Abstract
The nonpathogenic Bacillus subtilis and the pathogen Staphylococcus aureus are gram-positive model organisms that have to cope with the radical nitric oxide (NO) generated by nitrite reductases of denitrifying bacteria and by the inducible NO synthases of immune cells of the host, respectively. The response of both microorganisms to NO was analyzed by using a two-dimensional gel approach. Metabolic labeling of the proteins revealed major changes in the synthesis pattern of cytosolic proteins after the addition of the NO donor MAHMA NONOate. Whereas B. subtilis induced several oxidative stress-responsive regulons controlled by Fur, PerR, OhrR, and Spx, as well as the general stress response controlled by the alternative sigma factor SigB, the more resistant S. aureus showed an increased synthesis rate of proteins involved in anaerobic metabolism. These data were confirmed by nuclear magnetic resonance analyses indicating that NO causes a drastically higher increase in the formation of lactate and butanediol in S. aureus than in B. subtilis. Monitoring the intracellular protein thiol state, we observed no increase in reversible or irreversible protein thiol modifications after NO stress in either organism. Obviously, NO itself does not cause general protein thiol oxidations. In contrast, exposure of cells to NO prior to peroxide stress diminished the irreversible thiol oxidation caused by hydrogen peroxide.
Bacillus subtilis and Staphylococcus aureus are gram-positive model bacteria. Whereas B. subtilis is considered to be harmless, S. aureus is a facultative pathogen and the leading cause of nosocomial and community-acquired infections. Both microorganisms have to cope with high amounts of nitric oxide (NO) (up to the μM range) generated from coexisting denitrifying bacteria or from the innate immune response of the host, respectively (17, 31, 55, 68). Hence, the ability of B. subtilis and S. aureus to protect themselves against NO might be crucial for survival in their respective natural habitats.
The cytotoxic properties of the small, lipophilic, and freely diffusible radical NO are attributed to its high reactivity. Indirect effects of NO are caused by the reaction of the radical with oxygen or superoxide, resulting in the formation of a number of additional reactive nitrogen species, including nitrogen dioxide, peroxynitrite, and dinitrogen trioxide. These nitrogen species differ in reactivity, stability, and biological activity but result in a broad spectrum of antimicrobial activity (25). In general, reactive oxygen and nitrogen species can interact with numerous targets, including thiols, metal centers, tyrosine residues in proteins, nucleotide bases, and lipids (20, 69). NO directly affects the activity of enzymes by the reaction with bound free radicals or with metal centers (51, 72, 100). For example, the formation of metal-nitrosyl complexes in respiratory enzymes was shown to inhibit bacterial respiration (8, 13, 71, 88) and the formation of a dinitrosyl-iron complex of a protein essential for branched-chain amino acid biosynthesis was recently shown to be the main cause of NO-induced growth arrest in Escherichia coli (44). Moreover, reaction of NO with the tyrosyl radical formed in the catalytic turnover of ribonucleotide reductase is considered to be responsible for the suppression of DNA synthesis (53, 54).
Bacteria have specific and general defense strategies to counter environmental changes, including detoxification of stressors, as well as protection mechanisms and repair systems. These responses are based on sophisticated pathways of signal transduction, which subsequently trigger changes in gene expression. Proteomics is an excellent tool for the characterization of the adaptive network induced by stress and starvation stimuli (37, 97). Pulse-labeling with radioactive amino acids and separation of labeled protein extracts with two-dimensional polyacrylamide gel electrophoresis (2D PAGE) allows the generation of protein synthesis profiles that are particularly valuable to predict the physiological state of the cell and to allow comparative physiological proteomics involving multiple stress conditions. For B. subtilis, the proteome of growing cells (14, 24), as well as proteome signatures produced in response to stress or starvation conditions, including oxygen limitation (57), nutrient starvation (92, 94), and treatment with aromatic organic compounds and antibiotics, have been characterized (4, 23, 93). In particular, the responses of B. subtilis to peroxides and to the thiol-specific oxidant diamide have been extensively described at the proteome level (52, 61, 92). The analysis of the oxidative stress response was made more comprehensive by the introduction of a fluorescence-based proteomic approach that allows for the visualization of posttranslational modifications of the thiol group of cysteine residues (41). For S. aureus, the proteomes of exponentially growing and stationary-phase cells have been described (50), but the number of proteome signatures to stress and starvation conditions is still limited (98). The first comprehensive description of global changes in the synthesis of cytoplasmic proteins after a shift from aerobic to anaerobic growth conditions was recently provided by Fuchs et al. (28).
NO-induced changes in the proteome of B. subtilis and S. aureus have not been described thus far. In the present study, we characterized the response of both microorganisms toward NO by analyzing the protein synthesis profiles and by monitoring the protein thiol state. The data were compared to transcriptomic data for NO stress which were published recently (60, 83). In line with these data, our comprehensive proteome approach revealed major differences in the adaptation to NO stress in the two microorganisms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis strain 168 (1) and the S. aureus strain COL (87) were grown aerobically at 37°C with vigorous shaking. The composition of the synthetic medium used for B. subtilis (90) and S. aureus (29) has been described. Growth was monitored by measuring the optical density at 500 nm (OD500). The compound MAHMA NONOate [6-(2-hydroxy-1-methyl-2-nitrosohydrazino)-N-methyl-1-hexanamine; Sigma] was used as an NO donor. For each experiment, a 100 mM MAHMA NONOate solution was freshly prepared. The NO donor was dissolved in 10 mM NaOH, and the solution was kept at 4°C in the dark until used. A total of 100 ml of synthetic medium was inoculated with an exponentially growing overnight culture of S. aureus and B. subtilis to initial OD500s of 0.08 and 0.03, respectively. Cells were cultivated to an OD500 of 0.5, and MAHMA NONOate was added to final concentrations ranging from 100 μM to 1 mM.
[35S]methionine pulse-labeling and preparation of the cytoplasmic protein fraction.
Changes in protein synthesis after NO exposure were analyzed by [35S]methionine pulse-labeling. Radioactive methionine (15 μCi of l-[35S]methionine/ml) was added to exponentially growing (OD500 = 0.5) or stressed cells immediately before and 1, 5, 10, and 30 min (as well as 60 min in the case of S. aureus) after the addition of MAHMA NONOate to a final concentration of 100 μM for B. subtilis cells and 500 μM for S. aureus cells. In addition, as a control, cell cultures were treated with similar concentrations of the completely decomposed NO donor (dissolved in synthetic medium and incubated for 24 h at room temperature) for 10 min. After 5 min, the incorporation of radioactively labeled methionine was stopped by the addition of 0.1 mg of chloramphenicol/ml and 1 mM unlabeled l-methionine, and the cells were transferred to ice. Cells from 10 ml of culture medium were harvested by centrifugation for 5 min at 9,000 × g and 4°C. The resulting cell pellet was washed twice with TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]) and resuspended in 400 μl of TE buffer. In the case of S. aureus, the TE buffer was supplemented with 25 ng of lysostaphin/ml, and the cell suspension was incubated on ice for 10 min before cell disruption. Cells were disrupted on ice by sonication, and cell debris was removed by centrifugation for 30 min at 21,000 × g and 4°C. Protein extracts were stored at −20°C.
Fluorescence labeling of S-nitrosylated and reversibly oxidized proteins.
The fluorescence thiol modification assay was used to monitor the protein thiol state and to detect proteins with reversibly oxidized thiol groups following NO stress (41, 101). Samples (50 ml) were taken from exponentially growing B. subtilis and S. aureus cell cultures (OD500 = 0.5) immediately before and 10 min after the addition of 100 μM to 2 mM MAHMA NONOate. Cells were harvested by addition of ice-cold trichloroacetic acid (TCA; final concentration, 2% [wt/vol]) and subsequent centrifugation at 8,900 × g and 4°C. The resulting cell pellet was washed twice with deionized water acidified with TCA to pH 1.5 and resuspended in 450 μl of denaturing buffer (8 M urea, 1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 1 mM EDTA, 200 mM Tris [pH 8.0]) supplemented with 100 mM iodoacetamide. Cells were then disrupted by sonication, centrifuged for 5 min at 21,000 × g and 4°C, and incubated for 20 min at room temperature. Excess iodoacetamide was removed by the addition of four parts ice-cold acetone and storage for at least 1 h at −20°C. After centrifugation at 20°C, the samples were washed twice with acetone and dried in a vacuum centrifuge. The resulting protein pellet was dissolved in 150 μl of denaturing buffer, and the protein concentration was determined. Reversible thiol oxidations were then reduced with 10 nmol of Tris-(2-carboxyethyl)-phosphine (TCEP) per 50 μg of protein, and the newly generated thiol groups were labeled with the fluorescence dye BODIPY FL C1-IA [N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-yl)-methyl)-iodoacetamide] (Invitrogen, Eugene, OR) as already described (41). To exclusively label S-nitrosylated proteins, ascorbate instead of TCEP in the reduction step was used (45, 47). The protein extracts were incubated in ascorbate solution (prepared in denaturing buffer) in a ratio of 50 μg of protein to 50 nmol ascorbate for 1 h at room temperature in the dark prior to labeling.
2D PAGE and gel imaging.
