycan (9). Notably, a GlnA2 mutant of M. bovis is completely avirulent in the guinea pig model (4). In contrast, a GlnA2 mutant of M. tuberculosis was virulent in mice (19). Neither GlnA3 nor GlnA4 has been detected in culture, and little is known at present about their roles in M. tuberculosis biology and pathogenesis (15).
GS activity is effectively inhibited by l-methionine sulfoximine (MetSox) (31). In the presence of ATP, MetSox is phosphorylated by GS and irreversibly binds to the active site, thereby preventing the entry of glutamate. MetSox has activity against whole bacilli and can prevent the growth of several mycobacterial species (12, 14), indicating that GS activity could be a useful drug target. MetSox has wide activity against GS enzymes and demonstrates strong inhibition of all four M. tuberculosis GSs. However, MetSox itself cannot be used as an antitubercular agent due to its epileptogenic properties (28).
In most bacteria, GS activity is posttranscriptionally controlled by adenylylation (8) or oxidative modification (20, 21) in response to a multitude of extracellular and intracellular stimuli (1, 7). M. tuberculosis GlnA1 is a type I GS, and thus its activity is expected to be regulated by the adenylyl transferase GlnE. Consistent with this is the fact that both enzymes are located in the cell wall of M. tuberculosis (9). In Escherichia coli and other bacteria, modification of GS by the covalent addition of AMP leads to a reduction in enzyme activity (1, 30, 32). Since GlnE can catalyze both reactions (addition and removal of AMP), it has a critical role in modulating GS activity in the cell. In E. coli, GlnE has two distinct domains which catalyze the two reactions, the N-terminal deadenylylating domain and the C-terminal adenylylating domain (17). GlnE activity is in turn regulated by the PII signal transduction cascade.
We have previously shown that GlnE is essential in M. tuberculosis and that its expression is regulated by nitrogen availability (24, 26, 27). The essentiality of GlnE is surprising, given that it is not essential in other bacteria, including E. coli and streptomycetes (6). Given its central role in modulating GS activity, we therefore considered GlnE to be a potential antituberculosis drug target.
In this study we have further investigated the essentiality of GlnE. We demonstrate that GlnE mutants are not viable on a range of media, including nitrogen-limiting medium. We show that the key domain is the adenylylation domain, and within that domain we identify a key residue required for function. We show that the majority of GlnA1 is adenylylated under all conditions profiled and that this holds true for all of the mutants isolated. Finally we show that GlnE can be deleted, but only when GS activity is inhibited by MetSox and exogenous l-Gln is provided. Once generated, the GlnE deletion strain was able to survive in liquid culture in the absence of GS inhibition for extended periods, although it was not viable on solid medium.
MATERIALS AND METHODS
Bacterial strains, growth media, and antibiotics.
M. tuberculosis H37Rv (ATCC 25618) was grown in Middlebrook 7H9 medium plus 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC) supplement (Becton Dickinson) and 0.05% (wt/vol) Tween 80 or on Middlebrook 7H10 agar plus 10% (vol/vol) OADC. TSM medium, as described previously (26), contained 1.5 g K2HPO4 liter−1, 0.5 g KH2PO4 liter−1, 0.5 g MgSO4 liter−1, 0.5 mg CaCl2 liter−1, 0.1 mg ZnSO4 liter−1, 0.1 mg CuSO4 liter−1, and 50 mg FeCl3 liter−1 supplemented with 10% (vol/vol) OADC and 0.05% (wt/vol) Tween 80. For TSM-high ammonia, 30 mM (NH4)2SO4 was added, and for TSM-low ammonia, 0.1 mM (NH4)2SO4 was added; d-Gln, l-alanine, l-asparagine, l-Gln, and l-glutamate were added at 3 mM unless otherwise specified. Hygromycin was added at 100 μg/ml, kanamycin at 20 μg/ml, gentamicin at 10 μg/ml, and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 50 μg/ml.
Construction of unmarked deletion-integration strain.
The deletion delivery vector pGLN16 was constructed as follows. A NotI deletion in the glnE gene was made using vector pGLN1 (24). The glnE fragment was excised from the vector as a BamHI-HindIII fragment and cloned into p2NIL (24), and the PacI cassette containing Phsp60-sacB, PAg85a-lacZ, and hyg from pGOAL19 (25) was inserted to make pGLN16. The delivery vector was treated with UV light and electroporated into M. tuberculosis (25). A single-crossover strain (Glue11) was isolated after 3 weeks and transformed with the integrating vector pGLINT1 (24) to generate a merodiploid (Glue12). Double-crossover strains were generated as described previously by streaking out the single-crossover strain without antibiotic selection and then plating onto 2% (wt/vol) sucrose-50 μg/ml X-Gal for 4 weeks. White colonies were patch tested for hygromycin sensitivity and gentamicin resistance and then screened by PCR for the presence of the deletion band. One potential mutant (Glue13) was selected and subjected to Southern hybridization.
Generation of pLEDGE3 derivatives.
Plasmid pLEDGE3 was constructed by PCR amplification of glnE using primers glnEF7 (GAC TCT AGA GGA TCT ACC GGT) and glnER7 (GTT GGT CAG CGT GGA CAA GCT TAT AC) and inserting this into pUC18 as an XbaI-BamHI fragment. The antigen 85A promoter from pEM37 (5) was inserted upstream as a BamHI-BglII fragment into the BamHI site. The L5-based integrating cassette (Gm, L5 integrase, and attP) from pUC-Gm-INT (22) was inserted as a HindIII fragment. Three deletion derivatives were made using enzymes Eco47III (pLEDGE3D1), PpuMI (pLEDGE3D2), and XhoI (pLEDGE3D4). To generate specific amino acid substitution of conserved aspartic residues at positions 730 and 732, site-directed mutagenesis was performed. Primers SDM1 (GTT GGG CTA CGG GTC GGC TGC CGA CGT GAT GTT C) and SDM2 (GAA CAT CAC GTC GGC AGC CGA CCC GTA GCC CAA C) were used to generate pLEDGE3S-D730A, and primers SDM3 (CTA CGG GTC GGA TGC CGC CGT GAT GTT CGT C) and SDM4 (GAC GAA CAT CAC GGC GGC ATC CGA CCC GTA G) were used to generate pLEDGE3S-D732A.
