Abstract
A type IV secretion system (T4SS) is used by many symbiotic and pathogenic intracellular bacteria for the successful infection of and survival, proliferation, and persistence within hosts. In this study, the presence and function of the T4SS in Wolbachia strains were investigated by a combination of genetic screening and immunofluorescence microscopy. Two operons of virB-virD4 loci were found in the genome of Wolbachia pipientis strain wAtab3, from the Hymenoptera Asobara tabida, and strain wRi, infecting Drosophila simulans. One operon consisted of five vir genes (virB8, virB9, virB10, virB11, and virD4) and the downstream wspB locus. The other operon was composed of three genes (virB3, virB4, and virB6) and included four additional open reading frames (orf1 to orf4) orientated in the same direction. In cell culture and insect hosts infected with different Wolbachia strains, the bona fide vir genes were polycistronically transcribed, together with the downstream adjacent loci, notably, as virB8 to virD4 and wspB and as virB3, virB4, virB6, and orf1 to orf4. Two peptides encompassing conserved C and N termini of the Wolbachia VirB6 protein were used for the production of polyclonal antibodies. Anti-VirB6 antibodies could detect the corresponding recombinant protein by chemifluorescence on Western blots of total proteins from Escherichia coli transformants and Wolbachia strains cultured in cell lines. Using immunofluorescence microscopy, we further demonstrated that the VirB6 protein was produced by Wolbachia strains in ovaries of insects harboring wAtab3 or wRi and cell lines infected with wAlbB or wMelPop. As VirB6 is known to associate with other VirB proteins to form a membrane-spanning structure, this finding suggests that a T4SS may function in Wolbachia.
Wolbachia species are obligate intracellular alphaproteobacteria belonging to the order Rickettsiales (1, 21, 44). They are found in associations with numerous invertebrates, including arthropods and nematodes (53, 64). The latest investigations have reported that Wolbachia strains infect from 66 to 76% of species belonging to different orders of arthropods, thus identifying these bacteria as the most widespread intracellular symbionts yet described (26, 31). The interactions between Wolbachia strains and their hosts are very complex and range from parasitism to mutualism. In filarial nematodes, Wolbachia is required for host biology (5). In contrast, Wolbachia strains are mostly parasites which affect reproduction of arthropods; they alter host reproduction in a number of ways to enhance their own transmission through maternal inheritance from infected females to progeny (63). Some of these reproductive alterations include parthenogenesis induction in parasitoid wasps (54, 56), the feminization of genetic male isopods (8), and the induction of male killing (29) and cytoplasmic incompatibility (37, 55).
Despite the increasing number of investigations on Wolbachia-host interactions, little is known about the molecular determinants and mechanisms involved in the observed pleiotropic phenotypes. Research in this field is hampered by the uncultivable status of Wolbachia. However, the development of alternative cellular and molecular technologies has led to the suggestion that Wolbachia may excrete effectors that interfere with host development and reproduction programs (46, 58, 59). Analyzing the potential excretory machinery of Wolbachia may help in the identification of those effectors and orientate them to the corresponding potential targets in the host.
Bacteria have adapted series of machineries to mediate the transport of macromolecules across their envelopes. These transport apparatus play essential roles in bacterial life and adaptation in different environments, including the acquisition of nutrients, the expression of pathogenicity, or the establishment of symbiosis (9, 13, 22, 28, 67). Type IV secretion systems (T4SSs) are an example of these transport apparatus. T4SSs are broadly distributed among gram-negative and gram-positive bacteria (10, 24). T4SS family members have been grouped into three major subfamilies: (i) conjugation systems mediating DNA transfer from donor to recipient cells, (ii) effector translocator systems that transport molecules directly into the cytoplasm of eukaryotic cells, and (iii) DNA uptake or release systems mediating the exchange of DNA with the milieu by contact-independent mechanisms (10, 14, 20). By their versatility, T4SSs are involved in a wide range of biological processes. Conjugation through T4SSs is implicated in the transfer of genes involved in resistance to antibiotics or heavy metals, as well as in the acquisition of symbiotic or pathogenic islands, thus contributing to bacterial genome plasticity and evolution (10, 19, 22). Several pathogens, such as Helicobacter pylori, Legionella pneumophila, Anaplasma phagocytophilum, and Rickettsia spp., are known or suspected to secrete toxins via T4SSs of the effector translocator subfamily (1, 12, 38, 39, 60).
The Agrobacterium tumefaciens T4SS provides the hallmark of the T4SS-mediated transfer of bacterial effectors directly into eukaryotic cells, and it serves as a model system for basic research on the determinants and comprehensive mechanisms involved (27, 42). The bacterium A. tumefaciens uses the T4SS to transfer oncogenic DNA into susceptible plant cells, disturbing the plant's hormonal balance and causing the crown gall disease (69). This T4SS consists of a single operon encoding the mating pair formation (Mpf) VirB proteins and the coupling protein VirD4. The virB gene products elaborate a cell envelope-spanning structure required for substrate transfer, and the VirD4 coupling protein recruits the bacterial substrate to be delivered into the host cell (11).
In the past decade, virB and virD4 homologues clustered into a single operon have been found in many other bacteria by genetic screening and/or whole-genome sequencing (10, 13, 14). Split virB-virD operons in obligate intracellular bacteria of the order Rickettsiales, including Rickettsia spp., A. phagocytophilum, and Ehrlichia chaffeensis, have been described previously (2, 43). In Wolbachia pipientis, the presence of a T4SS was first evidenced by hybridization and PCR screening (40). Thereafter, the complete genome sequences of W. pipientis strains wMel from Drosophila melanogaster and wBm from the human filarial parasitic nematode Brugia malayi confirmed the existence of split virB and virD4 homologues (23, 66).
In the context of studying the potential role of the T4SS in Wolbachia, here we describe structural and functional analyses of virB-virD4 gene clusters in two strains that differ drastically in the phenotype induced in the host. The strain wRi induces strong cytoplasmic incompatibility, resulting in 100% offspring mortality in incompatible crosses between Drosophila simulans flies (45), whereas the strain wAtab3 is required for oogenesis in the wasp Asobara tabida, a rare case among arthropods of obligatory interdependence, as aposymbiotic females fail to produce eggs (18). Wolbachia virB and virD genes were cloned and sequenced, and their transcriptional activities were assayed. We developed antibodies against VirB6, an integral membrane protein constituting one of the T4SS structural core components (32, 33), and monitored the target protein in insect cell lines and insect tissues hosting different Wolbachia strains.
MATERIALS AND METHODS
Biological material.
A. tabida (Hymenoptera: Braconidae) is an endoparasitoid wasp associated with several Drosophila species. As females lay eggs in fly larvae, within which parasitic larvae feed and develop, parasitoids were bred on a Wolbachia-free line of Drosophila melanogaster originating from Ste. Foy-les-Lyon (France). A. tabida is naturally infected with three Wolbachia strains: wAtab1 and wAtab2 induce cytoplasmic incompatibility, and wAtab3 is required for host oogenesis (16, 18). The tri-infected line named Pi(123) originated from Pierrefeu, France, and is hereinafter referred to as A. tabida Pierrefeu. The line Pi(3), singly infected with wAtab3, was obtained using moderate rifampin antibiotic treatment (18). An A. tabida North American line infected with the three Wolbachia strains (wAtab1, wAtab2, and wAtab3) was also used. Aposymbiotic females were obtained from the North American line by antibiotic treatment (17). D. simulans flies naturally infected with the Wolbachia strain wRi, uninfected D. melanogaster flies, and A. tabida insects were reared on a standard-medium diet (15) at 20°C in a cycle of 12 h of light and 12 h of darkness and 70% relative humidity.