Proteins (500 μg of nonradioactive protein, 100 μg of radiolabeled protein) were separated with 2D PAGE in the pH range of 4 to 7 according to the method of Büttner et al. (14). For autoradiograms, gels were dried on filter paper and exposed to storage phosphor screens. The nonradioactive gels were stained with Sypro Ruby (Invitrogen) according the recommendations of the manufacturer. Fluorescent and radioactive gels were scanned with the Typhoon 9400 variable mode imager (Amersham Biosciences, Freiburg, Germany).
Data analysis.
Gel images were analyzed with Delta2D software (Decodon, Greifswald, Germany). The gel images from two independently analyzed cell cultures were included for every sampling time point. A fusion image was generated from all images of an experiment (56), and the detected spot centers of the fusion image were transferred to all other images to ensure constant spot detection. To determine proteins with changes in protein synthesis after NO stress, the protein synthesis ratios of the total normalized spot quantities of the autoradiograms of the NO-stressed samples, and the corresponding spot of the control sample were determined. Spots with synthesis ratios of ≥2.0 and ≤0.5 in both replicates at at least one time point after NO stress were considered as proteins with significant changes in the synthesis rate. Proteins whose synthesis rate was significantly increased after NO stress were defined as marker proteins. To detect proteins with thiol modifications, fluorescence images were overlaid with the Sypro Ruby-stained images of the same 2D gels, and the thiol modification ratios were calculated by dividing the fluorescence/protein ratios of the NO-stressed samples with the corresponding ratios of the control (41).
Protein identification.
Protein spots which could not be definitely assigned to already generated 2D gel proteome maps (4, 24, 28, 49, 52, 61, 92-94) were cut from preparative 2D gels and identified by automated tryptic digestion in the Ettan Spot Handling Workstation (Amersham-Biosciences, Uppsala, Sweden) and tandem mass spectrometry on a Proteome Analyzer 4700 (Applied Biosystems, Foster City, CA) as previously described (24).
Quantification of extracellular metabolites by 1H-NMR.
Filtered extracelluar samples (300 μl) were buffered to pH 7.0 by addition of 200 μl of a sodium hydrogen phosphate buffer (0.2 mM [pH 7.0]) made up with 25% D2O to provide an nuclear magnetic resonance (NMR)-lock signal. Samples were transferred to 5-mm NMR-glass tubes (length, 7 in.). Spectral referencing was relative to 1 mM sodium 3-trimethylsilyl-[2,2,3,3-D4]-1-propionic acid (TMSP) in phosphate buffer. All NMR spectra were obtained at 600.27 MHz at a nominal temperature of 298.5 K using a Bruker AVANCE-II 600 NMR spectrometer operated by TOPSPIN 2 software (both from Bruker Biospin GmbH, Rheinstetten, Germany). A modified 1D-NOESY pulse sequence was used with presaturation on the residual HDO signal during both the relaxation delay and the mixing time. A total of 128 free induction decays (FID scans) were collected into 64k data points using a spectral width of 20 ppm for a one-dimensional spectrum.
After Fourier transformation with 0.3-Hz line broadening and a single zero-filling, spectra were automatically phased and baseline corrected using the baseopt process, and the chemical shift scale was set by assigning the value of δ of 0.00 ppm corresponding to the signal from the added TMSP. Compound identification was done by matching the obtained spectra with a 1H-NMR spectra database using the program AMIX (Bruker Biospin) and comparing with spectra of standard compounds. Quantification was done by integration of designated peaks in the 1H-NMR spectrum and comparing them with the added standard TMSP.
RESULTS AND DISCUSSION
NO stress proteomic signature.
To investigate the response of B. subtilis and S. aureus to NO stress, exponentially growing cells were exposed to the NO donor MAHMA NONOate. One mole of MAHMA NONOate liberates two moles of authentic NO with a half-life of about 1 min in physiological buffer solutions (pH 7.4, 37°C) (48). The addition of 100 μM to 1 mM concentrations of the NO donor to exponentially growing cells in synthetic medium resulted in substantially decreased growth rates. The lag phase of growth was prolonged with increasing concentrations of the NO donor. Interestingly, S. aureus is more tolerant against NO stress than is B. subtilis. The transient growth arrest after NO stress in B. subtilis and S. aureus and the higher NO resistance of S. aureus compared to other microorganisms have also been observed by others (60, 83).
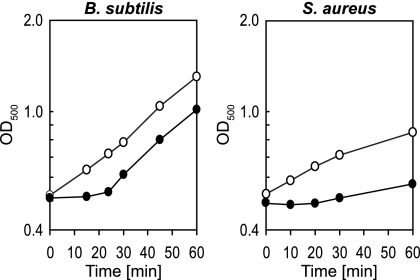
To study changes in gene expression at the proteome level, exponentially growing cells were exposed to 100 μM concentrations of the NO donor in the case of B. subtilis and 500 μM concentrations of the NO donor for S. aureus, achieving a comparable growth arrest of about 20 min (Fig. 1). Proteins were pulse-labeled by the addition of l-[35S]methionine to cells before, as well as 1, 5, 10, and 30 min after, stress. The proteins were separated by 2D PAGE, and the synthesis patterns of untreated control and NO-stressed cells were compared. Proteins represented by spots with a significant increase in radioactivity after NO exposure are synthesized at a higher level after stress and were defined as marker proteins. Movies were created from the individual overlaid 2D gel images in order to give an idea of the dynamics of the physiological changes. Identified marker proteins are labeled according to their regulatory groups in different colors, and the labels can be separately visualized. (Interactive movies are provided in the supplemental material).
FIG. 1.
Growth analysis of B. subtilis 168 and S. aureus COL cells after NO stress. Cells were grown in synthetic medium, and the cultures were initiated at an OD500 of 0.5 (0 min) (○). The growth of parallel cultures exposed to 100 μM (B. subtilis) or 500 μM (S. aureus) of the NO donor MAHMA NONOate is also shown (•).
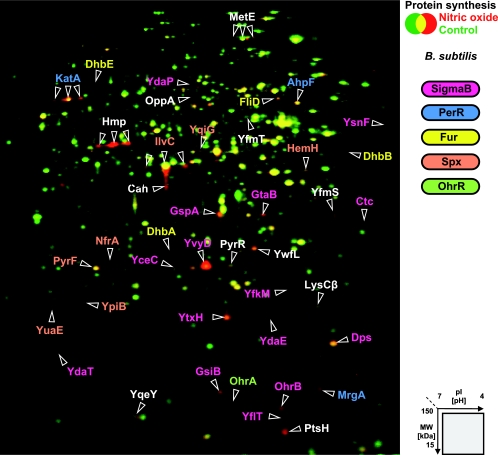
In total, we identified 41 proteins as markers for NO stress in B. subtilis (Fig. 2 and Table 1). About one-fourth of all marker proteins showed a higher synthesis level already a few minutes after imposition to NO stress. The remaining proteins were induced after 5 or 10 min. The synthesis rates of all marker proteins reached the control levels after 30 min, by which time the cells had already resumed growth.
FIG. 2.
Synthesis of cytoplasmic proteins of B. subtilis after NO stress. The 2D gel images of newly synthesized proteins (labeled with l-[35S]methionine) from exponentially growing cells (shown in green) and cells exposed to 100 μM concentrations of the NO donor MAHMA NONOate for 10 min (shown in red) were overlaid. Identified proteins with an increased synthesis rate 1 to 30 min after stress are labeled and color coded for their membership to specific regulons as indicated.
TABLE 1.