A library of mutated alleles was generated using hydroxylamine mutagenesis as previously described (10). Briefly, 20 μg of pLEDGE3 was suspended in 100 μl Tris-EDTA-5 M hydroxylamine-5 mM EDTA (pH 8.0), and the reaction mixture was incubated for 30 min at 81°C. Plasmid DNA was ethanol precipitated and resuspended in nuclease-free water.
Gene switching.
Gene switching was performed as described previously (27). Briefly, Glue13 was transformed with pUC-Hyg-INT, pLEDGE3, pLEDGE3D1, pLEDGE3D2, pLEDGE3D4, pLEDGE3S-D730A, pLEDGE3S-D732A, and the pLEDGE3 mutagenized library. Transformants were selected on hygromycin and patch tested for gentamicin sensitivity, which indicated that switching had occurred, i.e., that the resident plasmid, pGLINT1 containing gentamicin resistance, had been replaced by the incoming plasmid (carrying hygromycin resistance).
Growth curves and cell extracts.
Liquid cultures were diluted to give a starting optical density at 580 nm (OD580) of 0.01 in 3 ml of medium. Each tube contained a 12-mm magnetic stirrer and was incubated at 37°C on a Wheaton Biostir. OD580 readings were taken periodically for 18 days. Cell extracts were prepared as from liquid cultures. Cells were harvested, washed twice in 10 mM Tris (pH 8.0)-0.05% (vol/vol) Tween 80, resuspended in 1 ml of 10 mM Tris (pH 8.0), and added to lysing matrix B tubes (QBiogene). Cells were disrupted using the Fastprep (QBiogene) set at speed 6.0 for 30 seconds. Samples were centrifuged for 2 min, and the supernatant was recovered and filter sterilized.
GS activity assays.
The synthetic reaction was used to measure active GS, i.e., nonadenylylated enzyme. Phosphate release was measured as previously described (35). The assay mixture contained 0.76 mM ATP, 0.1 M l-glutamic acid, 0.05 M ammonia, 0.05 M MgCl2, 0.05 M imidazole buffer (pH 7), and 50 μl sample and was made up to 200 μl with sterile distilled water. Protein concentrations were assayed using a bicinchoninic acid kit (Pierce). Reactions were standardized against 1 unit of E. coli GS (Sigma), which will convert 1.0 μmol of l-glutamate to l-Gln in 15 min at pH 7.0 at 37°C (35). To determine the total amount of GS present in the sample, i.e., nonadenylylated and adenylylated enzyme, snake venom phosphodiesterase type IV (SVPD) from Crotalus atrox (Sigma) was added to a final concentration of 1 mg/ml in Tris buffer (pH 9.0)-5 mM MgCl2 and incubated at 37°C for 3 h. This removes the AMP group. Samples were then assayed for GS using the phosphate release assay as above.
RESULTS
glnE is essential in media containing alternative nitrogen sources.
We had previously shown that glnE is an essential gene by demonstrating that we could disrupt the chromosomal copy only when we provided a second functional copy of the gene elsewhere (24). In our case, we used the mycobacteriophage L5-based integrating vector pUC-Gm-INT (22) and tried to isolate mutants using the standard laboratory medium, Middlebrook 7H10 plus OADC, which contains 0.5 g/liter of glutamate and 40 mg/liter of ferric ammonium citrate. Salmonella enterica serovar Typhimurium GlnE mutants grow better in nitrogen-limited medium (2), so we wanted to determine if we could isolate mutants under low-ammonia conditions or by using alternative nitrogen sources. Our approach was to make use of a switching method for replacing the integrated vector carrying glnE with one without the gene and determining viability (see Fig. S1 in the supplemental material). Gene switching involves replacing the integrated glnE vector with an “empty” version of the vector carrying a different selection marker but no M. tuberculosis genes. We had previously shown that the integrated vector can be replaced very efficiently by an alternative version after transformation (27). Using this approach, we could rapidly test a number of different growth conditions rather than attempting the long process of trying to generate mutants of the wild-type and merodiploid strains under each independent condition.
Although we already had a strain with the chromosomal disruption, it carried a hygromycin resistance cassette, and we required this marker for subsequent experiments. Therefore, we made a new “del-int” strain (deletion on chromosome, integration of functional copy) with an unmarked deletion. The delivery vector pGLN16 was transformed into M. tuberculosis to generate a single-crossover strain (Glue11). This was then transformed with the pGLINT1 integrating vector (24) to make the merodiploid strain Glue12. A double-crossover strain with the deletion on the chromosome was then isolated (Glue13) and confirmed by Southern blotting (see Fig. S2 in the supplemental material).
The switching experiment was carried out using the vector pUC-Hyg-INT (27), which does not contain a functional copy of glnE. In order to test whether glnE mutants were viable under different conditions, we used TSM medium with various nitrogen sources. The del-int strain Glue13 was transformed with pUC-Hyg-INT (22), recovered, and plated onto different nitrogen sources under selection for the incoming vector. We used pLEDGE3, containing the glnE gene in pUC-Hyg-INT, as a positive control. We tested TSM plus low and high ammonia concentrations, as well as alanine, asparagine, glutamine, and glutamate. The transformation efficiency was 104 per μg of pLEDGE3; all transformants were hygromycin resistant and gentamicin sensitive, indicating that the original vector had been replaced, i.e., that switching had occurred. When pUC-Hyg-INT was used, no transformants were obtained on any of the media tested, indicating that we could not remove the only functional copy of the glnE gene. Thus, we have shown that the essentiality of glnE is not restricted to Middlebrook 7H9 medium and that mutants cannot be isolated under nitrogen-limiting conditions.
Adenylylation is the essential activity of GlnE.