Mosquito cell lines infected with Wolbachia, RML12 from Aedes aegypti infected with strain wMelPop (C. J. McMeniman, I. Iturbe-Ormaetxe, A. M. Lane, D. T. Voronin, R. Yamada, E. McGraw, and S. L. O'Neill, submitted for publication) and Aa23 infected with strain wAlbB, as well as uninfected cells, were grown in 25-cm2 culture flasks (Greiner Bio-One, Frickenhausen, Germany) at 26°C in equal volumes of Mitsuhashi-Maramorosch medium (Bioconcept, Switzerland) and Schneider's insect medium (Sigma, France), supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum and penicillin-streptomycin (50 U and 50 μg per ml, respectively [Gibco; Invitrogen]). The infection status was verified by PCR amplification of the wsp gene as described previously (41).
DNA purification.
Genomic DNA was isolated from adult A. tabida insects singly infected with Wolbachia strain wAtab3, adult D. simulans insects singly infected with Wolbachia wRi, and adult D. melanogaster insects singly infected with wMel by using the DNeasy tissue kit according to the recommendations of the manufacturer (Qiagen) with some modifications. Briefly, insects (30 mg) were crushed in 360 μl of buffer ATL (Qiagen). The mixture was treated with 2 mg of lysozyme (Euromedex) ml−1 for 3 h at 37°C and then with proteinase K from the DNeasy tissue kit for 12 h at 56°C as recommended by the manufacturer (Qiagen). The mixture was centrifuged twice at 16,000 × g for 1 min each time. The supernatant was transferred into a new tube and treated with 1 mg of RNase A ml−1 for 2 min at room temperature. Then 330 μl of buffer AL (Qiagen) was added, and the sample was incubated for 10 min at 70°C. After the addition of 330 μl of 96 to 100% ethanol, the sample was mixed thoroughly by being subjected to a vortex and was pipetted into the DNeasy mini spin column from the DNeasy tissue kit as recommended by the manufacturer (Qiagen). DNA was eluted with 100 μl of TE buffer (10 mM Tris-Cl, pH 7.5, and 1 mM EDTA, pH 8) and stored at −20°C until being used. Recombinant plasmid DNA was extracted from Escherichia coli transformants by using a QIAminiprep kit (Qiagen).
RNA extraction and reverse transcription.
Total RNA was isolated from 500 ovaries of A. tabida females singly infected with Wolbachia strain wAtab3, from 15 whole adult flies, and from 108 mosquito cells. Ovaries were dissected in a phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). Ovaries, insects, or cells were crushed in Trizol reagent (Invitrogen) with glass beads 212 to 300 μm in diameter (Sigma). After separation with chloroform, the aqueous phase was transferred into a new tube and the RNA was precipitated overnight with 2 M LiCl. Then samples were centrifuged at 16,000 × g and 4°C for 15 min. After the supernatant was discarded, pellets containing total RNA were washed twice with 70% ethanol by centrifugation at 16,000 × g and 4°C for 15 min each time and air dried for 30 min. The RNA obtained was dissolved in RNase-free water by incubation for 15 min at 37°C and then treated with DNase using a TURBO DNA-free kit as recommended by the manufacturer (Ambion). After the verification of the absence of contaminating DNA by PCR, aliquots of RNA were stored at −80°C until being used.
RNAs (1 to 3 μg) were reverse transcribed using SuperScript III reverse transcriptase (RT; Invitrogen) with random primers (Invitrogen). The procedure was done according to the manufacturer's protocols, but the final heat denaturation of the RT was omitted. Synthesized cDNA molecules were resuspended in 100 μl of H2O and treated with 3 U of RNase H (Promega) for 20 min at 37°C. cDNA molecules were purified using a QIAquick PCR purification kit (Qiagen) and eluted with 15 μl of nuclease-free water (Ambion).
PCR, cloning, and sequencing.
Oligonucleotide primers targeting vir genes and adjacent regions (Table 1) were designed according to the complete nucleotide sequence of the W. pipientis strain wMel genome (GenBank accession number AE017196) by using the software Oligo 5.1. To produce recombinant VirB6 protein, PCR primers were designed according to the nucleotide sequence of the virB6 gene of Wolbachia strain wAtab3, with the addition of restriction sites (Table 1). The oligonucleotides were synthesized by Invitrogen (France). PCR amplifications were done in a 25-μl reaction mixture containing the DNA template (30 to 90 ng) in 1× polymerase reaction buffer (Invitrogen), 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, a 200 nM concentration of each primer, and 0.5 U of Taq polymerase (Invitrogen). PCRs were performed in a T1 thermocycler (Biometra) under the following conditions: initial denaturation at 95°C for 1 min; 35 cycles of denaturation (94°C for 1 min), annealing (1 min at a melting temperature depending on the primers), and extension (72°C for 40 s to 2 min, depending on the fragment length); and a final extension at 72°C for 10 min. For nested PCRs, only 25 cycles were done. The PCR products were purified with a QIAquick PCR purification kit (Qiagen) and ligated into the PCR2.1-TOPO vector by using a TOPO-TA cloning kit (Invitrogen). After the transformation of competent E. coli DH5α cells, recombinant plasmids were extracted as described above and cloned inserts were subjected to sequencing at GenoScreen (Lille, France). All sequences were analyzed by the BLASTn program (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) and deposited in GenBank (Table 1).
TABLE 1.
Primers, genes, and regions
| Purpose | Gene(s) | Primer namea | Primer sequence (5′-3′) | Amplicon size (bp)/melting temp (°C) | GenBank accession no.b for wRi/wAtab3 or reference |
|---|---|---|---|---|---|
| PCR | ribA, virB8, virB9 | Vir2F | CGTCTGTTGGTGAATAATTTT | 1,721/52 | EF427901, EF423637/DQ887624, |
| Vir2Fb | TGGAAGAGGYATTGGTTTAAC | 1,191/53 | DQ887625 | ||
| Vir2R | TTTTTAACTTTTTCTCCTTCC | ||||
| virB9, virB10 | Vir3F | CGGTGGTTTTTAGTCAGGGTT | 1,979/56 | EF423637, EF427902/DQ887625, | |
| Vir3Fb | TGCSTTAKGAAGATGGTGAAG | 1,361/53 | EF427903 | ||
| Vir3R | TCATCATAAACAGACTTGCTT | ||||
| virB10 | FB10m | CCAAAAGAAACTCCTGTGTCA | 408/51 | EF427902/EF427903 | |
| RB10m | AAATGAATAACTGCCTATCAA | ||||
| virB10, virB11 | FB10e | TGATGCTGTTCTTGAAACTGC | 879/50 | EF427902, EF423636/EF427903, | |
| RB10e | CTTGAAATATGCCTTGTAATG | DQ887626 | |||
| virB10, virB11, virD4 | Vir4F | GGTAGAGCAGGAATAGCAGGA | 1,835/56 | EF427902, EF423636, | |
| Vir4Fb | TCCACAGGCAATATTAAATCA | 1,278/53 | AY833076/EF427903, | ||
| Vir4R | TATATAACTCTGGATGATGCC | DQ887626, DQ887627 | |||