Proteins with increased synthesis after NO stress in B. subtilis
| Category and protein | Description | Fold change in synthesis (NO/control)a
|
|||
|---|---|---|---|---|---|
| 1 min | 5 min | 10 min | 30 min | ||
| ResDE dependent | |||||
| Hmp | Flavohemoglobin | 1.6; 2.9 | 16.4; 22.1 | 63.8; 77.1 | 0.7; 0.9 |
| YwfI | Unknown; similar to unknown proteins | 1.4; 1.6 | 2.3; 2.7 | 4.1; 3.8 | 1.1; 0.9 |
| SigB dependent | |||||
| Ctc | General stress protein | 0.3; 7.2 | 3.1; 10.0 | 4.7; 8.8 | 0.5; 1.0 |
| Dps | Stress- and starvation-induced gene controlled by sigmaB | 1.4; 4.9 | 2.6; 6.9 | 1.3; 2.9 | 0.3; 0.6 |
| GsiB | General stress protein | 3.3; 3.8 | 7.7; 7.3 | 10.2; 5.6 | 1.4; 0.7 |
| GspA | General stress protein | 0.7; 1.4 | 2.3; 3.6 | 1.9; 2.4 | 0.5; 0.8 |
| GtaB | UTP-glucose-1-phosphate uridylyltransferase | 1.5; 3.1 | 6.0; 8.2 | 5.0; 3.6 | 1.3; 0.6 |
| NfrA | Flavin mononucleotide-containing NADPH-linked nitro/flavin reductase | 1.4; 1.2 | 2.0; 2.3 | 2.8; 2.5 | 1.0; 0.8 |
| OhrB | Unknown; similar to organic hydroperoxide resistance protein | 1.7; 4.4 | 5.0; 5.6 | 4.7; 4.5 | 0.6; 0.6 |
| YceC | Unknown; similar to tellurium resistance protein | 1.1; 2.9 | 1.9; 1.6 | 2.3; 2.3 | 1.0; 0.9 |
| YdaE | Unknown | 1.9; 2.9 | 3.9; 3.0 | 3.2; 2.0 | 0.6; 0.9 |
| YdaP | Unknown; similar to pyruvate oxidase | 2.0; 2.1 | 2.3; 2.9 | 1.5; 2.0 | 0.6; 0.6 |
| YdaT | Unknown | 2.6; 2.6 | 2.1; 2.3 | 2.3; 2.5 | 1.1; 0.8 |
| YfkM | Unknown; similar to unknown proteins | 1.9; 5.8 | 3.7; 7.5 | 3.1; 4.5 | 0.5; 1.0 |
| YflT | Unknown | 2.4; 4.7 | 7.4; 8.7 | 7.2; 6.0 | 0.6; 1.0 |
| YsnF | Unknown; similar to unknown proteins | 0.8; 3.0 | 1.0; 10.6 | 3.9; 5.8 | 0.8; 1.0 |
| YtxH | Unknown; similar to general stress protein | 1.4; 2.5 | 2.4; 4.7 | 2.5; 4.8 | 0.5; 0.9 |
| YvyD | Unknown; similar to sigma-54 modulating factor of gram-negative bacteria | 3.4; 13.9 | 18.3; 20.5 | 30.3; 39.2 | 0.7; 0.7 |
| PerR dependent | |||||
| AhpF | Alkyl hydroperoxide reductase (large subunit)/NADH dehydrogenase | 0.8; 1.7 | 4.2; 6.7 | 2.6; 3.5 | 0.9; 1.1 |
| KatA | Vegetative catalase 1 | 1.2; 1.5 | 1.8; 1.7 | 5.0; 9.1 | 0.8; 0.7 |
| MrgA | Metalloregulation DNA-binding stress protein | 1.7; 3.2 | 2.6; 2.0 | 3.0; 3.3 | 1.2; 0.9 |
| Fur dependent | |||||
| DhbA | 2,3-Dihydro-2,3-dihydroxybenzoate dehydrogenase | 1.2; 3.9 | 2.9; 1.4 | 2.8; 2.9 | 0.9; 1.0 |
| DhbB | Isochorismatase | 51.2; 34.1 | 22.6; 23.4 | 12.0; 3.2 | 3.6; 1.6 |
| DhbE | Isochorismate synthase | 1.5; 3.0 | 3.2; 2.2 | 0.8; 0.9 | 0.7; 0.8 |
| FliD | Flagellar hook-associated protein 2 (HAP2) | 0.5; 0.4 | 0.7; 0.8 | 2.6; 2.3 | 1.0; 1.1 |
| Spx dependent | |||||
| HemH | Ferrochelatase | 0.6; 0.7 | 1.6; 2.2 | 2.6; 2.0 | 1.2; 1.1 |
| IlvC | Ketol-acid reductoisomerase | 0.7; 0.9 | 2.8; 2.7 | 3.2; 2.1 | 1.0; 0.8 |
| NfrA | Flavin mononucleotide-containing NADPH-linked nitro/flavin reductase | 1.4; 1.2 | 2.0; 2.3 | 2.8; 2.5 | 1.0; 0.8 |
| PyrF | Orotidine 5′-phosphate decarboxylase | 2.3; 2.0 | 2.2; 0.4 | 1.8; 2.2 | 1.5; 0.2 |
| YpiB | Unknown; similar to unknown proteins | 1.4; 3.1 | 2.6; 3.9 | 5.6; 5.2 | 1.1; 1.0 |
| YqiG | Unknown; similar to NADH-dependent flavin oxidoreductase | 2.0; 5.1 | 4.8; 4.0 | 2.5; 1.8 | 1.2; 0.9 |
| YuaE | Unknown | 1.4; 1.0 | 1.9; 1.7 | 3.0; 2.1 | 1.2; 0.7 |
| OhrR dependent | |||||
| OhrA | Organic hydroperoxide resistance protein OhrA | 2.4; 2.6 | 2.2; 1.6 | 2.9; 2.0 | 1.2; 0.8 |
| Others | |||||
| Cah | Cephalosporin C deacetylase | 0.4; 0.8 | 0.8; 2.2 | 4.3; 2.9 | 1.1; 0.7 |
| LysCβ | Aspartokinase II (alpha and beta subunits) fragment | 1.0; 1.3 | 2.0; 3.4 | 3.0; 1.8 | 1.3; 1.6 |
| MetE | Cobalamin-independent methionine synthase | 4.7; 1.7 | 4.5; 3.4 | 0.9; 1.1 | 0.6; 0.7 |
| OppA | Lipooligopeptide ABC transporter (binding protein; initiation of sporulation, competence development) | 5.7; 2.4 | 2.4; 1.0 | 2.3; 1.8 | 1.7; 0.8 |
| PtsH | Histidine-containing phosphocarrier protein of the PTS (HPr protein) | 3.9; 8.4 | 8.8; 11.1 | 9.6; 10.3 | 1.2; 3.9 |
| PyrR | Transcriptional attenuation of the pyrimidine operon/uracil phosphoribosyltransferase activity | 1.3; 1.7 | 1.9; 2.1 | 2.6; 2.8 | 0.6; 0.7 |
| YfmS | Unknown; similar to methyl-accepting chemotaxis protein | 2.0; 7.6 | 3.5; 5.8 | 5.3; 8.4 | 0.8; 1.2 |
| YfmT | Unknown; similar to benzaldehyde dehydrogenase | 0.5; 0.4 | 0.7; 1.4 | 3.4; 2.6 | 0.9; 0.9 |
| YqeY | Unknown; similar to unknown proteins | 1.3; 1.7 | 2.1; 2.2 | 3.8; 1.6 | 1.3; 1.1 |
At the indicated times after addition of the NO donor to the cell culture, radioactively labeled methionine was added for 5 min to study protein synthesis within this time frame (for details, see Materials and Methods). The fold change in the protein synthesis rate was calculated by dividing the normalized intensity of the protein spot in the 2D gel image at the indicated time point after NO stress with the corresponding spot in the image of the unstressed control. Values are induction ratios from two independent experiments. Protein synthesis ratios of at least 2.0 are indicated in boldface. The time points with synthesis ratios of at least 2.0 from two independent experiments are italicized.
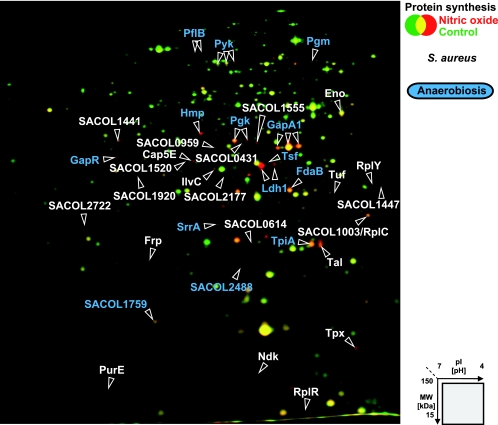
In S. aureus, a total of 35 proteins were identified as marker proteins for NO stress (Fig. 3 and Table 2). The synthesis of most of the proteins increased 5 to 10 min after stress exposure. Only four proteins were already synthesized at higher rates a few minutes after stress exposure. These included lactate dehydrogenase Ldh1 and pyruvate formate lyase PflB, which showed overall the highest induction rates after NO stress. After 30 min, when cells had already resumed growth, the synthesis of half of the marker proteins had reached control levels.
FIG. 3.
Synthesis of cytoplasmic proteins of S. aureus after NO stress. The 2D gel images of newly synthesized proteins (labeled with l-[35S]methionine) from exponentially growing cells (shown in green) and cells exposed to 500 μM concentrations of the NO donor MAHMA NONOate for 5 min (shown in red) were overlaid. Identified proteins with increased synthesis after 1 to 60 min after stress are labeled. Proteins whose synthesis was also induced after a shift from aerobic to anaerobic growth conditions are colored blue (28).
TABLE 2.