Analysis of the domain structure of GlnE proteins by use of BLAST software confirms that there are two Pfam adenylylation domains and predicts that the N-terminal domain is responsible for the deadenylylation activity (17). The C-terminal domain contains the DNA polymerase motif, indicative of a nucleotide binding site (16), and therefore is responsible for the adenylylation activity (17). Our original deletion of glnE would disrupt both the deadenylylation and adenylylation domains of the protein. Since we hypothesized that GlnE is required to inactivate the large amounts of GlnA1 synthesized, we predicted that the deadenylylation activity might be unnecessary. We used our switching strategy to determine which domain(s) was essential for growth.
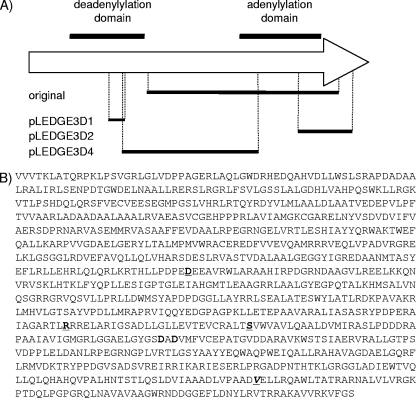
We constructed three deletion derivatives of pLEDGE3 (Fig. 1A) and determined whether they were functional using the gene switching method. pLEDGE3D1 was an Eco47II deletion resulting in a 147-bp in-frame deletion of the deadenylylation domain. pLEDGE3D2 was a PpuMI deletion resulting in a 476-bp deletion leading to the loss of the 600-bp C-terminal adenylylation domain (as the deletion was out of frame). pLEDGE3D4 was an XhoI deletion resulting in a 1,182-bp in-frame deletion covering both domains. Each plasmid was transformed into the del-int strain Glue13 (ΔglnE [glnE+int Gm]), and switching was assessed by selecting for hygromycin resistance. Plasmid pLEDGE3, containing full-length glnE, was used as a positive control. Hygromycin-resistant transformants were obtained with pLEDGE3 (control) and pLEDGE3D1 (deletion in the N-terminal domain); 24 of each were patched onto gentamicin, and all were sensitive, confirming the loss of the resident pGLINT1 plasmid and successful switching. The expected genotypes were confirmed by Southern analysis (not shown). No transformants were obtained with pLEDGE3D2 (C-terminal domain deletion) or pLEDGE3D4 (deletion of both domains), indicating that these alleles cannot functionally complement. These data demonstrate that the adenylylation activity is critical but that the deadenylylating activity is dispensable.
FIG. 1.
Functional analysis of GlnE. (A) The proposed deadenylylation and adenylylation domains and the location of the conserved nucleotidyl transferase motif in GlnE are indicated. The three domain deletions are shown below. The original NotI deletion in pGLN16 is also marked. This is the region deleted in the chromosome of strain Glue14. The drawing is not to scale. (B) Primary amino acid sequence of GlnE from M. tuberculosis. The potential nucleotidyl transferase motif found in the adenylylation domain of GlnE is boxed. The two conserved aspartates in this motif which were individually mutated to alanines are shown in bold. Residues which were mutated in the temperature-sensitive strains, TS1 and TS2, are underlined.
Identification of key residues in the adenylylation domain of GlnE.
Our data demonstrated that the C-terminal adenylylation domain is essential for in vitro growth. This region contains the motif GGAELGYGSDADVMFV (highly conserved residues are in bold), noted as being present in a superfamily of nucleotidyltransferases, and is the proposed active site for adenylylation (Fig. 1B) (16). Within this motif, the two conserved aspartates (D730 and D732) are predicted to bind a divalent cation, which has been demonstrated as essential for catalytic activity in at least one member of the superfamily (16). In order to determine if this was a key catalytic motif in the M. tuberculosis enzyme, we replaced each aspartate residue individually with an alanine residue and tested the resultant alleles for functionality. Vectors pLEDGE3M1 (D730A) and pLEDGE3M2 (D732A) were transformed into strain Glue13 as before to replace the resident vector. Viable, hygromycin-resistant, gentamicin-sensitive transformants were obtained with pLEDGE3M2 only, indicating that this mutation does not completely abrogate the enzyme activity of GlnE. In contrast, no transformants were isolated with pLEDGE3M1, indicating that this mutation does compromise its activity. These data indicate that the predicted motif is the correct assignment and that the aspartate at residue 730 is critical for GlnE activity.
Isolation of a temperature-sensitive GlnE mutant.
We were unable to construct complete (both domains) deletion mutants of GlnE under any of the conditions that we tested. In order to further our understanding of the role of this enzyme, we attempted to construct a temperature-sensitive GlnE strain of M. tuberculosis. We generated a library of randomly mutated glnE alleles in pLEDGE3 using hydroxylamine mutagenesis (10). We transformed M. tuberculosis Glue13 (ΔglnE [glnE+int Gm]) with the library and selected for switched strains on hygromycin. A total of 4.1 × 105 transformants were isolated, each of which could potentially carry a different mutant glnE allele.
One hundred sixty strains were screened for temperature sensitivity on solid medium. Since M. tuberculosis does not grow at 42°C, growth was scored at 30°C and 37°C. We isolated two strains (TS1 and TS2) which showed very poor growth at 37°C but normal growth at 30°C on plates. In fact, growth at 37°C was slower in these strains than it was at 30°C, in contrast to the case for wild-type M. tuberculosis and the other 158 strains, which grew much more slowly at 30°C than at 37°C. However, growth of TS1 and TS2 was not completely prevented at 37°C, indicating that the temperature-sensitive phenotype was incomplete. This was not surprising, since a higher temperature is often required to completely abrogate enzyme activity. However, given the retarded growth, we are able to conclude that the mutation(s) in glnE affects activity.
In order to characterize the mutations responsible for this phenotype, we amplified and sequenced the glnE genes from strains TS1 and TS2. GlnE from strain TS1 had three substitutions (D438Y, R657S, and S684K), and GlnE from strain TS2 had one substitution (V921Q). None of the substitutions were found within the nucleotidyl transferase motif.
Growth and GS activity with various nitrogen sources.
We wanted to determine what effect domain deletion or mutation had on growth and on the level of adenylylation of GS in the cells. Since all strains carried integrated vectors derived from pLEDGE3, carrying GlnE expressed from PAg85a, we used strain Glue14 (chromosomal glnE deletion, pLEDGE3 integrated) as the control.