| virD4 | VirD4F | TTGTTGGCATCATCCAGAGTT | 1,530/51 | AY833076/DQ887627 | |
| VirD4Fb | TGGTTGACGATGTGCAGAAAA | 1,024/53 | |||
| VirD4R | GCAGAAAGAACTTTATTTGGG | ||||
| virD4, wspB | U21 | ACCCAATTAAATCAAAGAAAA | 622/49 | AY833076, EU095939/ | |
| RRT5 | TAGACAAATTTTTGATACCTG | DQ887627, EU095938 | |||
| virD4, wspB | FRT5 | GATAAGTTTGAAGATGATGAC | 1,029/50 | AY833076, EU095939/ | |
| RwspB | TAAGCTTTGCTGGCAAAATGG | DQ887627, EU095938 | |||
| wspB, WD0010 | F2wspB | AGGTAGCAAAGTAACTGAGGA | 470/50 | EU095939/EU095938 | |
| R2wspB | ATGGTGGTATTGATGGTTGA | ||||
| WD0856, virB6 | VirB6F | ACGACCAGAAGAAAATTTACA | 1,246/50 | EF423639/DQ887628 | |
| VirB6R | GTTTTTGTCCTTTATGTTCAT | ||||
| virB6 | VirB6Fb | CTTGTCCGGTTGTTCATTTTT | 1,316/50 | EF423639/DQ887628 | |
| VirB6Rb | AATGTAGAGTTCTCAAAGCAG | ||||
| FvirB6NdeI | ACGGAATTCATATGATAGCTGCAATTGTT | 1,014/52 | |||
| RvirB6BamHI | GGCGGATCCTTTCAGCCTTTTCTG | ||||
| virB6, virB4 | VirB4F | TGCCCGTTTTTCTCACTGCTT | 1,285/50 | EF423639, EF423640/DQ887628, | |
| VirB4R | TAGATAATCCAGTTTTTGCAC | DQ887629 | |||
| virB4 | VirB4Fb | AAAAACATCCCGATAGACACT | 1,338/50 | EF423640/DQ887629 | |
| VirB4Rb | TTTTTACAACTTCCTTACGAG | ||||
| virB4, virB3 | VirB3F | CGACAGAGCGTTATCCAATGA | 1,576/49 | EF423640, EF423641/DQ887629, | |
| VirB3R | GTCCATAACCAGTTTCAAATA | DQ887630 | |||
| virB3, WD0860 | VirB3Fb | GAGCATTGGAGGTCTTGTAAG | 1,334/50 | EF423641/DQ887630 | |
| VirB3Rb | TAAGAGGATATAAAACCCCAT | ||||
| RT-PCR | virB8, virB9 | FRT1 | TGGGTAACGATGCTCTTCAGG | 377/50 | |
| RRT1 | TTTTTAACTTTTTCTCCTTCC | ||||
| virB9, virB10 | FRT2 | TTGTAAAAGGCTGTTATTTGG | 339/50 | ||
| RRT2 | GATCAGTTATTATTCTTTCAG | ||||
| virB10, virB11 | FRT3 | AAGCAATCGGGAAATCAATCG | 383/50 | ||
| RRT3 | GCTTCTTCGCTAAGTTTTTGT | ||||
| virB11, virD4 | FRT4 | GATCTGCGAGTGAAAATGCTT | 282/50 | ||
| RRT4 | TATATAACTCTGGATGATGCC | ||||
| virD4, wspB | FRT5 | GATAAGTTTGAAGATGATGAC | 350/50 | ||
| RRT5 | TAGACAAATTTTTGATACCTG | ||||
| WD0853, WD0854 | F853 | CAATCTCTTTGTCTTCCTTAT | 263/48 | ||
| R854 | TTCATCTTCAATCCTCTTCTC | ||||
| WD0854, WD0855 | F854 | TCCAAGATGTCCATGCACCATTAACT | 603/53 | 66 | |
| R855 | CTTTGGCGTGTTAGGTAATTTAGCCT | 66 | |||
| WD0855, WD0856 | F855 | ATTTGATTTCCTCACCCTTAC | 282/51 | ||
| R856 | TGACGTACCTGACTGTTGAGA | ||||
| WD0856, virB6 | FRT6 | CCCATTTCTGCTCGTTGACA | 225/50 | ||
| RRT6 | CTAAGAAGTTGAGTGGTGGAG | ||||
| virB6, virB4 | FRT7 | TCTGCAAGCACTGGATATGAG | 245/48 | ||
| RRT7 | TCAATGCTGTAGTAGCTAGAA | ||||
| virB4, virB3 | FRT8 | CTTTTGTCATCAAGGTTGTGC | 360/48 | ||
| RRT8 | TTGGTGTGAGTTATATATTTG | ||||
| virB3, WD0860 | FRT9 | GAGCATTGGAGGTCTTGTAAG | 379/48 | ||
| RRT9 | ATATCATCTGAAACTGCAATG | ||||
| Nested RT-PCR | virB8, virB9 | FB8/B9n | ATTTACACGGAAAGATGGAAG | 272/47 | |
| RB8/B9n | TACTTCATTAGGGCTATACAC | ||||
| virB9, virB10 | FB9/B10n | GGCGTAACAATTCAGAAGATG | 101/50 | ||
| RB9B10n | AAGCACTAAAATAATGACCAT | ||||
| virB10, virB11 | FB10/B11n | CAATGAAATGAACTATGCTGC | 169/47 | ||
| RB10/B11n | CAAATGGTTAAGATCGAATAC | ||||
| virB11, virD4 | FB11/D4n | GCGTACTTGAGTTTTGCTTCT | 110/50 | ||
| RB11/D4n | AGGGAAAGGCGTCAAACTAGG | ||||
| virB6, virB4 | FB6/B4n | CTGGATATGAGAGTAAAAAGT | 222/47 | ||
| RB6B4n | AGCTAGAATAGACTTAAAAGG |
F, forward; R, reverse. Restriction site designations incorporated into primer names are underlined, as are the corresponding sequences.
GenBank accession numbers correspond to sequences obtained in this study.
For the cloning of virB6 into an expression vector, PCR amplification was done with the high-fidelity Vent DNA polymerase (Ozyme) by employing an initial denaturation at 95°C for 3 min and two cycles of denaturation at 94°C, annealing at 40°C, and extension at 72°C for 1 min each, followed by a second round of denaturation at 95°C for 1 min 30 s; 35 cycles of denaturation (94°C for 1 min), annealing (52°C for 1 min), and extension (72°C for 1 min); and a final extension at 72°C for 10 min. The PCR amplicon was purified as described above, and a single adenosine was added to the 3′ ends of the double-stranded DNA molecules. Products were purified and cloned into the PCR2.1-TOPO vector as described above. The recombinant plasmid was extracted from E. coli, double-restricted with NdeI and BamHI, and subjected to gel electrophoresis. The expected virB6 fragment was excised from the gel, purified using a QIAquick gel extraction kit (Qiagen), ligated into the restricted pIVEX2.4d vector (Roche Applied Science, Germany), and introduced into E. coli DH5α by transformation. Five transformant colonies were selected, and the corresponding pIVEX2.4d-virB6 recombinant plasmids were purified and submitted to sequencing. One recombinant plasmid containing the expected virB6 sequence was used to chemically transform E. coli BL21-AI (Invitrogen).
Production of anti-VirB6 peptides and recombinant VirB6 protein and immunoblotting.
Two peptides encompassing residues 43 to 57 (FNSTGSFSRASNPDC) and residues 687 to 701 (LDESYKNEQPDKSYE) from the VirB6 sequence of wAtab3 were synthesized by Covalab, France. The antisera were obtained by the immunization of rabbits with both oligopeptides coupled with keyhole limpet hemocyanin, and specific antibodies were purified from the antisera by affinity column chromatography using a mixture of the corresponding peptides fixed on the column (Covalab, France).
To produce VirB6 protein, the transformant E. coli BL21-AI/pIVEX2.4d-virB6 was grown overnight at 37°C in modified Luria-Bertani medium (10 g of Bacto tryptone [Difco] liter−1, 5 g of yeast extract [Difco] liter−1, 5 g of NaCl liter−1) containing 50 μg of carbenicillin ml−1. Cultures (250 ml) were induced by l-arabinose at a 0.02% final concentration. Cell cultures were harvested by centrifugation at 6,500 × g for 20 min at 4°C. The pellet was resuspended in sample buffer (62.5 mM Tris-HCl [pH 6.8], 1% [wt/vol] sodium dodecyl sulfate [SDS], 100 mM dithioerythritol), the suspension was boiled at 95°C for 5 min, and a 25-μl sample was used to determine the protein concentration by the Bradford method with an assay kit from Bio-Rad. Then 10% glycerol and 0.001% bromophenol blue were added to the protein extract, and the mixture was heated for 5 min at 95°C. A 10-μg sample of proteins was separated by electrophoresis on a 1-mm-thick slab gel containing 12% polyacrylamide and 0.32% SDS in the upper buffer only (34). After electrophoresis, proteins were stained with Coomassie blue and the overexpressed protein band was excised and used for sequencing by liquid chromatography-tandem mass spectrometry at the Institut de Biologie et Chimie des Proteines, Université Lyon 1, Lyon, France.