Proteins with increased synthesis after NO stress in S. aureus
| Category and TIGR locus | Protein | Description | GI no. | Fold change in synthesis (NO/control)a
|
||||
|---|---|---|---|---|---|---|---|---|
| 1 min | 5 min | 10 min | 30 min | 60 min | ||||
| Nitric oxide detoxification | ||||||||
| SACOL0220 | Hmp | Flavohemoprotein, putative | 57286684 | 1.9; 1.9 | 7.5; 7.3 | 9.4; 8.8 | 3.6; 0.9 | 1.4; 1.5 |
| Energy metabolism | ||||||||
| SACOL0204 | PflB | Formate acetyltransferase | 57285406 | 15.1; 9.9 | 29.3; 18.5 | 32.1; 16.9 | 12.9; 0.7 | 1.9; 1.5 |
| SACOL0222 | Ldh1 | l-Lactate dehydrogenase | 57286685 | 12.1; 9.9 | 15.7; 13.3 | 15.7; 16.1 | 3.8; 0.7 | 1.8; 0.9 |
| SACOL0837 | GapR | Gap transcriptional regulator | 57284299 | 2.6; 3.2 | 4.1; 4.8 | 4.4; 6.7 | 1.8; 2.6 | 0.8; 1.3 |
| SACOL0838 | GapA1 | Glyceraldehyde 3-phosphate dehydrogenase | 57284300 | 2.1; 1.8 | 4.7; 3.9 | 4.5; 3.4 | 2.5; 1.4 | 1.4; 1.8 |
| SACOL0839 | Pgk | Phosphoglycerate kinase | 57284301 | 1.7; 1.8 | 5.2; 4.6 | 7.6; 7.1 | 6.5; 2.2 | 1.4; 1.6 |
| SACOL0840 | TpiA | Triosephosphate isomerase | 57284302 | 1.1; 1.1 | 2.4; 2.5 | 3.1; 2.0 | 3.0; 1.3 | 1.0; 1.2 |
| SACOL0841 | Pgm | Phosphoglycerate mutase, 2,3-bisphosphoglycerate-independent | 57284303 | 0.9; 1.1 | 1.4; 1.4 | 2.6; 2.9 | 4.0; 2.9 | 1.9; 1.8 |
| SACOL0842 | Eno | Enolase | 57284304 | 1.1; 1.0 | 1.4; 1.5 | 2.1; 2.0 | 1.4; 1.1 | 1.4; 0.8 |
| SACOL1535 | SrrA | DNA-binding response regulator SrrA | 57284634 | 1.6; 3.5 | 1.8; 3.7 | 2.3; 5.1 | 2.1; 1.7 | 0.8; 1.3 |
| SACOL1745 | Pyk | Pyruvate kinase | 57284754 | 0.7; 0.9 | 1.5; 1.1 | 2.1; 2.0 | 2.0; 1.6 | 1.9; 1.5 |
| SACOL1831 | Tal | Transaldolase | 57286264 | 1.8; 1.5 | 3.6; 3.9 | 4.2; 2.8 | 2.9; 3.2 | 1.0; 1.2 |
| SACOL2177 | Alcohol dehydrogenase, zinc-containing | 57286389 | 1.0; 1.5 | 1.7; 1.7 | 2.6; 2.3 | 2.1; 2.6 | 1.6; 1.7 | |
| SACOL2488 | Oxidoreductase, short-chain dehydrogenase/reductase family | 57285174 | 1.6; 1.6 | 2.6; 2.3 | 2.4; 2.8 | 1.5; 2.7 | 1.0; 1.4 | |
| SACOL2622 | FdaB | Fructose-bisphosphate aldolase, class I | 57286527 | 2.0; 1.7 | 3.1; 3.6 | 4.0; 4.9 | 2.9; 3.1 | 1.3; 1.5 |
| Amino acid and protein biosynthesis | ||||||||
| SACOL0431 | Trans-sulfuration enzyme family protein | 57286805 | 0.9; 1.5 | 3.0; 2.5 | 4.4; 2.7 | 3.2; 2.4 | 0.8; 1.4 | |
| SACOL0545 | RplY | Ribosomal protein L25 | 57284228 | 1.5; 1.5 | 3.8; 1.6 | 3.6; 2.7 | 3.9; 1.8 | 1.7; 1.3 |
| SACOL0594 | Tuf | Translation elongation factor Tu | 57285610 | 1.1; 0.8 | 1.0; 0.8 | 0.6; 1.0 | 1.0; 0.8 | 2.8; 3.1 |
| SACOL1276 | Tsf | Translation elongation factor Ts | 57286013 | 1.9; 1.6 | 1.8; 2.0 | 3.3; 2.9 | 1.5; 1.0 | 1.5; 1.1 |
| SACOL2045 | IlvC | Ketol-acid reductoisomerase | 57284914 | 2.7; 3.9 | 1.9; 1.5 | 2.3; 2.1 | 1.7; 2.2 | 1.3; 1.6 |
| SACOL2223 | RplR | Ribosomal protein L18 | 57285004 | 0.7; 1.5 | 2.1; 2.4 | 1.3; 2.5 | 1.8; 3.4 | 0.6; 1.7 |
| Others | ||||||||
| SACOL0140 | Cap5E | Capsular polysaccharide biosynthesis protein Cap5E | 57285343 | 1.2; 2.3 | 2.2; 4.7 | 2.2; 2.7 | 1.7; 4.1 | 1.0; 2.1 |
| SACOL0614 | Conserved hypothetical protein | 57285629 | 1.3; 0.9 | 2.3; 2.4 | 2.9; 3.3 | 3.1; 3.0 | 1.1; 1.4 | |
| SACOL0959 | NADH-dependent flavin oxidoreductase; Oye family | 57285833 | 1.1; 1.2 | 1.6; 1.6 | 2.0; 2.5 | 2.0; 2.9 | 1.0; 1.6 | |
| SACOL1073 | PurE | Phosphoribosylaminoimidazole carboxylase; catalytic subunit | 57285859 | 1.5; 2.4 | 3.4; 4.1 | 3.7; 5.0 | 2.3; 2.8 | 1.1; 1.8 |
| SACOL1441 | Tellurite resistance protein, putative | 57286092 | 0.6; 1.0 | 1.6; 2.0 | 1.8; 2.9 | 2.0; 2.7 | 0.6; 1.5 | |
| SACOL1447 | Conserved hypothetical protein | 57286098 | 1.8; 1.6 | 2.3; 1.6 | 2.0; 2.7 | 2.4; 2.3 | 1.0; 1.5 | |
| SACOL1509 | Ndk | Nucleoside diphosphate kinase | 57284610 | 0.8; 0.8 | 0.5; 0.5 | 0.4; 0.6 | 0.8; 0.5 | 2.1; 2.3 |
| SACOL1520 | Pyridine nucleotide-disulfide oxidoreductase | 57284620 | 1.1; 1.2 | 2.1; 1.8 | 3.1; 1.4 | 2.2; 2.3 | 1.5; 2.1 | |
| SACOL1555 | Peptidase, M20/M25/M40 family | 57284653 | 1.6; 1.8 | 2.5; 2.5 | 2.1; 2.2 | 1.3; 1.2 | 1.0; 1.1 | |
| SACOL1759 | Universal stress protein family | 57286194 | 0.9; 1.6 | 1.3; 1.8 | 2.0; 3.2 | 2.2; 3.1 | 0.8; 1.3 | |
| SACOL1762 | Tpx | Thiol peroxidase. putative | 57286197 | 2.4 1.6 | 4.9; 3.7 | 5.7; 5.6 | 4.3; 3.7 | 1.4; 1.3 |
| SACOL1920 | d-Isomer-specific 2-hydroxyacid dehydrogenase family protein | 5728483 | 0.8; 1.0 | 1.2; 2.2 | 1.7; 2.9 | 2.3; 2.9 | 1.0; 1.4 | |
| SACOL2534 | Frp | NAD(P)H-flavin oxidoreductase | 57285217 | 0.7; 1.1 | 1.3; 1.3 | 1.7; 2.0 | 2.2; 4.0 | 1.3; 2.2 |
| SACOL2722 | N-Acetyltransferase family protein | 57285276 | 0.7; 1.3 | 1.2; 2.1 | 1.6; 4.2 | 2.3; 4.4 | 0.8; 2.2 | |
At the indicated times after addition of the NO donor to the cell culture, radioactively labeled methionine was added for 5 min to study protein synthesis within this time frame (for details, see Materials and Methods). The fold change in the protein synthesis rate was calculated by dividing the normalized intensity of the protein spot in the 2D gel image at the indicated time point after NO stress with the corresponding spot in the image of the unstressed control. Values are induction ratios from two independent experiments. Protein synthesis ratios of at least 2.0 are indicated in boldface. The time points with synthesis ratios of at least 2.0 from two independent experiments are italicized.
Surprisingly, the ketol-acid reductoisomerase IlvC and the flavohemoglobin Hmp are the only two proteins that were induced after NO stress both in B. subtilis and in S. aureus, indicating that different signaling pathways play a role in the adaptation to this stressor.
Increased Hmp synthesis after NO stress in B. subtilis and S. aureus.
The protein with the highest increase in synthesis after NO stress in B. subtilis (70-fold) and one of the highest in S. aureus (9-fold) was identified as the flavohemoglobin Hmp (Tables 1 and 2). In the presence of oxygen, the Hmp enzyme acts as an NO scavenger and is responsible for the direct detoxification of NO to nitrate (35, 79). Furthermore, it has been shown that hmp mutants of B. subtilis and S. aureus, as well as of Salmonella enterica and E. coli, are hypersensitive to NO-related killing (59, 83, 84, 89). Thus, the upregulation of Hmp is a central feature of NO stress adaptation in both B. subtilis and S. aureus.
In B. subtilis, hmp gene expression is controlled by the two-component system ResDE involved in aerobic and anaerobic gene expression (57, 62, 66, 91). Other than Hmp, there are three additional NO marker proteins: YwfI (unknown function); YvyD (similar to sigma-54 modulating factor of gram-negative bacteria); and the ferrochelatase HemH, whose gene expression is induced under anaerobic conditions and reduced in a ResDE mutant after anaerobic induction (65, 102). However, a comparison of B. subtilis proteins whose amounts were increased in cells shifted from aerobic to anaerobic growth conditions with those increased in NO-stressed cells showed no further overlap (57). Interestingly, hmp, yvyD, and hemH are controlled by additional regulators. In addition to ResDE, the repressor NsrR (formerly YhdE) was shown to be involved in the transcriptional regulation of B. subtilis hmp (63). Under aerobic conditions, NO or the nitrosonium cation donor sodium nitroprusside inactivates NsrR, ensuring the expression of hmp, independent of the ResDE system (62, 63, 84). The activity of NsrR is possibly modulated by the interaction of NO with the Fe-S cluster of the protein (7, 63, 85). Transcription of yvyD is controlled by two alternative sigma factors: SigB and SigH (9, 22), while hemH expression was shown to be dependent on Spx (67). In fact, many members of the SigB and Spx regulons were synthesized at a higher rate after NO (see below). Taken together, the results indicate that the increased synthesis of these proteins in the presence of NO under aerobic conditions might occur independently of ResDE.