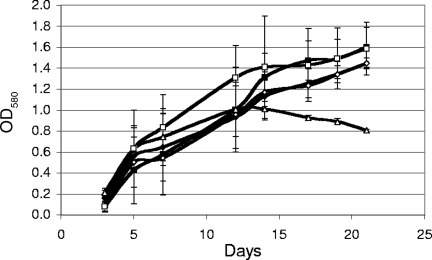
We first profiled growth and GS activity in the control strain. Growth of Glue14 was measured in medium containing one of the following nitrogen sources: a high concentration of ammonia, a low concentration of ammonia, glutamine, or glutamate. Growth also was measured in 7H9 medium. Glue14 was able to grow well in all the media tested (Fig. 2). There were no significant differences between the growth rates or maximal OD580s reached between 7H9 and TSM containing a high concentration of ammonia, glutamate, or glutamine. In contrast, growth with a low concentration of ammonia was limited and appeared to cease at a lower OD580, although the initial rate of growth was similar. Thus, the low concentration of ammonia does seem to result in a restriction of growth, presumably due to ammonia depletion during growth.
FIG. 2.
Growth of the M. tuberculosis glnE del-int strain (Glue14) in liquid medium with different nitrogen sources. Media were 7H9 (black diamonds), 30 mM (high) ammonia (black squares), 0.1 mM (low) ammonia (open triangles), 3 mM glutamate (open diamonds), or 3 mM glutamine (open squares). Data are the averages and standard deviations from three independent cultures.
We also determined the amounts of active and total GS in the cells. GS activity was measured in cell extracts by using the biosynthetic assay (35). Using this assay, we were able to measure the amount of active, unmodified GS. This represents the basic glutamine biosynthetic ability of the cells. We also wanted to assess the total amount of GS (adenylylated and nonadenylylated) and so to determine how much of the enzyme had been modified by GlnE. In order to measure the total amount of GS (modified and unmodified), we treated samples with SVPD to remove the adenylyl group from Tyr-406, making it fully active, and then assayed enzyme activity using the biosynthetic assay.
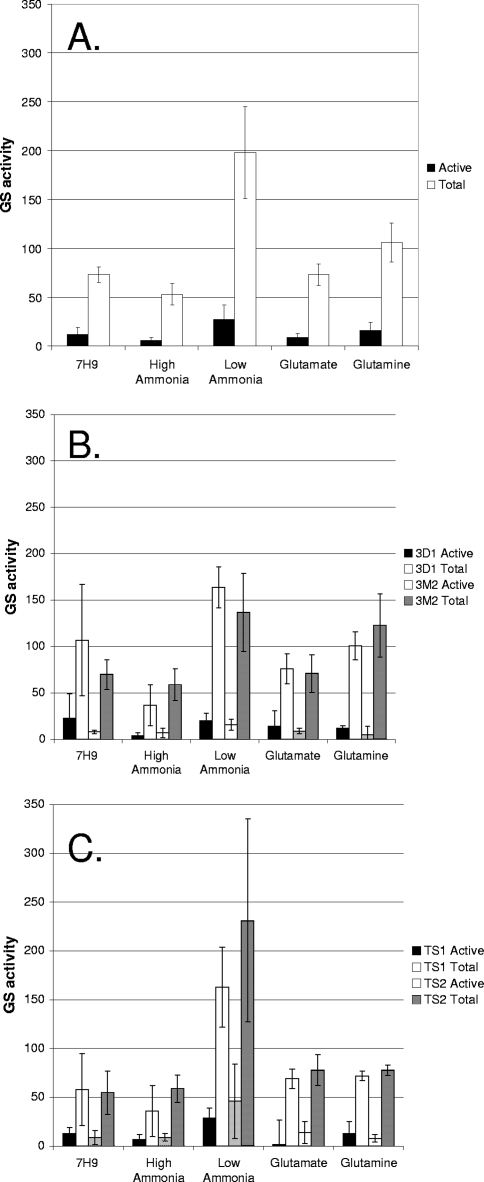
We measured GS activity in the control strain, Glue14, grown in Middlebrook 7H9 medium or in TSM with a high concentration of ammonia, a low concentration of ammonia, glutamate, or glutamine (see Fig. 4A). The level of active GS was significantly higher in the low-ammonia medium (P < 0.05) than in all the other media; the largest changes were a 4.5-fold increase over the value with a high concentration of ammonia and a 3-fold increase over the value with glutamate. Similarly, total GS was highest in low-ammonia medium, being 3.7-fold and 2.7-fold higher than in high-ammonia and glutamate media, respectively. In all conditions, the majority of GS (84 to 89%) was in the inactive, modified state. The relative levels of total GS mirrored those of active GS, so that while total GS varied between media, the proportion of active GS did not vary substantially (11 to 16%).
FIG. 4.
GS activity in M. tuberculosis strains grown in different nitrogen sources. Active GS and total GS were assayed in cell extracts from strains grown in liquid media with different nitrogen sources. (A) Control strain Glue14 carrying wild-type GlnE. (B) Strains 3D1, carrying plasmid pLEDGE3D1 (GlnE deadenylylation domain deletion), and 3M2, carrying plasmid pLEDGE3M2 (D732A mutation) (C). Temperature-sensitive strains TS1 and TS2. Data are the averages and standard deviations from three independent cultures, assayed in duplicate (n = 6). GS activity is given in μmol min−1 mg−1.
Effect of the GlnE deletions and mutations on growth and GS activity.