To extract proteins from Wolbachia, the mosquito cell line RML12 infected with strain wMelPop and uninfected cells were used. Five 175-cm2 flasks containing 35 ml of medium were used to grow cells using the medium and temperature described above. Cells were recovered when they reached 80% confluence (107 cells/ml). To isolate Wolbachia, cell cultures were centrifuged, the pellet was resuspended in 6 ml of fresh medium, and the suspension was divided into two 3-ml fractions, which were then homogenized thoroughly by being subjected to a vortex for 20 min in the presence of 3-mm borosilicate glass beads. Lysates were combined and centrifuged at 300 × g for 5 min, and the supernatant enriched with Wolbachia bacteria was centrifuged at 8,150 × g and 4°C for 15 min. Pellets containing bacteria were resuspended in 200 μl of 1× PBS, and the suspension was layered onto 800 μl of 250 mM sucrose in a 1.5-ml Eppendorf tube and centrifuged at 21,600 × g for 5 min at 4°C. Supernatant was removed, the pellet of Wolbachia cells was suspended in 50 μl of Laemmli buffer (35) containing 4% SDS, and the suspension was boiled for 5 min at 95°C and then used in SDS-polyacrylamide gel electrophoresis (PAGE). When flies were used as starting materials, five D. simulans adults infected with strain wRi and five uninfected individuals were directly homogenized in 50 μl of Laemmli buffer, and the homogenates were boiled and centrifuged briefly to pellet debris before being used.
For Western blotting, separated proteins were transferred onto a nitrocellulose membrane (PerkinElmer) according to the method of Kyhse-Andersen (34). The transfer was done at 150 mA for 2 h. For dot blotting, crude boiled insect extracts were transferred onto a nitrocellulose membrane by using a Bio-Dot apparatus (Bio-Rad) with standard protocols. Membranes were soaked in 4% dried milk in 1× TBST buffer (20 mM Tris [pH 7.5], 130 mM NaCl, 0.1% Tween 20) for 1 h at room temperature. Membranes were washed three times in 1× TBST buffer and incubated overnight at 4°C with anti-VirB6 at a 1:200 dilution in 1× TBST. Incubation was followed by three washes with 1× TBST, and then the membranes were incubated for 45 min at room temperature with the conjugate goat anti-rabbit immunoglobulin G-peroxidase at a 1:15,000 dilution in 1× TBST. Membranes were washed three times as described above. Hybridizing bands were revealed using an ECL kit as recommended by the manufacturer (Amersham).
Immunofluorescence microscopy.
To obtain samples of ovaries, D. simulans females infected with wRi, A. tabida Pierrefeu females singly infected with wAtab3, A. tabida North American females tri-infected with Wolbachia wAtab1, wAtab2, and wAtab3, and uninfected A. tabida North American females were dissected in 1× PBS. Recovered ovaries were rinsed once with 1× PBS before the fixation procedure. Cells of mosquito lines RML12 and Aa23 that were uninfected or infected with the Wolbachia strains wMelPop and wAlbB were also used for bacterial detection by immunofluorescence. A total of 104 to 105 cells of each cell line were separately transferred into Trac bottles containing a glass slide (Fisher Scientific) and grown in 1 ml of medium until approximately 80% confluence was achieved. The medium was removed, and cells attached to the glass were washed once with 1× PBST (PBS containing 1% Triton X-100) before the application of a fixative. Samples of cells and ovaries were fixed with 4% formaldehyde in 1× PBST for 20 min at room temperature. After being washed three times in 1× PBST (5 min each time), samples were incubated with anti-VirB6 (diluted 1:200 in 1× PBST and 1% bovine serum albumin Σ) for 1 h at room temperature. Samples were washed in 1× PBST three times for 10 min each time and incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (diluted 1:500 in 1× PBST and 1% bovine serum albumin). At the end of the 1-h incubation time, samples were washed as described above and then mounted in 80% glycerol with 1× PBS. The samples were observed by using a fluorescence microscope (Axio ImagerZ1; Zeiss).
Nucleotide sequence accession numbers.
The gene sequences obtained in this study have been deposited in GenBank under accession numbers AY833076, DQ887624, DQ887625, DQ887626, DQ887627, DQ887628, DQ887629, DQ887630, EF427901, EF427902, EF427903, EF423636, EF423637, EF423639, EF423640, EF423641, EU095938, and EU095939.
RESULTS
Identification of the virB and virD4 loci in Wolbachia strains wAtab3 and wRi.
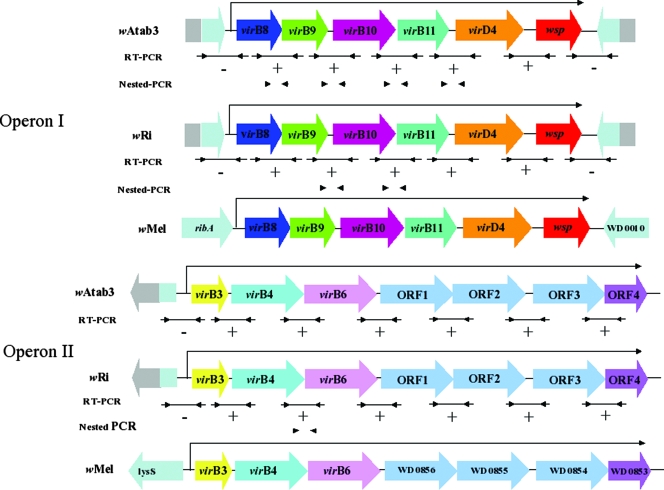
Based on the genome sequence of Wolbachia strain wMel (66), oligonucleotide primers targeting virB and virD4 genes were designed and used in PCR amplifications. Eight orthologs of virB3, virB4, virB6, virB8, virB9, virB10, virB11, and virD4 were successfully amplified using genomic DNA extracted from A. tabida and D. simulans singly infected with Wolbachia strains wAtab3 and wRi, respectively. Sequences of genes and intergenic regions indicated that the genomic organizations of these T4SS loci in the two strains are similar (Fig. 1 and Table 1). The vir genes are arranged into two separate clusters; one cluster (herein designated cluster I) contains the genes virB8, virB9, virB10, virB11, and virD4, and the other cluster (herein designated cluster II) is composed of virB3, virB4, and virB6 (Fig. 1). This gene composition and clustering are similar to those of the T4SS determinants present in other Wolbachia strains, including wMel (66), wBm (23), wAna (http://www.ncbi.nlm.nih.gov), and wVul (GenBank accession numbers AY967767 and AY967766), as well as in the closest relatives, Anaplasma spp., Ehrlichia spp., and Rickettsia spp. (2, 43). Genes virB1, virB2, virB5, and virB7 of A. tumefaciens seem to be absent in most Rickettsiales investigated so far.
FIG. 1.
Genomic organization and expression of the T4SS in Wolbachia strains wAtab3 and wRi. ORFs are represented as open boxes, with arrowheads indicating their orientations. Lines with facing arrows at the ends indicate the regions subjected to RT-PCR transcriptional analysis. Arrows indicate positions of primers. Facing arrowheads indicate nested PCR. + or − indicates positive or negative transcription. The wMel transcription pattern is depicted based on data from Wu et al. (66).
All the Vir protein sequences of wRi and wAtab3 showed higher levels of similarity (90 to 100%) to their homologues in Wolbachia strains wMel and wAna from insects than to Vir protein sequences of Wolbachia strains wVul from the isopod Armadillidium vulgare and wBm from the nematode B. malayi (81 to 90%) (see Table ST1 in the supplemental material). Alignments with available VirB and VirD4 protein sequences from other alphaproteobacteria showed the highest levels of amino acid similarity to Vir proteins from members of the closest genera, Anaplasma spp. and Ehrlichia spp. (33 to 78%), and the more distantly related Rickettsia spp. (11 to 28%), as expected (data not shown). These results are in accordance with the fact that all these four genera belong to the same order, Rickettsiales (21).