As in B. subtilis, so the S. aureus hmp is regulated by the ResDE orthologous system, SrrAB (formerly SrhSR) (83). For the NO-dependent induction of hmp expression under aerobic conditions, an SrrAB independent regulatory mechanism has been postulated (83).
Increased synthesis of enzymes involved in anaerobic metabolism after NO stress in S. aureus.
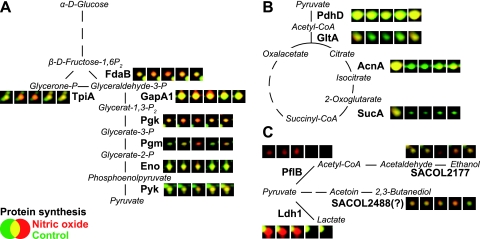
Besides Hmp induction, S. aureus responds to NO stress with the increased synthesis of at least 34 additional marker proteins. These proteins were assigned to different functional groups such as energy metabolism and protein biosynthesis (Table 2). The detailed comparison shows that about one-third of the NO marker proteins are also induced under anaerobic conditions (28) and mainly belong to glycolysis (Eno, FdaB, GapA1, Pgk, Pgm, and TpiA) and fermentation pathways (Ldh1, PflB, SACOL2177, and SACOL2488) (Fig. 3 and Fig. 4). Fermentation enzymes such as lactate dehydrogenase Ldh1 and pyruvate formate lyase PflB are among the earliest and most strongly induced enzymes upon NO challenge. Two additional NO marker proteins are possibly involved in fermentation reactions: SACOL2177 is a zinc-containing alcohol dehydrogenase, and the oxidoreductase SACOL2488 probably catalyzes the reversible reduction of acetoin to 2,3-butanediol in the presence of NADH (Swiss-Prot Protein knowledgebase at http://www.expasy.org/sprot/). Just as under anaerobic conditions (28), we additionally observed a decrease in the synthesis of PdhD, one subunit of the pyruvate dehydrogenase complex and several enzymes of the tricarboxylic acid cycle (AcnA, GltA, and SucA) (Fig. 4). A similar expression pattern was also found for aerobically grown S. aureus cells with a defect in electron transport chain (hemB mutant), indicating that the activation of anaerobic metabolism is independent of the presence of oxygen (49, 86). Remarkably, NO is already known to inhibit aerobic respiration by reversible binding to cytochromes (5, 10, 60, 88, 100). Thus, NO-induced inhibition of aerobic respiration is most likely the reason for the induction of anaerobic metabolism in S. aureus. The decline of the synthesis rates of most of the proteins to prestress levels with the recovery of growth demonstrates the transient nature of this response once the stress has abated (Table 2; see also the 2D movies in the supplemental material). In a recent publication, the induction of a few genes encoding proteins involved in energy and fermentative metabolism has been demonstrated at the transcriptional level, including the l-lactate dehydrogenase ldh1 and 1,6-fructosebisphosphate aldolase fdaB (83). As the protein expression analysis makes more clear, the induction of anaerobic gene expression by NO, even in the presence of oxygen, would be a major difference compared to B. subtilis, where low oxygen tension is a prerequisite for the induction of genes involved in anaerobic respiration and fermentation (64, 82). The main reason for this might be the strict oxygen sensitivity of Fnr, which is a main regulator in the adaptation to anaerobic conditions in B. subtilis (81, 82). An Fnr homolog is not encoded in the S. aureus genome. The ability to switch to anaerobic metabolism during NO stress might be of decisive advantage to S. aureus and critical for its higher resistance under this condition.
FIG. 4.
Proteins with changed synthesis after NO stress in S. aureus COL involved in glycolysis (A), the tricarboxylic acid cycle (B), and fermentation (C). Details of the 2D dual-channel images generated from radioactively labeled cytoplasmic protein extracts of exponentially growing cells (colored in green) and of cells exposed to 500 μM concentrations of the NO donor MAHMA NONOate (colored in red) for 1, 5, 10, 30, and 60 min are shown.
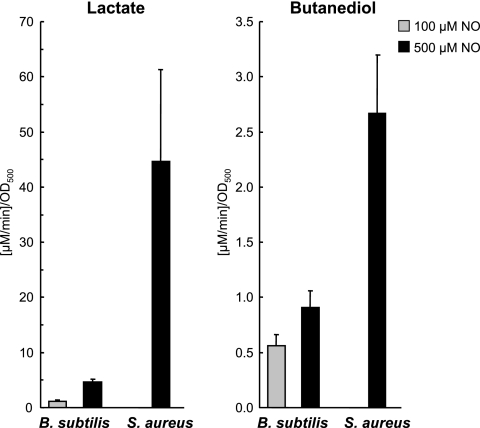
In order to underline these data, we performed additional experiments on the formation of fermentation products. After NO treatment, S. aureus produced drastically higher amounts of lactate and also butanediol compared to B. subtilis (Fig. 5). Increased formation of acetate and ethanol was not observed either. As suggested from the strong induction of fermentation enzymes at the protein level in S. aureus, the data verify a higher activity of fermentative pathways leading to formation of lactate and butanediol. The absence of increased production of acetate and ethanol after NO treatment is most likely explained by the presence of oxygen, leading to inactivation of the upstream pyruvate format lyase.
FIG. 5.
Formation of lactate and butanediol in S. aureus and B. subtilis 20 min after exposition to 100 μM (only for B. subtilis) or 500 μl (for B. subtilis and S. aureus) of the NO donor MAHMA NONOate. Cells were grown in synthetic medium to an OD500 of 0.5 and exposed to the NO donor. For metabolite analyses, samples were taken immediately before and 20 min after addition of the NO donor. Cells were separated from the supernatant by filtration, and the obtained supernatants were used for further analyses. Lactate and butanediol were detected and quantified by 1H-NMR. The graphs show the increase of the concentration per minute and the OD500 within 20 min after the addition of the NO donor for lactate and butanediol. The values are given as means ± the standard deviation of three parallel studies of two independently analyzed cell cultures.
The response regulator SrrA was detected among the NO marker proteins (Fig. 3 and Table 2). It is part of the two-component system SrrAB, which is an ortholog of the ResDE system in B. subtilis. Throup et al. (96) demonstrated that SrrAB is essential for the increased synthesis of several fermentation enzymes, as well as the repression of tricarboxylic acid cycle enzymes under anaerobic conditions. In addition, an SrrAB mutant was described as being hypersensitive to NO stress (83). Taken together, the results implicate a crucial role of SrrAB in the switch to anaerobic metabolism during NO stress adaptation.
A number of NO marker proteins in S. aureus catalyze reactions with unknown substrate specificity or unknown function (Table 2). Most of them show similarities to oxidoreductases. Since these proteins were specifically upregulated after NO stress but not after oxidative stress conditions induced by hydrogen peroxide, the superoxide-generating agent paraquat, or the thiol-specific oxidant diamide (101), these proteins may well play important roles in the NO stress resistance response in S. aureus.
NO induces general and specific stress responses in B. subtilis.
In B. subtilis, ca. 40% (15 proteins) of the NO marker proteins belong to the general stress regulon and are controlled by the alternative sigma factor SigB (76-78, 80). The remaining marker proteins can be assigned to oxidative stress regulons controlled by Fur (four NO marker proteins), PerR (three NO marker proteins), Spx (seven NO marker proteins), and OhrR (one NO marker proteins) (2, 3, 12, 15, 27, 39, 67, 70).
The NO proteome signature of B. subtilis matches well with a recently performed transcriptome study (60), thus indicating that increased transcription of the respective genes resulted in increased protein synthesis. SigB-dependent general stress proteins equip the cells with an unspecific protection that aids survival during prolonged periods of stress and starvation (36, 37, 80). Moore et al. (60) demonstrated that the strong induction of SigB-dependent genes after NO treatment in B. subtilis is mediated by the energy branch of the SigB regulatory cascade, which is possibly activated due to the NO-induced block of the respiratory chain and an accompanied decrease of ATP. The additional NO-induced regulons globally controlled by Fur, PerR, OhrR, or Spx in B. subtilis are also upregulated after specific oxidative stress conditions induced by inorganic and organic peroxides, superoxide, or the thiol-specific oxidant diamide (38, 52, 61, 92, 104). The corresponding proteins protect the cell against an oxidative threat mainly by direct detoxification of the stressor and repair of damage. The contribution of the individual oxidative stress as well as the general stress proteins in resistance to NO stress is largely unknown. However, for example, the NO-induced and PerR-dependent peroxiredoxin AhpC is known to detoxify reactive nitrogen species (11, 16, 58).