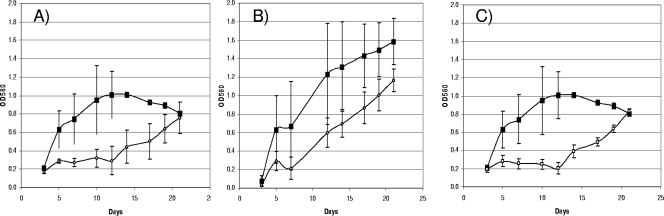
We wanted to determine whether deletion of the deadenylylation domain or the D732A mutation of the adenylylation domain had any effect on the status of GS or growth of the mutant. The strains carrying either the deletion or the mutation were subjected to the same growth and GS activity assays as for Glue14. No difference in growth rates or final OD580 was seen in any of the media (data not shown) for the deletion strain, indicating that it was able to utilize all the nitrogen sources tested. In contrast, the strain carrying GlnE-D720A showed reduced growth rates in the low-ammonia- and glutamine-containing media. Growth in the low-ammonia medium was very slow, although the strain eventually reached the same final OD580 as the control strain (Fig. 3A). Growth with glutamine was reduced, but to a lesser extent.
FIG. 3.
Growth of M. tuberculosis strains in liquid medium. (A and B) The control strain, Glue14 (black squares), and the recombinant strain carrying the mutated glnE allele (pLEDGE3M2) (open diamonds) were grown with 0.1 mM (low) ammonia (A) or 3 mM glutamine (B). (C) The control strain (Glue14 [glnE+int]) (black squares) and the temperature-sensitive strain TS2 (open squares) were grown with 0.1 mM (low) ammonia. Data are the averages and standard deviations from three independent cultures.
GS assays were carried out to measure active and total GS (Fig. 4B). The levels of GS were similar in both mutant strains, and there were no significant differences from the control strain. We observed the same pattern of expression in the different media, with higher GS levels in the low-ammonia medium and a large proportion of the GS being inactive.
Growth and GS activity of TS strains.
We had originally observed that the TS1 and TS2 strains were markedly affected in growth on 7H10 solid medium at 37°C, although growth did not completely cease. We looked at the ability of both strains to grow at 37°C in liquid medium using the same nitrogen sources as before. Surprisingly, both mutants were able to grow in the majority of the media tested and showed no significant reduction in growth rate compared to the control strain. The only significant difference was seen with strain TS2, which manifested a growth defect in low-ammonia medium. As with the GlnE-D732A strain, the growth rate was reduced, but the cells were able to grow slowly and eventually reached the same final OD580 as the control strain. It was apparent that there was an extended lag period before the cells were able to resume growth (Fig. 3C).
GS assays demonstrated that the mutant GlnE proteins still retained adenylylation activity, since modified GS still represented the major species. There was a trend toward an increase in the proportion of active GS in low-ammonia medium in both strains, from 13% in the control strain to 18% and 20% in TS1 and TS2, respectively, although this was not statistically significant. This compared to 12% in the both the deletion and mutation strains. This is consistent with a proposed reduction in GlnE activity resulting in less inactivation of GS (Fig. 4C).
Generation and characterization of a ΔGlnE strain.
We have demonstrated that glnE is essential in all media tested to date (37, 46). Our hypothesis is that GlnE is required to inactivate the large amount of GlnA1 synthesized in M. tuberculosis. In the absence of GlnE, unrestrained GlnA1 activity could potentially severely deplete the intracellular pool of glutamate and ATP. We reasoned that if this were the case, then we should be able to delete glnE in the absence of GS activity. Inhibition of GS activity is normally lethal, but this can be relieved by the addition of exogenous l-Gln (19).
We therefore attempted to construct a glnE mutant in the presence of the GS inhibitor MetSox and l-Gln. We confirmed that growth of wild-type M. tuberculosis was completely abolished on agar plates containing 200 μM Met-Sox and that addition of 3 mM l-Gln restored growth (data not shown). We transformed Glue13 with the empty vector pUC-Hyg-INT as before and selected for hygromycin-resistant strains in the presence or absence of 200 μM MetSox and 3 mM l-Gln. As before, no hygromycin-resistant, gentamicin-sensitive strains were recovered in the absence of the supplements. However, we obtained 107 transformants in the presence of MetSox and l-Gln. A number of these strains were patch tested for gentamicin sensitivity to confirm the loss of the resident plasmid; all 24 were gentamicin sensitive. We confirmed the replacement of the glnE plasmid by Southern blotting (not shown). This confirmed that we had generated a true deletion strain for glnE in the absence of GS activity. One strain (Glue21) was selected for further study.
Survival of the GlnE deletion mutant in the absence of GS inhibition.
It is possible that GlnE is required only at certain stages of the bacterial life cycle and is not always essential for survival and/or growth. In the absence of a conditional mutant, in which GlnE expression could be switched off at different times during growth, it had not previously been possible to examine this possibility. Having isolated a MetSox-dependent mutant, we now had the ability to conduct such studies by removing MetSox and thus GS inhibition in the GlnE mutant and determining the outcome.
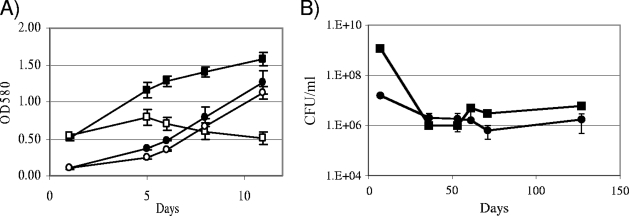
We looked at the growth of the wild-type and ΔGlnE strains in liquid medium in the presence and absence of MetSox and l-Gln (Fig. 5A). The wild-type strain showed restricted growth in the presence of MetSox and l-Gln, with both a slightly lower growth rate and a reduced final OD580. After plateauing, the OD580 started decreasing, indicating that cell death and lysis was occurring. We also saw a similar phenomenon on solid medium, where after extended periods of incubation on MetSox and Gln, both the wild-type and mutant strains started to lyse on the plate (not shown). It is probable that this is due to depletion of the l-Gln in the medium during growth.
FIG. 5.
Growth of M. tuberculosis strains in the presence or absence of GS inhibition. (A) Wild-type (squares) and Glue21 (ΔglnE) (circles) growth in liquid culture. Strains were grown in 7H9 medium with (open symbols) or without (filled symbols) 200 μM MetSox and 3 mM l-Gln (MSG). Data are the averages and standard deviations from three independent cultures. (B) Long-term survival of wild-type and Glue21 strains. Stationary-phase cultures were kept for 4 months and CFU/ml calculated at each time point. Data are the averages and standard deviations from three independent cultures.