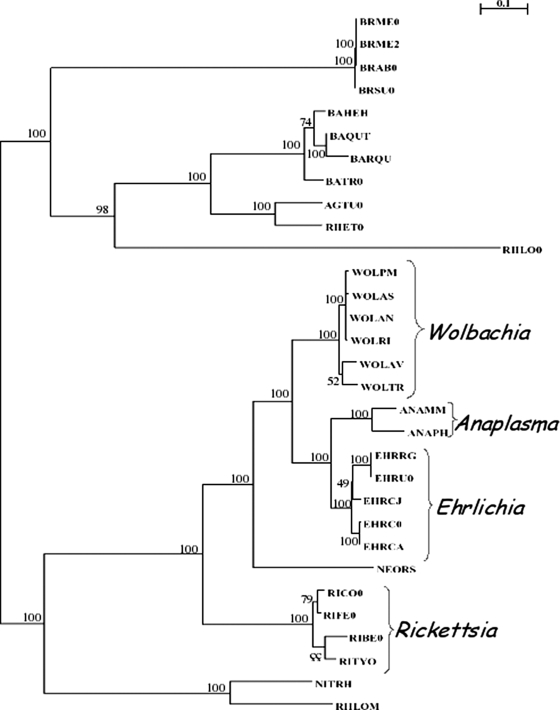
To further analyze the relationships among VirB and VirD orthologs of wAtab3 and wRi and those of other alphaproteobacteria, we constructed a phylogenetic tree based on concatenated amino acid sequences. Concatenated VirB and VirD amino acid sequences of Wolbachia strains clustered into the same group, as did those of Anaplasma spp., Ehrlichia spp., and Rickettsia spp. (Fig. 2). The four groups were promiscuously located. The VirB-VirD phylogeny presented in Fig. 2 and the species phylogeny (65) are congruent, suggesting that no recent lateral acquisition of vir genes from distant bacteria by Rickettsiales has occurred.
FIG. 2.
T4SS tree topology of strains belonging to the Rickettsiales and other alphaproteobacteria. This tree was inferred from concatenated alignments of amino acid sequences corresponding to VirB3, VirB4, VirB8, VirB9, and VirB11. The topology and branch lengths are according to the results of a maximum-likelihood analysis of the five protein sequences with 100 bootstraps replicates. BRME0, Brucella melitensis biovar suis; BRME2, Brucella melitensis biovar abortus; BRAB0, Brucella abortus biovar 1 strain 9-941; BRSU0, Brucella suis 1330; BAHEH, Bartonella henselae strain Houston-1; BAQUT, Bartonella quintana strain Toulouse; BARQU, Bartonella quintana; BATR0, Bartonella tribocorum; AGTU0, Agrobacterium tumefaciens strain C58; RHET0, Rhizobium etli; WOLPM, Wolbachia strain wMel; WOLAS, Wolbachia strain wAtab3; WOLAN, Wolbachia strain wAna; WOLRI, Wolbachia strain wRi; WOLAV, Wolbachia strain wVul; WOLTR, Wolbachia strain wBm; ANAMM, Anaplasma marginale strain St. Marie; ANAPH, Anaplasma phagocytophilum HZ; EHRRG, Ehrlichia ruminantium strain Gardel; EHRU0, Ehrlichia ruminantium strain Welgevonden; EHRCJ, Ehrlichia canis strain Jake; EHRC0, Ehrlichia chaffeensis; EHRCA, Ehrlichia chaffeensis strain Arkansas; NEORS, Neorickettsia sennetsu strain Miyayama; RICO0, Rickettsia conorii strain Malish; RIFE0, Rickettsia felis URRWXCAL2; RIBE0, Rickettsia bellii RML369-C; RITYO, Rickettsia typhi strain Wilmington; NITRH, Nitrobacter hamburgensis X14; RHLOM, Mesorhizobium loti.
As in the wMel genome, in both wRi and wAtab3, cluster I is positioned downstream of the ribA gene, encoding the GTP cyclohydrolase II, and upstream of the wspB gene, encoding an outer membrane protein, whereas cluster II is located downstream of the lysS gene, encoding lysyl-tRNA synthetase, and upstream of a gene encoding a hypothetical membrane protein (the WD0856 gene). The intergenic regions between virD4 and wspB and between virB6 and WD0856 are short to harbor potential promoters for these genes, and in addition, the wspB and WD0856 genes in the two strains are in the same orientation as the respective neighboring vir loci, which may suggest coregulation.
Transciptional analysis of virB and virD genes in Wolbachia strains.
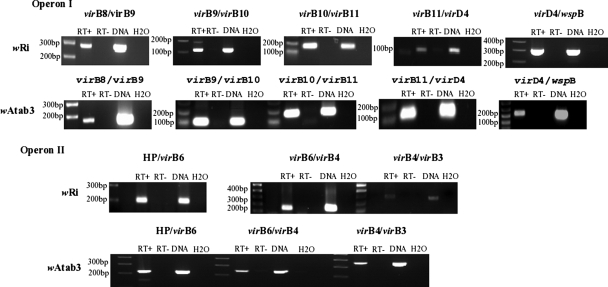
To analyze the cotranscription of vir genes of the two clusters in strains wAtab3 and wRi, RT-PCR was used with oligonucleotide primers that overlapped contiguous genes (Fig. 1). Results showed that the cDNA amplicons obtained had the expected sizes, as did the control DNA amplicons (Fig. 3). No amplification was achieved in the absence of RT, whatever the quantity of RNA used, up to 9 ng, thus excluding genomic DNA contamination. The sequencing of RT-PCR products ascertained the presence of the expected overlapping vir genomic regions (data not shown). These results indicate that virB8 to virB11 and virD4 of cluster I are cotranscribed, that virB3, virB4, and virB6 of cluster II are cotranscribed, and that these clusters constitute two operons.
FIG. 3.
Transcriptional analysis of virB-virD4 operons in Wolbachia strains wRi and wAtab3. RT-PCR was used to analyze cotranscription. The primers used and their positions are shown in Table 1 and Fig. 1. RT+ and RT− indicate the presence and absence of RT in the reaction mixtures. The DNA control lanes (containing 30 ng of genomic DNA from insects singly infected with wRi or with wAtab3) show the specificity of amplification with each pair of primers. HP, ORF1.
As some genes adjacent to vir loci were reported previously to be cotranscribed in Wolbachia wMel from D. melanogaster (66), further transcriptional examination of contiguous genes in each cluster was performed using RNA extracted from insect tissues and oligonucleotide primers as depicted in Fig. 1. We found that wspB of cluster I and the four open reading frames (ORFs) of cluster II were cotranscribed with the genes of their respective operons (Fig. 1 and 3). Successful transcription of bona fide virB and virD genes and adjacent loci was also achieved using RNA extracted from mosquito RML12 and Aa23 cells infected with Wolbachia wMelPop and wAlbB (data not shown).
All together, these results clearly showed that T4SS genes of several Wolbachia strains from arthropods, notably, wAtab3 and wRi, as well as wAlbB and wMelPop, are conserved and are transcriptionally expressed both in cellulo and in vivo.
Immunodetection of Wolbachia VirB6 protein in infected tissues.
A comparative analysis of amino acid sequences of VirB6 showed high levels of similarity (up to 99% identity) among the proteins of the three Wolbachia strains from arthropods (wMel, wAtab3, and wRi). Polyclonal anti-VirB6 antibodies were produced by the immunization of rabbits with two peptides corresponding to conserved and antigenic C- and N-terminal Wolbachia VirB6 sequences (see Materials and Methods). Therefore, the anti-VirB6 peptides produced may detect these strains in the immunochemistry experiments.