Regulons controlled by SigB, Fur, PerR (a Fur homolog), and Spx, respectively, have been also described in S. aureus (6, 42, 43, 74, 75). Surprisingly, none of the identified NO marker proteins in S. aureus could be assigned to these regulons, which points to major differences in the response of these two microorganisms. The absence of increased SigB-dependent protein synthesis after NO exposure in S. aureus was not surprising since SigB activity was shown to be increased in cells grown in synthetic medium and could not be further increased after heat or ethanol stress (29). Moreover, a B. subtilis-like induction of the SigB regulatory cascade by NO cannot be expected because of the lack of signal transduction components responsible for SigB activation in response to energy limitation (75).
The Fe(II)-containing forms of the repressors Fur and PerR in B. subtilis are thought to be inactivated by NO due to the formation of iron-nitrosyl complexes, thereby derepressing transcription (19, 60). The regulatory site of PerR in B. subtilis binds Fe(II) or Mn(II) (40), whereas PerR:Mn is not inactivated by NO (60). In S. aureus, PerR preferentially binds Mn(II) (18, 42), which might explain the observed lack of PerR-dependent gene expression on transcriptome (83) and proteome levels in response to NO.
Genes of the Fur regulon are transcriptionally induced after treatment of S. aureus cells with NO (83). The failure to detect proteins of the Fur regulon with our approach may be explained by the fact that their pIs are out of the analyzed pH range of 4 to 7 and/or that they do not belong to the soluble cytoplasmic protein fraction (43).
NO does not cause protein thiol modifications but diminishes irreversible protein thiol oxidation in vivo.
The thiol groups of cysteine residues are preferred targets of reversible and irreversible oxidation reactions by reactive oxygen species and are believed to be readily modified by reactive nitrogen species, too (21, 25, 26, 30, 73). Recently, with a fluorescence-based assay many intracellular cysteine-containing proteins of B. subtilis were shown to be reversibly or even irreversibly modified at their thiol groups after oxidative stress induced by various oxidative agents, including the antibiotic nitrofurantoin (41). Furthermore, we applied the thiol modification assay to S. aureus cells and demonstrated that oxidative stress induced by hydrogen peroxide or diamide causes thiol oxidation of many intracellular proteins (101).
Here, we used the fluorescence thiol modification assay to detect proteins whose thiol groups were covalently modified by an NO moiety (S nitrosylation). In brief, untreated control cells and cells treated with NO are disrupted in a denaturing buffer containing iodoacetamide. With this alkylation step all accessible thiol groups (unmodified cysteines) are chemically blocked to preserve the native thiol state. Second, S-nitrosylated protein thiols are reduced with ascorbate (45, 46). The newly generated thiol groups are then labeled with a thiol-specific fluorescence dye. The labeled protein mixture is separated by 2D PAGE, and the fluorescence image is overlaid with the image of the total protein stain.
The resulting dual-channel images for exponentially growing cells showed that only a few proteins in B. subtilis, but none of the proteins in S. aureus, were significantly labeled with the fluorescence dye (data not shown). The labeled proteins in B. subtilis have similarities to thioredoxins (YdfQ) and thioredoxin reductases (YumC), as well as a protein with similarity to nitroreductases (YodC). The results indicate that certain proteins of B. subtilis might be S nitrosylated already in growing cells. Interestingly, thioredoxin of endothelial cells is regulated by S nitrosylation, and the thioredoxin system is suggested to play a role in S-nitrosothiol homeostasis (33, 34). Gel-free mass spectrometry is currently being carried out to confirm the existence of S nitrosylations in growing B. subtilis cells.
To detect proteins particularly susceptible to S nitrosylations after NO stress, we applied the fluorescence thiol modification assay to B. subtilis and S. aureus cells exposed to different concentrations of the NO donor. We could not detect an increase in fluorescence labeling of protein thiols after NO stress even after treatment of the cells with up to a 2 mM concentration of the NO donor for 10 min. Since S-nitrosothiols can be readily converted to disulfides (21, 26), we also used the phosphine derivative TCEP as a reductant to reduce all reversibly modified cysteines (41). Again, we did not detect an increase in fluorescently labeled proteins, indicating no accumulation of specifically S-nitrosylated or generally thiol oxidized proteins upon exogenous NO treatment. The precise mechanism of S nitrosylation in cells is not completely understood. Reactive nitrogen species (e.g., NO2 and N2O3) generated from NO or compounds releasing the nitrosonium cation (e.g., S-nitrosocysteine) are predicted to be responsible for S nitrosylations in vivo (103). Possibly, under pure exogenous NO treatment as performed in the present study there is little if any formation of these compounds, and the degree of S-nitrosylated proteins was below the detection limit of the method applied.
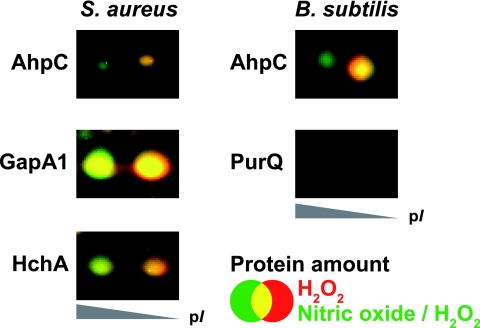
Gusarov and Nudler (32) recently described a cytoprotective system in B. subtilis based on NO. These authors showed that NO protects cells from H2O2-induced DNA damage and provided evidence that this is due to inhibition of the DNA-damaging Fenton reaction and activation of the catalase KatA. In B. subtilis and S. aureus, hydrogen peroxide causes irreversible oxidation of the active-site cysteines of certain proteins to sulfinic or sulfonic acid (41, 98). In the case of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) of S. aureus, this leads to the loss of enzymatic function (98). To analyze whether NO can also protect proteins from irreversible thiol oxidation, we monitored the acidic isoelectric shift of peroxide-sensitive proteins in the 2D gel. The shift in the isoelectric point of the protein is due to the additional negative charge of the sulfinic or sulfonic acid form generated after peroxide treatment (41, 98). Interestingly, the amount of irreversibly oxidized proteins significantly decreases if the cells were incubated with 100 μM concentrations of the NO donor for 10 min prior to peroxide addition (Fig. 6). The effect was even stronger when higher concentrations (up to 1 mM concentrations of NO donor) were added. The results show that NO diminishes the peroxide-induced irreversible thiol oxidation of proteins.
FIG. 6.
Details of the 2D dual-channel image generated from the images of Sypro Ruby-stained cytoplasmic proteins of exponentially growing cells exposed to 10 mM H2O2 for 10 min (shown in red) and cells incubated with 100 μM (B. subtilis) or 500 μM (S. aureus) of the NO donor MAHMA NONOate prior to H2O2 addition (shown in green). Note that a lower amount of the proteins shows an acidic isoelectric point shift in cells preincubated with NO, indicating diminished irreversible thiol oxidation. AhpC, alkyl hydroperoxide reductase subunit C; HchA, chaperone protein HchA (Hsp31); GapA1, glyceraldehyde 3-phosphate dehydrogenase; PurQ, phosphoribosylformylglycinamidine synthase 1.
The antioxidant potential of NO has been discussed in eukaryotic systems, where NO reduces the oxidizing capacity of reactive oxygen and nitrogen species and modulates cellular and physiological processes that prevent cell and tissue injury (95, 99). Our results strongly indicate that NO can also function as a protein protection determinant in bacteria. The model of NO-mediated cytoprotection against DNA damage in B. subtilis and S. aureus (32) should therefore be extended to include its ability to protect against peroxide-induced irreversible protein thiol oxidation.
Different mechanisms are conceivable for how exogenous NO might prevent H2O2-induced irreversible oxidation of protein thiols. NO itself or derivatives that might be generated prior to H2O2 addition could (i) mediate an upregulation or activation of antioxidant systems, thereby decreasing the amount of H2O2; (ii) react with the sulfenic acid intermediate which is transiently formed during oxidation to sulfinic or sulfonic acid by H2O2 and thereby blocking the irreversible oxidation of sulfur; or (iii) directly react with H2O2 and form products that are less thiol-toxic. To clarify whether the observed comparable thiol-protective role of NO against H2O2 in B. subtilis and S. aureus is solely attributed to an activation of catalase (32) or to what extent other processes might play an additional role, ongoing studies are needed.
Concluding remarks.
In the present study, the NO stress-induced expression profiles of two model organisms, the nonpathogenic B. subtilis and its closely related counterpart the pathogen S. aureus, were compared by 2D gel-based proteomics. The most striking similarity between these two model organisms was the substantial induction of synthesis of the NO detoxification enzyme Hmp after stress exposure. In contrast, major differences became obvious, particularly in the increased synthesis of members of the SigB-controlled general stress regulon, as well as of regulons involved in oxidative stress resistance in B. subtilis which were completely absent in S. aureus. The protein synthesis signature observed for S. aureus obviously showed strong similarities to that found under anaerobic conditions. NO disrupts the respiratory chain by binding to cytochromes (5, 8, 10, 13, 71, 88, 100). Unlike B. subtilis, the extensive switch to the anaerobic metabolism even under high oxygen tension might therefore be a beneficial consequence of NO stress in the more tolerant S. aureus. This is likely due to differences in the underlying signaling network of these organisms.
Monitoring the cytoplasmic protein thiol state revealed no accumulation of thiol modifications after NO exposure in either species. In contrast, NO was found to mediate protection against peroxide-induced irreversible thiol oxidation.