In liquid culture, the ΔGlnE mutant grew at the same rate in medium with or without MetSox and l-Gln supplementation, although it showed an increased lag phase and a slightly lower growth rate than the wild-type strain. Thus, surprisingly, in liquid medium complete inhibition of GS is not required for its viability. We looked at the long-term survival of strains in liquid culture in the absence of GS inhibition. Cells from late stationary phase were monitored over 4 months for viability by colony counts (Fig. 5B). The number of viable cells dropped in both cultures initially until they reached 106 per ml for both strains, where they remained stable for the next 100 days. Thus, no increase in cell death was seen in the mutant strain in the absence of GS inhibition.
In order to determine whether GS activity was different in cells grown in liquid versus contrast medium, we assayed GS activity from the wild type. There were no significant differences between either total or active GS levels when cells were harvested from either solid or liquid medium, with active GS in the solid medium at 20 ± 8 and total GS at 53 ± 8. This excluded the possibility that the growth in liquid medium was due to normal levels of GS being lower in liquid medium than on solid agar.
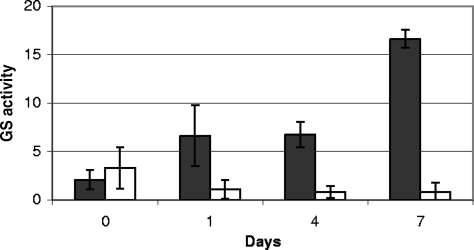
Another possibility to account for the growth of cells in liquid medium is that the cells accumulate intracellular pools of MetSox and these reserves are sufficient to maintain the GS in the inactive state. We therefore assayed active GS from the wild-type and ΔGlnE strains after passage from liquid medium with MetSox and Gln into medium without supplementation (Fig. 6). In the wild-type strain, active GS slowly increased over 7 days, suggesting that newly synthesized GS is not inactivated by residual MetSox. After 7 days, the levels of active GS were similar to those seen in culture previously. Surprisingly, in the GlnE mutant, no increase in active GS was seen over the same time period, and the levels remained just detectable. This therefore explains the viability of the cells in the absence of GlnE expression, since high levels of active GS are not being produced.
FIG. 6.
Active GS in M. tuberculosis strains after removal of MetSox. Strains were grown in 7H9 medium with 200 μM MetSox and 3 mM l-Gln for 7 days, washed, and subcultured in fresh 7H9 medium without supplementation for up to 7 days. Active GS was assayed in cell extracts prepared using the biosynthetic reaction. Data are the averages and standard deviations from three independent cultures, assayed in duplicate (n = 6). GS activity is given in μmol min−1 mg−1.
DISCUSSION
Essentiality of GlnE.
The essentiality of GlnE in M. tuberculosis is an oddity among bacterial species. We wanted to understand why GlnE is essential in M. tuberculosis when it is not in other bacteria. Essentiality was previously demonstrated in Middlebrook 7H9 medium, which contains a large quantity of glutamate (24). GlnE mutants of other bacteria show growth defects on certain nitrogen sources (2, 6), so we tested whether we could obtain mutants when the cells were grown on alternative nitrogen sources. In particular, the availability of nitrogen (high or low level) and/or the ability to utilize glutamate or glutamine might be expected to have an effect on the requirement for GlnE activity. In the absence of glutamate, one might expect that the lack of modification and inactivation of GS would not be critical, since there is no exogenous substrate. In contrast to our prediction, we found that glnE was required for growth in all nitrogen sources. Thus, the modification of GS seems to be a prerequisite for mycobacterial growth, and we have discounted the possibility that our inability to isolate a mutant was due to the choice of medium in our original work.
Domain identification.
GlnE is comprised of two domains (adenylylation and deadenylylation) and an interregion linker (18). In addition to the GS binding regions in the Pfam domains, there are independent PII, PII-UMP, and glutamine binding sites on the enzyme. In high-glutamine environments, glutamine and PII bind to GlnE, causing the linker region to bring the two domains together and allow adenylylation. In contrast, when there is a low glutamine level, PII-UMP binds to GlnE, causing a conformation change which allows deadenylylation (18).
We have shown that the adenylylation domain encodes the key activity in M. tuberculosis, since deletion of the deadenylylation domain had no effect on growth or GS activity in a variety of media. In addition, we show that we could isolate a deletion mutant when all GS activity was inhibited. Taken together, these data suggest that our original hypothesis that inactivation of GS is required is correct. Recent work showed that a triple mutant (GlnA1, GlnE, and GlnA2 deletion), in which the major GS is deleted together with GlnE, was viable (19), further supporting our hypothesis.
Motif analysis and mutagenesis.
Mutations induced by hydroxylamine resulted in base substitutions from guanine to adenine as expected. Two temperature-sensitive strains, TS1 and TS2 were isolated. No complete structural data are available for M. tuberculosis GlnE, so it is not possible to predict the effect of these mutations on the enzyme activity. However, it is likely that mutations affect protein folding rather than interfere with the catalytic activity per se. The temperature-sensitive phenotype was most pronounced in the nitrogen-limited medium (low ammonia level); in this medium growth was the most restricted. This is probably due to the fact that GS levels (active and total) were highest in this medium; thus, the reduced inactivation of the GS would have the greatest effect on these cells. This is supported by the data showing that the levels of active GS were at the highest in the two TS mutants grown in low-ammonia medium.
GS activity and GlnE expression.
It was surprising that the majority of the GS in the cells was inactive. We predicted that the proportion of active GS might change in response to the available nitrogen source, since this provides the quickest response to environmental changes. In fact, the proportion of inactive GS was fairly constant (and low), and the increase in GS activity seen in low-ammonia medium was due to an increase in total GS levels. There was a small increase in the proportion of active GS in the TS2 strain, which was accompanied by reduced growth. It is likely that the two are related and that increased GS activity causes a reduction in growth rate due to perturbation of the intracellular balance of glutamate to glutamine and decreased ATP levels. Previous work with M. tuberculosis grown in Sauton's medium indicated that GS was not adenylylated in the absence of glutamine (23); from these data it was not clear whether the GS assayed was intracellular or extracellular. In our work, we did not find any conditions under which GS was unmodified, and this ties in with our observations that GlnE is essential in the same conditions. However, it remains possible that the differences in our data result from the different media used, since Sauton's medium contains both asparagine and ferric ammonium citrate as nitrogen sources, whereas we used single nitrogen sources in our work. Interestingly, we were unable to isolate a GlnE mutant using asparagine as a sole nitrogen source.