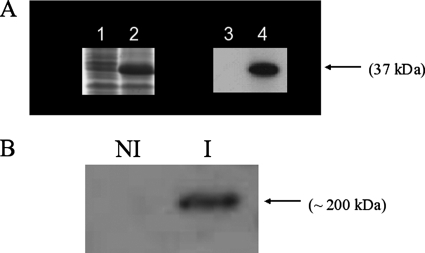
To examine the quality of anti-VirB6 to be used for detecting the corresponding Wolbachia protein, an analysis was done by Western blotting of crude protein extracts from E. coli harboring a recombinant form of VirB6 from the Wolbachia wAtab3 strain. The results revealed a chemiluminescent specific band of the expected size in the induced E. coli culture overproducing recombinant VirB6 protein, and no signal was detected in the uninduced culture (Fig. 4A). The identity of the VirB6 protein was also confirmed by the sequencing of the corresponding band extracted from the polyacrylamide gel (data not shown). In addition, the production of VirB6 protein was investigated using the Wolbachia protein extract purified from infected mosquito cells. The anti-VirB6 antibodies detected a unique and specific band of approximately 200 kDa exclusively in the fraction of Wolbachia-infected cells (Fig. 4B). The detected 200-kDa protein was more than twice the expected size of Wolbachia VirB6 protein (90 kDa). SDS-PAGE and Western blotting have previously revealed VirB6 and VirD4 proteins with unusually high molecular masses in Agrobacterium (25). The authors of the previous study suggested that this finding resulted from aggregates formed during boiling, as seen for some membrane proteins with high molecular masses. Other authors have reported difficulties and failures in detecting Agrobacterium VirB6 protein in Western blot assays due to the fact that VirB6 protein may form an SDS-resistant oligomer that does not enter the gel (32, 33). We took advantage of the aggregation behavior of VirB6 protein to perform a dot blot assay using adult flies. Results showed strong signals in the extracts from flies harboring Wolbachia strain wRi, whereas weak signals corresponding to nonspecific bindings were seen in the samples from uninfected flies (see Fig. S1 in the supplemental material).
FIG. 4.
Western blots of VirB6 proteins. (A) SDS-PAGE profile of E. coli BL21-AI harboring pIVEX2.4d-virB6 (lanes 1 and 2) and the corresponding immunoblot with anti-VirB6 (lanes 3 and 4). Lanes 1 and 3, uninduced culture; lanes 2 and 4, culture induced by l-arabinose. (B) Western blot of Wolbachia protein revealed by anti-VirB6. NI, extract from Aedes aegypti RML12 cells not infected with Wolbachia; I, extract from Wolbachia-infected Aedes aegypti RML12 cells.
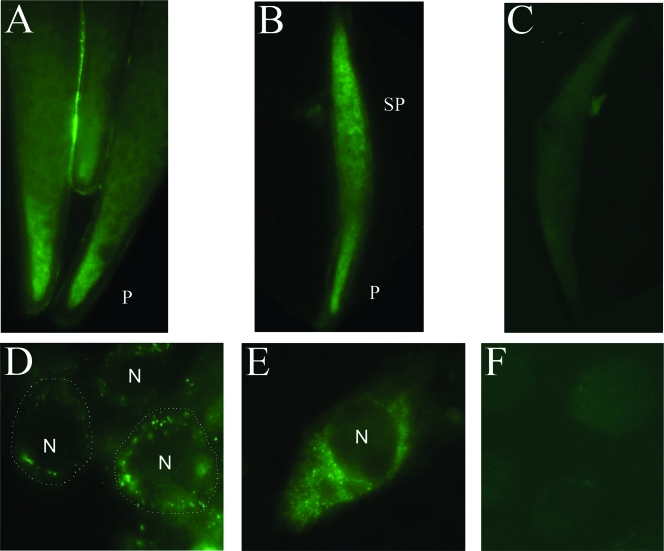
To detect VirB6 expression in vivo, we first used ovaries of A. tabida Pierrefeu females monoinfected with Wolbachia strain wAtba3. The analysis showed specific signals mainly in the cytoplasm of posterior oocytes hosting wAtab3 (Fig. 5A), which is in accordance with the usual location of this strain as shown previously (18). To ascertain that the signals corresponded to Wolbachia VirB6, we performed the same experiment with ovaries of the A. tabida North American line triply infected with Wolbachia and with ovaries of uninfected A. tabida North American females. In contrast to A. tabida Pierrefeu females, females of the North American line treated with antibiotic to remove Wolbachia still have the ability to produce a certain number of eggs (17) and thus provided us with a negative control. The results confirmed strong specific signals in the poles of infected oocytes, albeit more extended toward the subpole areas of the oocytes than in the singly infected lines, indicating high Wolbachia cell density due to tri-infection (Fig. 5B). No signal was detected in uninfected ovaries, as expected (Fig. 5C). Similar specific immunofluorescent signals were also obtained using ovaries from D. simulans flies singly infected with strain wRi (data not shown). Moreover, we applied immunofluorescence microscopy approaches to detect Wolbachia in the mosquito cell cultures by using anti-VirB6. Results showed fluorescent signals in the cytoplasm of cells infected with Wolbachia wMelPop (Fig. 5D and E) and wAlbB (data not shown), whereas no signal was seen in uninfected cells (Fig. 5F) used as a negative control.
FIG. 5.
Immunodetection of Wolbachia by anti-VirB6 peptides in cellulo and in insecta. Tissues were labeled with anti-VirB6 and fluorescent conjugates. (A to C) Oocytes of A. tabida singly infected with strain wAtab3 (A) and infected with the three strains wAtab1, wAtab2, and wAtab3 (B) and of an uninfected A. tabida line generated by antibiotic treatment (C). P and SP indicate polar and subpolar regions, respectively. (D) Low-magnification image showing Wolbachia wMelPop in a group of Aedes aegypti RML12 cells with boundaries manually delineated by circles. (E) High-magnification image of an enlarged Wolbachia-infected RML12 cell demonstrating the cell shape. (F) Uninfected RML12 cell. N designates the nucleus.
These results indicate that the synthetic anti-VirB6 peptides produced in this study can specifically detect the VirB6 protein expressed by different Wolbachia strains (notably, wAtab3 and wRi, as well as wAlbB and wMelPop) in host tissues.
DISCUSSION
The vir genes of the T4SS were investigated for their presence and expression in Wolbachia strains by PCR and RT-PCR amplifications, as well as by Western blotting and immunofluorescence microscopy using polyclonal antibodies to VirB6, a transmembranal core protein of the VirB-VirD4 apparatus. We have identified and sequenced two vir gene clusters from Wolbachia strains wAtab3 and wRi, which had infected A. tabida and D. simulans, respectively. One cluster comprises virB8, virB9, virB10, virB11, and virD4 located upstream from the wspB gene, and the other contains virB3, virB4, and virB6 found upstream from the WD0856 homologue. The deduced amino acid sequences corresponding to these vir genes showed high levels of similarity (81 to 100% identity) to previously described vir loci in other Wolbachia strains (23, 40, 66).
The virB8 to virB11 and virD4 genes of wAtab3 and wRi were polycistronically transcribed, probably through a promoter located between the upstream gene ribA and virB8, as in Wolbachia strain wTai (40). Here, this operon was found to extend to the downtream wspB gene, as in wMel (66). Two other wsp genes are present in the Wolbachia genome, of which one (wspA) is usually used in strain identification and phylogeny analyses (68), although currently these analyses are completed by multilocus sequence typing (3, 4, 47). The wsp locus encodes an outer surface protein (Wsp) whose homologue in Wolbachia strain wDm from the nematode Dirofilaria immitis is able to inhibit the apoptosis of human granulocytes (6). A gene for an antigenic outer membrane protein, a member of the p44 multigene family, is also cotranscribed with the virB-virD4 cluster in A. phagocytophilum (43). The fact that wspB is cotranscribed with the virB8 to virD4 genes suggests its possible role in Wolbachia T4SS functioning. This role remains to be established, as wspB is interrupted by insertion sequence elements in some Wolbachia strains (49).