Supplementary Material
Acknowledgments
We are indebted to Robert S. Jack for critical review of the manuscript. We thank Thomas Meier for excellent technical assistance.
This study was supported by grants from the European Union (LSHG-CT-2004-503468), BMBF (031U107A/-207A; 031U213B), and the Deutsche Forschungsgemeinschaft (HE 1887/7-4, SFB/TRR34/1-2006, GK212/3-00) to M.H. and S.E.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 1845826-58232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 451613-16129. [DOI] [PubMed] [Google Scholar]
- 4.Bandow, J. E., H. Brötz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltran, B., A. Mathur, M. R. Duchen, J. D. Erusalimsky, and S. Moncada. 2000. The effect of nitric oxide on cell respiration: a key to understanding its role in cell survival or death. Proc. Natl. Acad. Sci. USA 9714602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 1864085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borisov, V. B., E. Forte, A. A. Konstantinov, R. K. Poole, P. Sarti, and A. Giuffre. 2004. Interaction of the bacterial terminal oxidase cytochrome bd with nitric oxide. FEBS Lett. 576201-204. [DOI] [PubMed] [Google Scholar]
- 9.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 1844881-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, G. C. 2001. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim. Biophys. Acta 150446-57. [DOI] [PubMed] [Google Scholar]
- 11.Bryk, R., P. Griffin, and C. Nathan. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407211-215. [DOI] [PubMed] [Google Scholar]
- 12.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29189-198. [DOI] [PubMed] [Google Scholar]
- 13.Butler, C., E. Forte, F. Maria Scandurra, M. Arese, A. Giuffre, C. Greenwood, and P. Sarti. 2002. Cytochrome bo3 from Escherichia coli: the binding and turnover of nitric oxide. Biochem. Biophys. Res. Commun. 2961272-1278. [DOI] [PubMed] [Google Scholar]
- 14.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mäder, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 222908-2935. [DOI] [PubMed] [Google Scholar]
- 15.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 928190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, L., Q. W. Xie, and C. Nathan. 1998. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol. Cell 1795-805. [DOI] [PubMed] [Google Scholar]
- 17.Choi, P. S., Z. Naal, C. Moore, E. Casado-Rivera, H. D. Abruna, J. D. Helmann, and J. P. Shapleigh. 2006. Assessing the impact of denitrifier-produced nitric oxide on other bacteria. Appl. Environ. Microbiol. 722200-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosgrove, K., G. Coutts, I. M. Jonsson, A. Tarkowski, J. F. Kokai-Kun, J. J. Mond, and S. J. Foster. 2007. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 1891025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Autreaux, B., D. Touati, B. Bersch, J. M. Latour, and I. Michaud-Soret. 2002. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. USA 9916619-16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Groote, M. A., and F. C. Fang. 1999. Antimicrobial properties of nitric oxide. In F. C. Fang (ed.), Nitric oxide and infection. Kluwer Academic/Plenum Publishers, New York, NY.
- 21.Di Simplicio, P., F. Franconi, S. Frosali, and D. Di Giuseppe. 2003. Thiolation and nitrosation of cysteines in biological fluids and cells. Amino Acids 25323-339. [DOI] [PubMed] [Google Scholar]
- 22.Drzewiecki, K., C. Eymann, G. Mittenhuber, and M. Hecker. 1998. The yvyD gene of Bacillus subtilis is under dual control of sigmaB and sigmaH. J. Bacteriol. 1806674-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duy, N. V., U. Mäder, N. P. Tran, J. F. Cavin, T. Tam le, D. Albrecht, M. Hecker, and H. Antelmann. 2007. The proteome and transcriptome analysis of Bacillus subtilis in response to salicylic acid. Proteomics 7698-710. [DOI] [PubMed] [Google Scholar]
- 24.Eymann, C., A. Dreisbach, D. Albrecht, J. Bernhardt, D. Becher, S. Gentner, T. Tam le, K. Büttner, G. Buurman, C. Scharf, S. Venz, U. Völker, and M. Hecker. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 42849-2876. [DOI] [PubMed] [Google Scholar]
- 25.Fang, F. C. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2820-832. [DOI] [PubMed] [Google Scholar]
- 26.Forman, H. J., J. M. Fukuto, and M. Torres. 2004. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am. J. Physiol. Cell Physiol. 287C246-C256. [DOI] [PubMed] [Google Scholar]
- 27.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 1834134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs, S., J. Pané-Farré, C. Kohler, M. Hecker, and S. Engelmann. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 1894275-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261558-566. [DOI] [PubMed] [Google Scholar]
- 30.Ghezzi, P., and V. Bonetto. 2003. Redox proteomics: identification of oxidatively modified proteins. Proteomics 31145-1153. [DOI] [PubMed] [Google Scholar]
- 31.Goretski, J., O. C. Zafiriou, and T. C. Hollocher. 1990. Steady-state nitric oxide concentrations during denitrification. J. Biol. Chem. 26511535-11538. [PubMed] [Google Scholar]
- 32.Gusarov, I., and E. Nudler. 2005. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc. Natl. Acad. Sci. USA 10213855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haendeler, J. 2006. Thioredoxin-1 and posttranslational modifications. Antioxid. Redox Signal 81723-1728. [DOI] [PubMed] [Google Scholar]
- 34.Haendeler, J., J. Hoffmann, V. Tischler, B. C. Berk, A. M. Zeiher, and S. Dimmeler. 2002. Redox regulatory and antiapoptotic functions of thioredoxin depend on S nitrosylation at cysteine 69. Nat. Cell Biol. 4743-749. [DOI] [PubMed] [Google Scholar]
- 35.Hausladen, A., A. J. Gow, and J. S. Stamler. 1998. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 9514100-14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hecker, M., J. Pané-Farré, and U. Völker. 2006. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61215-236. [DOI] [PubMed] [Google Scholar]
- 37.Hecker, M., and U. Völker. 2004. Towards a comprehensive understanding of Bacillus subtilis cell physiology by physiological proteomics. Proteomics 43727-3750. [DOI] [PubMed] [Google Scholar]
- 38.Helmann, J. D., M. F. Wu, A. Gaballa, P. A. Kobel, M. M. Morshedi, P. Fawcett, and C. Paddon. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herbig, A. F., and J. D. Helmann. 2002. Metal ion uptake and oxidative stress, p. 405-414. In A. L. Sonenschein, J. A. Hoch, and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 40.Herbig, A. F., and J. D. Helmann. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41849-859. [DOI] [PubMed] [Google Scholar]
- 41.Hochgräfe, F., J. Mostertz, D. Albrecht, and M. Hecker. 2005. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol. Microbiol. 58409-425. [DOI] [PubMed] [Google Scholar]
- 42.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 693744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyduke, D. R., L. R. Jarboe, L. M. Tran, K. J. Chou, and J. C. Liao. 2007. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. USA 1048484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaffrey, S. R., H. Erdjument-Bromage, C. D. Ferris, P. Tempst, and S. H. Snyder. 2001. Protein S nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3193-197. [DOI] [PubMed] [Google Scholar]
- 46.Jaffrey, S. R., and S. H. Snyder. 2001. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001PL1. [DOI] [PubMed] [Google Scholar]
- 47.Jeffery, C. J. 1999. Moonlighting proteins. Trends Biochem. Sci. 248-11. [DOI] [PubMed] [Google Scholar]
- 48.Kim, S. O., K. Merchant, R. Nudelman, W. F. Beyer, Jr., T. Keng, J. DeAngelo, A. Hausladen, and J. S. Stamler. 2002. OxyR: a molecular code for redox-related signaling. Cell 109383-396. [DOI] [PubMed] [Google Scholar]
- 49.Kohler, C., C. von Eiff, G. Peters, R. A. Proctor, M. Hecker, and S. Engelmann. 2003. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J. Bacteriol. 1856928-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohler, C., S. Wolff, D. Albrecht, S. Fuchs, D. Becher, K. Büttner, S. Engelmann, and M. Hecker. 2005. Proteome analyses of Staphylococcus aureus in growing and non-growing cells: a physiological approach. Int. J. Med. Microbiol. 295547-565. [DOI] [PubMed] [Google Scholar]
- 51.Kwon, N. S., D. J. Stuehr, and C. F. Nathan. 1991. Inhibition of tumor cell ribonucleotide reductase by macrophage-derived nitric oxide. J. Exp. Med. 174761-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 1851967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lepoivre, M., F. Fieschi, J. Coves, L. Thelander, and M. Fontecave. 1991. Inactivation of ribonucleotide reductase by nitric oxide. Biochem. Biophys. Res. Commun. 179442-448. [DOI] [PubMed] [Google Scholar]
- 54.Lepoivre, M., J. M. Flaman, and Y. Henry. 1992. Early loss of the tyrosyl radical in ribonucleotide reductase of adenocarcinoma cells producing nitric oxide. J. Biol. Chem. 26722994-23000. [PubMed] [Google Scholar]
- 55.Lewis, R. S., S. Tamir, S. R. Tannenbaum, and W. M. Deen. 1995. Kinetic analysis of the fate of nitric oxide synthesized by macrophages in vitro. J. Biol. Chem. 27029350-29355. [DOI] [PubMed] [Google Scholar]