There is conflicting information available on the regulation of GlnA1 in response to ammonia levels. We had previously seen a higher level of total GS activity in cell extracts and culture filtrates from cells grown with high levels of ammonia compared to low levels of ammonia in large-scale cultures (29). In contrast Harth and Horwitz (13) showed that GlnA1 was upregulated, both at the mRNA level and at the enzymatic activity level, in low-ammonia conditions. In our current work, we saw higher levels of GS in low-ammonia conditions, in agreement with Harth and Horwitz (13). However, there are several differences between our current methodology and our previous work which may account for this. In particular, we used the biosynthetic assay in this study, rather than the transferase assay as used previously (29); the latter distinguishes between active and inactive GS by the use of different covalent metal ions, as opposed to the use of SVPD. In addition, we used different culture conditions here, i.e., 10-ml stirred cultures as opposed to 100-ml rolling cultures. The two growth conditions are quite different in terms of growth rate and oxygen availability, which could have significant consequences in determining the activity of basic metabolic pathways. In other bacterial species GS activity and expression are controlled by nitrogen availability and by the intracellular concentrations of other key molecules, including glutamine, ATP, and 2-ketoglutarate. Our data and those of others suggest that expression of GlnA1 in M. tuberculosis is also regulated by more than the nitrogen source alone.
Persistence of the GlnE mutant in the absence of GS inhibition.
Given that we were unable to isolate a GlnE mutant in the presence of GS activity, it was surprising that once we obtained the deletion strain, it was able to grow and survive for extended periods in the absence of GS inhibition. One possibility was that the inhibition of GS is required only on solid medium; this was supported by the increased severity of the temperature-sensitive mutation on solid medium and the inability of the mutant strain to grow on plates without GS inhibition (MetSox). This could stem from an increased requirement for glutamine during metabolism. However, given that levels of active and total GS were the same in solid and liquid media, this is unlikely.
Our data indicate that there was no new synthesis of GS in the GlnE mutant strain after subculture from MetSox medium, strongly suggesting that this is the reason for its ability to grow and persist in liquid. There was a low level of active GS present, which could be sufficient for glutamine biosynthesis, but the higher levels of active GS seen in the wild type were not found. If GS has a secondary function, aside from glutamine biosynthesis, then the presence of large amounts of GS already in the cell, albeit inactive, may repress expression of new active GS. If a reversible or competitive inhibitor were used, which could be washed out of the cells, then it would be possible to sassess the effect of deletion of GlnE on growing cells. Unfortunately, no specific inhibitors currently exist, so it is not possible to assay this.
Role of inactive GS.
At present, it is not clear why M. tuberculosis produces such large quantities of GS or why it is exported at a high level. Previous work has suggested that nonpathogenic mycobacteria produce much lower levels of GS and have little, if any, adenylylated GS, in contrast to pathogenic mycobacteria. It has been suggested that GS may modulate the extracellular pH, for example in macrophages, by modulating external ammonia levels in the phagosome or that the poly-l-Gln found in large quantities in the cell wall only in pathogenic species is important for pathogenesis (11). However, the synthetic reaction requires an ATP source, which may not be present in the extracellular milieu, and it is still possible that GS plays an alternative, as-yet-undefined role.
Conclusion.
We have demonstrated that the nucleotidyltransferase locus in the adenylylation domain of GlnE is critical for the control of GS and, ultimately, essential for M. tuberculosis viability. However, the finding that we could generate a viable GlnE null mutant in the presence of MetSox and glutamine and its ability to survive in the absence of GS inhibition cast doubt on whether the enzymes involved in this regulatory cascade are ideal drug targets. However, the abundance of inactive GS produced by the bacilli leads to the hypothesis that it may have a secondary function, and this novel phenomenon is of interest for the further study in M. tuberculosis.
Supplementary Material
Acknowledgments
This work was funded by Wellcome Trust grant 063894, St. Bartholomew's and the Royal London Charitable Foundation grant RAB 03/PJ/08, and European Union project LSHP-CT-2005-018923.
We thank Shekharam Chandra for advice regarding GS assays and Tanjore Balganesh for helpful discussion.