In wAtab3 and wRi, we showed that the clustered virB3, virB4, and virB6 genes were also cotranscribed as an operon, together with four downstream ORFs. This polycistronic transcription pattern is similar to that demonstrated previously for Wolbachia wMel (66). Among the four adjacent cotranscribed ORFs, three are annotated as genes for putative membrane-spanning proteins (WD0854 to WD0856) in the wMel genome whereas the products of the corresponding coding sequences in the wBm genome are classified as VirB6 paralogs (Wbm0793 to Wbm0795); thus, the proteins corresponding to these ORFs are possibly some of the T4SS components. The fourth ORF encodes a typical hypothetical protein.
The architectural organization into two separate operons of tandemly arranged virB and virD genes in Wolbachia is similar to that found in members of closely related bacterial genera of the order Rickettsiales, including Anaplasma spp., Ehrlichia spp., and Rickettsia spp. (2, 43). This pattern is in contrast to the single virB-virD locus found in other free-living or facultative intracellular bacteria, like A. tumefaciens, Mesorhizobium loti, Bordetella pertussis, and Bartonella spp. (51, 57, 61, 62). Surprisingly, orthologs of virB1, virB2, virB5, and virB7 seem to be absent in most members of the order Rickettsiales investigated so far. VirB2 and VirB5 are pilus components (36, 50), and VirB7 is a pilus-associated protein (48), whereas VirB1 is not an essential component of transport systems (7). Because Wolbachia and closely related genera are strict intracellular bacteria, they may be able to excrete molecules across their cell walls through the core T4SS membrane-spanning structure directly into the cytoplasm of the host cell, or the related pilus-encoding genes may be present in the genomes but have extremely low levels of similarity to the corresponding virB genes that prevent them from being detected by a simple sequence comparison.
An increasing amount of literature has shown the importance of T4SSs in intracellular bacteria for the successful infection of and survival, proliferation, and persistence within their hosts (10, 13, 52). For instance, the secretion of AnkA protein by A. phagocytophilum through a T4SS was found to facilitate infection (38). Therefore, the T4SS is expected to play a significant role in the wide range of phenotypes induced by Wolbachia strains in their invertebrate hosts. Of primary significance, in this study, genes encoding T4SS machinery were transcribed in insect tissues infected with Wolbachia strains wAtab3 and wRi, as well as in cultured cell lines harboring wAlbB and wMelPop. To further investigate the functionality of the Wolbachia T4SS, polyclonal antibodies raised against VirB6 were produced and tested in different tissues infected with Wolbachia. The A. tumefaciens VirB6 protein (343 amino acids [aa], corresponding to GenBank accession number AAK90538) is considerably smaller than the Wolbachia protein (854 aa). The level of similarity between the two proteins is relatively low (11%), and identical amino acids are found mainly in stretches of 1 to 2 aa scattered along the central part of the Wolbachia VirB6 protein. VirB6 is a polytopic integral membrane protein that forms complexes with the core structure of T4SS. Several studies have shown the crucial role of VirB6 in both the assembly and functioning of T4SS apparatus (30, 32, 33). An examination of ovaries recovered from A. tabida and D. simulans females hosting Wolbachia strains wAtab3 and wRi, respectively, showed the presence of VirB6 protein, as identified by immunofluorescence methods. Moreover, specific fluorescent VirB6 signals were observed in the cytoplasm of mosquito cell lines infected with Wolbachia strain wMelPop from the D. melanogaster w1118 line and wAlbB from Aedes albopictus. To our knowledge, this was the first study to combine both transcriptional and immunolocalization analyses to monitor the expression of vir loci of Wolbachia in cellulo and in vivo.
In this study, we detected the presence of the virB and virD genes in the genomes of two strains of Wolbachia, wRi and wAtab3, and determined their expression in host tissues. These two strains are involved in two different phenotypes; wRi induces cytoplasmic incompatibility in D. simulans, whereas wAtab3 is directly involved in the production of oocytes in A. tabida. Could the T4SS help to elucidate the answers to some fundamental questions about the interactions between Wolbachia strains and their hosts? What kinds of molecules are secreted across the bacterial membrane via the T4SS? Candidates are proteins with ankyrin domains that are encoded in Wolbachia genomes (23, 66). Are the excreted molecules involved in the interactions between Wolbachia strains and their hosts? Are they essential for the persistence of the bacteria in eukaryotic cells? Our investigations open new avenues for exploring T4SS function at the protein level in Wolbachia.
Supplementary Material
Acknowledgments
We thank F. Vavre and N. Krammer for insect-rearing facilities and encouraging suggestions, D. Charif for T4SS tree construction, J. C. Diaz and S. Belin for help in Western blotting and advice, B. Loppin and G. Orsi for access to a fluorescence microscope, and P. Potier and F. Wisniewski for helpful discussions.
This work was supported in part by grants ANR-06-BLAN-0316 and ANR-06-SEST-07. E.R. has a fellowship from the French Ministère de l'Education Nationale, de la Recherche et des Nouvelles Technologies.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, C. L., and T. L. Karr. 2001. Wolbachia: evolutionary novelty in rickettsial bacteria. BMC Evol. Biol. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Pontén, U. C. Alsmark, R. M. Podowski, A. K. Näslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396133-140. [DOI] [PubMed] [Google Scholar]
- 3.Baldo, L., J. C. Dunning Hotopp, K. A. Jolley, S. R. Bordenstein, S. A. Biber, R. R. Choudhury, C. Hayashi, M. C. Maiden, H. Tettelin, and J. H. Werren. 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 727098-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldo, L., and J. H. Werren. 2007. Revisiting Wolbachia supergroup typing based on WSP: spurious lineages and discordance with MLST. Curr. Microbiol. 5581-87. [DOI] [PubMed] [Google Scholar]
- 5.Bandi, C., J. W. McCall, C. Genchi, S. Corona, L. Venco, and L. Sacchi. 1999. Effects of tetracycline on the filarial worms Brugia pahangi and Dirofilaria immitis and their bacterial endosymbionts Wolbachia. Int. J. Parasitol. 29357-364. [DOI] [PubMed] [Google Scholar]
- 6.Bazzocchi, C., S. Comazzi, R. Santoni, C. Bandi, C. Genchi, and M. Mortarino. 2007. Wolbachia surface protein (WSP) inhibits apoptosis in human neutrophils. Parasite Immunol. 2973-79. [DOI] [PubMed] [Google Scholar]
- 7.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 1763646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouchon, D., T. Rigaud, and P. Juchault. 1998. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc. Biol. Sci. 2651081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brüggemann, H., C. Cazalet, and C. Buchrieser. 2005. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Cur. Opin. Microbiol. 91-9. [DOI] [PubMed] [Google Scholar]
- 10.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for DNA substrate. Science 3041170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 9314648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David, J., and M. F. Clavel. 1967. Influence of temperature during the course of development on various biometric characteristics of adult Drosophila melanogaster Meigen. J. Insect. Physiol. 13717-729. [DOI] [PubMed] [Google Scholar]
- 16.Dedeine, F., F. Vavre, D. D. Shoemaker, and M. Boulétreau. 2004. Intra-individual coexistence of a Wolbachia strain required for host oogenesis with two strains inducing cytoplasmic incompatibility in the wasp Asobara tabida. Evolution Int. J. Org. Evolution 582167-2174. [DOI] [PubMed] [Google Scholar]
- 17.Dedeine, F., M. Boulétreau, and F. Vavre. 2005. Wolbachia requirement for oogenesis: occurrence within the genus Asobara (Hymenoptera, Braconidae) and evidence for intraspecific variation in A. tabida. Heredity 95394-400. [DOI] [PubMed] [Google Scholar]
- 18.Dedeine, F., F. Vavre, F. Fleury, B. Loppin, M. E. Hochberg, and M. Boulétreau. 2001. Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc. Natl. Acad. Sci. USA 986247-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Cruz, F., and J. Davies. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8128-133. [DOI] [PubMed] [Google Scholar]