- 56.Luhn, S., M. Berth, M. Hecker, and J. Bernhardt. 2003. Using standard positions and image fusion to create proteome maps from collections of two-dimensional gel electrophoresis images. Proteomics 31117-1127. [DOI] [PubMed] [Google Scholar]
- 57.Marino, M., T. Hoffmann, R. Schmid, H. Mobitz, and D. Jahn. 2000. Changes in protein synthesis during the adaptation of Bacillus subtilis to anaerobic growth conditions. Microbiology 146(Pt. 1)97-105. [DOI] [PubMed] [Google Scholar]
- 58.Master, S. S., B. Springer, P. Sander, E. C. Boettger, V. Deretic, and G. S. Timmins. 2002. Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology 1483139-3144. [DOI] [PubMed] [Google Scholar]
- 59.Membrillo-Hernandez, J., M. D. Coopamah, M. F. Anjum, T. M. Stevanin, A. Kelly, M. N. Hughes, and R. K. Poole. 1999. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274748-754. [DOI] [PubMed] [Google Scholar]
- 60.Moore, C. M., M. M. Nakano, T. Wang, R. W. Ye, and J. D. Helmann. 2004. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J. Bacteriol. 1864655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150497-512. [DOI] [PubMed] [Google Scholar]
- 62.Nakano, M. M. 2002. Induction of ResDE-dependent gene expression in Bacillus subtilis in response to nitric oxide and nitrosative stress. J. Bacteriol. 1841783-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakano, M. M., H. Geng, S. Nakano, and K. Kobayashi. 2006. The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J. Bacteriol. 1885878-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52165-190. [DOI] [PubMed] [Google Scholar]
- 65.Nakano, M. M., and P. Zuber. 2001. Anaerobiosis, p. 393-404. In A. L. Sonenschein, J. A. Hoch, and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 66.Nakano, M. M., P. Zuber, P. Glaser, A. Danchin, and F. M. Hulett. 1996. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol. 1783796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakano, S., E. Kuster-Schock, A. D. Grossman, and P. Zuber. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 10013603-13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nalwaya, N., and W. M. Deen. 2005. Nitric oxide, oxygen, and superoxide formation and consumption in macrophage cultures. Chem. Res. Toxicol. 18486-493. [DOI] [PubMed] [Google Scholar]
- 69.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 978841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ollinger, J., K. B. Song, H. Antelmann, M. Hecker, and J. D. Helmann. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 1883664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pacelli, R., D. A. Wink, J. A. Cook, M. C. Krishna, W. DeGraff, N. Friedman, M. Tsokos, A. Samuni, and J. B. Mitchell. 1995. Nitric oxide potentiates hydrogen peroxide-induced killing of Escherichia coli. J. Exp. Med. 1821469-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Padmaja, S., and R. E. Huie. 1993. The reaction of nitric oxide with organic peroxyl radicals. Biochem. Biophys. Res. Commun. 195539-544. [DOI] [PubMed] [Google Scholar]
- 73.Paget, M. S., and M. J. Buttner. 2003. Thiol-based regulatory switches. Annu. Rev. Genet. 3791-121. [DOI] [PubMed] [Google Scholar]
- 74.Pamp, S. J., D. Frees, S. Engelmann, M. Hecker, and H. Ingmer. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 1884861-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pané-Farré, J., B. Jonas, K. Forstner, S. Engelmann, and M. Hecker. 2006. The sigmaB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296237-258. [DOI] [PubMed] [Google Scholar]
- 76.Petersohn, A., H. Antelmann, U. Gerth, and M. Hecker. 1999. Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis. Microbiology 145(Pt. 4)869-880. [DOI] [PubMed] [Google Scholar]
- 77.Petersohn, A., J. Bernhardt, U. Gerth, D. Höper, T. Koburger, U. Völker, and M. Hecker. 1999. Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 1815718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 1835617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poole, R. K. 2005. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 33176-180. [DOI] [PubMed] [Google Scholar]
- 80.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41757-774. [DOI] [PubMed] [Google Scholar]
- 81.Reents, H., I. Gruner, U. Harmening, L. H. Bottger, G. Layer, P. Heathcote, A. X. Trautwein, D. Jahn, and E. Hartig. 2006. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol. Microbiol. 601432-1445. [DOI] [PubMed] [Google Scholar]
- 82.Reents, H., R. Munch, T. Dammeyer, D. Jahn, and E. Härtig. 2006. The Fnr regulon of Bacillus subtilis. J. Bacteriol. 1881103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Richardson, A. R., P. M. Dunman, and F. C. Fang. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61927-939. [DOI] [PubMed] [Google Scholar]
- 84.Rogstam, A., J. T. Larsson, P. Kjelgaard, and C. von Wachenfeldt. 2007. Mechanisms of adaptation to nitrosative stress in Bacillus subtilis. J. Bacteriol. 1893063-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 9814895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seggewiss, J., K. Becker, O. Kotte, M. Eisenacher, M. R. Yazdi, A. Fischer, P. McNamara, N. Al Laham, R. Proctor, G. Peters, M. Heinemann, and C. von Eiff. 2006. Reporter metabolite analysis of transcriptional profiles of a Staphylococcus aureus strain with normal phenotype and its isogenic hemB mutant displaying the small-colony-variant phenotype. J. Bacteriol. 1887765-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shafer, W. M., and J. J. Iandolo. 1979. Genetics of staphylococcal enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect. Immun. 25902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stevanin, T. M., N. Ioannidis, C. E. Mills, S. O. Kim, M. N. Hughes, and R. K. Poole. 2000. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo′ or bd, from nitric oxide. J. Biol. Chem. 27535868-35875. [DOI] [PubMed] [Google Scholar]
- 89.Stevanin, T. M., R. K. Poole, E. A. Demoncheaux, and R. C. Read. 2002. Flavohemoglobin Hmp protects Salmonella enterica serovar Typhimurium from nitric oxide-related killing by human macrophages. Infect. Immun. 704399-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 1392041-2045. [DOI] [PubMed] [Google Scholar]
- 91.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 1781374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tam le, T., H. Antelmann, C. Eymann, D. Albrecht, J. Bernhardt, and M. Hecker. 2006. Proteome signatures for stress and starvation in Bacillus subtilis as revealed by a 2-D gel image color coding approach. Proteomics 64565-4585. [DOI] [PubMed] [Google Scholar]
- 93.Tam le, T., C. Eymann, D. Albrecht, R. Sietmann, F. Schauer, M. Hecker, and H. Antelmann. 2006. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ. Microbiol. 81408-1427. [DOI] [PubMed] [Google Scholar]
- 94.Tam le, T., C. Eymann, H. Antelmann, D. Albrecht, and M. Hecker. 2007. Global gene expression profiling of Bacillus subtilis in response to ammonium and tryptophan starvation as revealed by transcriptome and proteome analysis. J. Mol. Microbiol. Biotechnol. 12121-130. [DOI] [PubMed] [Google Scholar]
- 95.Thomas, D. D., L. Ridnour, S. Donzelli, M. G. Espey, D. Mancardi, J. S. Isenberg, M. Feelisch, D. D. Roberts, and D. A. Wink. 2006. The chemistry of protein modifications elucidated by nitric oxide and related nitrogen oxides, p. 25-58. In I. Dalle-Donne, A. Scaloni, and A. Butterfield (ed.), Redox proteomics: from protein modifications to cellular dysfunction and diseases. John Wiley & Sons, Inc., Hoboken, NJ.
- 96.Throup, J. P., F. Zappacosta, R. D. Lunsford, R. S. Annan, S. A. Carr, J. T. Lonsdale, A. P. Bryant, D. McDevitt, M. Rosenberg, and M. K. Burnham. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 4010392-10401. [DOI] [PubMed] [Google Scholar]
- 97.VanBogelen, R. A., E. E. Schiller, J. D. Thomas, and F. C. Neidhardt. 1999. Diagnosis of cellular states of microbial organisms using proteomics. Electrophoresis 202149-2159. [DOI] [PubMed] [Google Scholar]
- 98.Weber, H., S. Engelmann, D. Becher, and M. Hecker. 2004. Oxidative stress triggers thiol oxidation in the glyceraldehyde-3-phosphate dehydrogenase of Staphylococcus aureus. Mol. Microbiol. 52133-140. [DOI] [PubMed] [Google Scholar]
- 99.Wink, D. A., K. M. Miranda, M. G. Espey, R. M. Pluta, S. J. Hewett, C. Colton, M. Vitek, M. Feelisch, and M. B. Grisham. 2001. Mechanisms of the antioxidant effects of nitric oxide. Antioxid. Redox Signal 3203-213. [DOI] [PubMed] [Google Scholar]
- 100.Wink, D. A., and J. B. Mitchell. 1998. Chemical biology of nitric oxide: insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic. Biol. Med. 25434-456. [DOI] [PubMed] [Google Scholar]
- 101.Wolf, C., F. Hochgräfe, H. Kusch, D. Albrecht, M. Hecker, and S. Engelmann. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics, in press. [DOI] [PubMed]
- 102.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 1824458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang, Y., and N. Hogg. 2006. S nitrosylation of cysteine thiols as a redox signal, p. 169-188. In I. Dalle-Donne, A. Scaloni, and A. Butterfield (ed.), Redox proteomics: from protein modifications to cellular dysfunction and diseases. John Wiley & Sons, Inc., Hoboken, NJ.
- 104.Zuber, P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 1861911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.