Footnotes
Published ahead of print on 9 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Almassy, R. J., C. A. Janson, R. Hamlin, N. H. Xuong, and D. Eisenberg. 1986. Novel subunit-subunit interactions in the structure of glutamine synthetase. Nature 323304-309. [DOI] [PubMed] [Google Scholar]
- 2.Bancroft, S., S. G. Rhee, C. Neumann, and S. Kustu. 1978. Mutations that alter the covalent modification of glutamine synthetase in Salmonella typhimurium. J. Bacteriol. 1341046-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, 3rd, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 4.Collins, D. M., T. Wilson, S. Campbell, B. M. Buddle, B. J. Wards, G. Hotter, and G. W. De Lisle. 2002. Production of avirulent mutants of Mycobacterium bovis with vaccine properties by the use of illegitimate recombination and screening of stationary-phase cultures. Microbiology 1483019-3027. [DOI] [PubMed] [Google Scholar]
- 5.Downing, K. J., R. A. McAdam, and V. Mizrahi. 1999. Staphylococcus aureus nuclease is a useful secretion reporter for mycobacteria. Gene 239293-299. [DOI] [PubMed] [Google Scholar]
- 6.Fink, D., D. Falke, W. Wohlleben, and A. Engels. 1999. Nitrogen metabolism in Streptomyces coelicolor A3(2): modification of glutamine synthetase I by an adenylyltransferase. Microbiology 1452313-2322. [DOI] [PubMed] [Google Scholar]
- 7.Gill, H. S., G. M. Pfluegl, and D. Eisenberg. 2002. Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation. Biochemistry 419863-9872. [DOI] [PubMed] [Google Scholar]
- 8.Ginsburg, A. 1972. Glutamine synthetase of Escherichia coli: some physical and chemical properties. Adv. Protein Chem. 261-79. [Google Scholar]
- 9.Gu, S., J. Chen, K. M. Dobos, E. M. Bradbury, J. Belisle, and X. Chen. 2003. Comprehensive proteomic profiling of the membrane constitutents of a Mycobacterium tuberculosis strain. Mol. Cell Proteome 21284-1296. [DOI] [PubMed] [Google Scholar]
- 10.Guilhot, C., B. Gicquel, and C. Martin. 1992. Temperature-sensitive mutants of the Mycobacterium plasmid pAL5000. FEMS Microbiol. Lett. 77181-186. [DOI] [PubMed] [Google Scholar]
- 11.Harth, G., D. L. Clemens, and M. A. Horwitz. 1994. Glutamine synthetase of Mycobacterium tuberculosis: extracellular release and characterization of its enzymatic activity. Proc. Natl. Acad. Sci. USA 919342-9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harth, G., and M. A. Horwitz. 1999. An inhibitor of exported Mycobacterium tuberculosis glutamine synthetase selectively blocks the growth of pathogenic mycobacteria in axenic culture and in human monocytes: extracellular proteins as potential novel drug targets. J. Exp. Med. 1891425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harth, G., and M. A. Horwitz. 1997. Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J. Biol. Chem. 27222728-22735. [DOI] [PubMed] [Google Scholar]
- 14.Harth, G., and M. A. Horwitz. 2003. Inhibition of Mycobacterium tuberculosis glutamine synthetase as a novel antibiotic strategy against tuberculosis: demonstration of efficacy in vivo. Infect. Immun. 71456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harth, G., S. Maslesa-Galic, M. V. Tullius, and M. A. Horwitz. 2005. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 581157-1172. [DOI] [PubMed] [Google Scholar]
- 16.Holm, L., and C. Sander. 1995. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 20345-347. [DOI] [PubMed] [Google Scholar]
- 17.Jaggi, R., W. C. van Heeswijk, H. V. Westerhoff, D. L. Ollis, and S. G. Vasudevan. 1997. The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction. EMBO J. 165562-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, P., E. A. Mayo, and A. J. Ninfa. 2007. Escherichia coli glutamine synthetase adenylyltransferase: kinetic characterization of regulation by PII, PII-UMP, glutamine, and alpha-ketoglutarate. Biochemistry 464133-4136. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S., B. Y. Jeon, S. Bardarov, M. Chen, S. L. Morris, and W. R. Jacobs. 2006. Protection elicited by two glutamine auxotrophs of Mycobacterium tuberculosis and in vivo growth phenotypes of the four unique glutamine synthetase mutants in a murine model. Infect. Immun. 746491-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine, R. L., L. Mosoni, B. S. Berlett, and E. R. Stadtman. 1996. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 9315036-15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liaw, S. H., J. J. Villafranca, and D. Eisenberg. 1993. A model for oxidative modification of glutamine synthetase, based on crystal structures of mutant H269N and the oxidized enzyme. Biochemistry 327999-8003. [DOI] [PubMed] [Google Scholar]
- 22.Mahenthiralingam, E., B. I. Marklund, L. A. Brooks, D. A. Smith, G. J. Bancroft, and R. W. Stokes. 1998. Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals no essential role for the protein in the growth and virulence of Mycobacterium intracellulare. Infect. Immun. 663626-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehta, R., J. T. Pearson, S. Mahajan, A. Nath, M. J. Hickey, D. R. Sherman, and W. M. Atkins. 2004. Adenylylation and catalytic properties of Mycobacterium tuberculosis glutamine synthetase expressed in Escherichia coli versus mycobacteria. J. Biol. Chem. 27922477-22482. [DOI] [PubMed] [Google Scholar]
- 24.Parish, T., and N. G. Stoker. 2000. glnE is an essential gene in Mycobacterium tuberculosis. J. Bacteriol. 1825715-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 1461969-1975. [DOI] [PubMed] [Google Scholar]
- 26.Pashley, C. A., A. C. Brown, D. Robertson, and T. Parish. 2006. Identification of the Mycobacterium tuberculosis GlnE promoter and its response to nitrogen availability. Microbiology 1522727-2734. [DOI] [PubMed] [Google Scholar]
- 27.Pashley, C. A., and T. Parish. 2003. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 229211-215. [DOI] [PubMed] [Google Scholar]
- 28.Proler, M., and P. Kellaway. 1962. The methionine sulfoximine syndrome in the cat. Epilepsia 3117-130. [DOI] [PubMed] [Google Scholar]
- 29.Read, R., C. A. Pashley, D. Smith, and T. Parish. 2007. The role of GlnD in ammonia assimilation in Mycobacterium tuberculosis. Tuberculosis 87384-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee, S. G., S. C. Park, and J. H. Koo. 1985. The role of adenylyltransferase and uridylyltransferase in the regulation of glutamine synthetase in Escherichia coli. Curr. Top. Cell Regul. 27221-232. [DOI] [PubMed] [Google Scholar]
- 31.Ronzio, R. A., and A. Meister. 1968. Phosphorylation of methionine sulfoximine by glutamine synthetase. Proc. Natl. Acad. Sci. USA 59164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stadtman, E. R. 2001. The story of glutamine synthetase regulation. J. Biol. Chem. 27644357-44364. [DOI] [PubMed] [Google Scholar]
- 33.Tullius, M. V., G. Harth, and M. A. Horwitz. 2003. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 713927-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tullius, M. V., G. Harth, and M. A. Horwitz. 2001. High extracellular levels of Mycobacterium tuberculosis glutamine synthetase and superoxide dismutase in actively growing cultures are due to high expression and extracellular stability rather than to a protein-specific export mechanism. Infect. Immun. 696348-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woolfolk, C. A., B. Shapiro, and E. R. Stadtman. 1966. Regulation of glutamine synthetase. I. Purification and properties of glutamine synthetase from Escherichia coli. Arch. Biochem. Biophys. 116177-192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.