- 20.Ding, Z., and P. J. Christie. 2003. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185760-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 512145-2165. [DOI] [PubMed] [Google Scholar]
- 22.Duttan, C., and A. Pan. 2002. Horizontal gene transfer and bacterial diversity. J. Biosci. 2727-33. [DOI] [PubMed] [Google Scholar]
- 23.Foster, J., M. Ganatra, I. Kamal, J. Ware, K. Makarova, N. Ivanova, A. Bhattacharyya, V. Kapatral, S. Kumar, J. Posfai, T. Vincze, J. Ingram, L. Moran, A. Lapidus, M. Omelchenko, N. Kyrpides, E. Ghedin, S. Wang, E. Goltsman, V. Joukov, O. Ostrovskaya, K. Tsukerman, M. Mazur, D. Comb, E. Koonin, and B. Slatko. 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 30599-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microb. Mol. Biol. Rev. 67277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hapfelmeier, S., N. Domke, P. C. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 1824505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilgenboecker, K., P. Hammerstein, P. Schlattmann, A. Telschow, and J. H. Werren. 2008. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 281215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooykaas, P. J. J., and A. G. M. Beijersbergen. 1994. The virulence system of Agrobacterium tumefaciens. Annu. Rev. Phytopathol. 32157-179. [Google Scholar]
- 28.Hubber, A., A. C. Vergunst, J. T. Sullivan, P. J. Hooykaas, and C. W. Ronson. 2003. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 54561-574. [DOI] [PubMed] [Google Scholar]
- 29.Hurst, G. D., J. H. Graf von der Schulenburg, T. M. Majerus, D. Bertrand, I. A. Zakharov, J. Baungaard, W. Volkl, R. Stouthamer, and M. E. Majerus. 1999. Invasion of one insect species, Adalia bipunctata, by two different male-killing bacteria. Insect Mol. Biol. 8133-139. [DOI] [PubMed] [Google Scholar]
- 30.Jakubowski, S. J., V. Krishnamoorthy, and P. J. Christie. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 1852867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeyaprakash, A., and M. A. Hoy. 2000. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol. 9393-405. [DOI] [PubMed] [Google Scholar]
- 32.Judd, P. K., D. Mahli, and A. Das. 2005. Molecular characterization of the Agrobacterium tumefaciens DNA transfer protein VirB6. Microbiology 513483-3492. [DOI] [PubMed] [Google Scholar]
- 33.Judd, P. K., R. B. Kumar, and A. Das. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55115-124. [DOI] [PubMed] [Google Scholar]
- 34.Kyhse-Andersen, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10203-209. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lai, E. M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 1802711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laven, H. 1967. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature 216383-384. [DOI] [PubMed] [Google Scholar]
- 38.Lin, M., A. den Dulk-Ras, P. J. Hooykaas, and Y. Rikihisa. 2007. Anaplasma phagocytophilum AnkA secreted by the type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 92644-2657. [DOI] [PubMed] [Google Scholar]
- 39.Malek, J. A., J. M. Wierzbowski, W. Tao, S. A. Bosak, D. J. Saranga, L. Doucette-Stamm, D. R. Smith, P. J. McEwan, and K. J. McKernan. 2004. Protein interaction mapping on a functional shotgun sequence of Rickettsia sibirica. Nucleic Acids Res. 321059-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masui, S., T. Sasaki, and Y. Ishikawa. 2000. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J. Bacteriol. 1826529-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mavingui, P., V. Tran Van, E. Labeyrie, E. Rancès, F. Vavre, and P. Simonet. 2005. Efficient procedure of purification of obligate intracellular Wolbachia pipientis and representative amplification of its genome by multiple-displacement amplification. Appl. Environ. Microbiol. 716910-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCullen, C. A., and A. N. Binns. 2006. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 22101-127. [DOI] [PubMed] [Google Scholar]
- 43.Ohashi, N., N. Nzi, Q. Lin, and Y. Rikihisa. 2002. Characterization and transcriptional analysis of gene clusters for type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 702128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Neill, S. L., R. Giordano, A. M. E. Colbert, T. L. Karr, and H. M. Robertson. 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. USA 892699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Neill, S. L., and T. L. Karr. 1990. Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature 348178-180. [DOI] [PubMed] [Google Scholar]
- 46.Pannebakker, B. A., B. Loppin, C. P. Elemans, L. Humblot, and F. Vavre. 2007. Parasitic inhibition of cell death facilitates symbiosis. Proc. Natl. Acad. Sci. USA 104213-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paraskevopoulos, C., S. R. Bordenstein, J. Wernegreen, J. H. Werren, and K. Bourtzis. 2006. Toward a Wolbachia multilocus sequence typing system: discrimination of Wolbachia strains present in Drosophila species. Curr. Microbiol. 53388-395. [DOI] [PubMed] [Google Scholar]
- 48.Sagulenko, E., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 1835813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanogo, Y. O., S. L. Dobson, S. R. Bordenstein, and R. J. Novak. 2007. Disruption of the Wolbachia surface protein gene wspB by a transposable element in mosquitoes of the Culex pipiens complex (Diptera, Culicidae). Insect Mol. Biol. 16143-154. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. C. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 1817485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulein, R., and C. Dehlio. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 461053-1067. [DOI] [PubMed] [Google Scholar]
- 52.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3178-185. [DOI] [PubMed] [Google Scholar]
- 53.Stevens, L., R. Giordano, and R. F. Fialho. 2001. Male killing, nematode infections, bacteriophage infection, and virulence of cytoplasmic bacteria in the genus Wolbachia. Annu. Rev. Ecol. Syst. 32519-545. [Google Scholar]
- 54.Stouthamer, R., J. A. Breeuwer, R. F. Luck, and J. H. Werren. 1993. Molecular identification of microorganisms associated with parthenogenesis. Nature 36166-68. [DOI] [PubMed] [Google Scholar]
- 55.Stouthamer, R., J. A. J. Breeuwer, and G. D. D. Hurst. 1999. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 5371-102. [DOI] [PubMed] [Google Scholar]
- 56.Stouthamer, R., R. F. Luck, and W. D. Hamilton. 1990. Antibiotics cause parthenogenetic Trichogramma (Hymenoptera/Trichogrammatidae) to revert to sex. Proc. Natl. Acad. Sci. USA 872424-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. De Bruijn, and C. W. Ronson. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 1843086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, M. J., C. Bandi, and A. Hoerauf. 2005. Wolbachia bacterial endosymbionts of filarial nematodes. Adv. Parasitol. 60245-284. [DOI] [PubMed] [Google Scholar]
- 59.Tram, U., and W. Sullivan. 2002. Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 291124-1126. [DOI] [PubMed] [Google Scholar]
- 60.Vogel, J. P., and R. R. Isberg. 1999. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 230-34. [DOI] [PubMed] [Google Scholar]
- 61.Ward, J. E., D. E. Akiyoshi, D. Regier, A. Datta, M. P. Gordon, and E. W. Nester. 1988. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J. Biol. Chem. 2635804-5814. [PubMed] [Google Scholar]
- 62.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. USA 902970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Werren, J. H. 1997. Biology of Wolbachia. Annu. Rev. Entomol. 42587-609. [DOI] [PubMed] [Google Scholar]
- 64.Werren, J. H., and D. M. Windsor. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. Biol. Sci. 2671277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams, K. P., B. W. Sobral, and A. W. Dickerman. 2007. A robust species tree for the Alphaproteobacteria. J. Bacteriol. 1894578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 20327-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong, Q., S. H. Shao, L. L. Cui, R. H. Mu, X. L. Ju, and S. R. Dong. 2007. Type IV secretion system in Helicobacter pylori: a new insight into pathogenicity. Chin. Med. J. (England) 1202138-2142. [PubMed] [Google Scholar]
- 68.Zhou, W., F. Rousset, and S. O'Neill. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. Lond. B 265509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zupan, J. R., and P. Zambryski. 1995. Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol. 1071041-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.