Abstract
The virulence of the uropathogenic Escherichia coli Dr+ IH11128 strain is associated with the presence of Dr fimbrial structures and a DraD invasin which can act as a fimbrial capping domain at the bacterial cell surface. However, a recent study suggests that the DraD protein is surface exposed in two forms: fimbria associated and fimbria nonassociated (prone to interaction with the N-terminal extension of the DraE protein located on the fimbrial tip). The actual mechanism of DraD surface secretion is presently unknown. We identified a previously unrecognized type II secretory pathway (secreton) in the uropathogenic E. coli Dr+ strain which is well conserved among gram-negative bacteria and used mainly for secretion of virulence determinants. An active secreton is composed of 12 to 15 different proteins, among which GspD functions as an outer-membrane channel to permit extrusion of proteins in a folded state. Therefore, we inactivated the pathway by inserting the group II intron into a gspD gene of the type II secretion machinery by site-specific recombination. DraD secretion by the E. coli Dr+ and gspD mutant strains was determined by immunofluorescence microscopy (with antibodies raised against DraD) and an assay of cell binding between bacteria and HeLa cells. The specificity of DraD-mediated bacterial binding for the integrin receptor was confirmed by examination of the adhesion of DraD-coated beads to HeLa cells in the presence and absence of α5β1 monoclonal antibodies. The investigations that we performed showed that type II secretion in E. coli Dr+ strains leads to DraD translocation at the bacterial cell surfaces.
Gram-negative bacteria secrete a wide range of proteins necessary for the biogenesis of adhesive surface organelles termed pili and flagella, nutrient acquisition, expression of virulence factors, and efflux of drugs and other toxins. Export of these proteins at the bacterial cell surface requires a complex transport system that moves secreted proteins from the cytoplasm into the extracellular environment (17).
The export systems of gram-negative bacteria can be divided into two main groups: the Sec-dependent general secretory pathway (GSP) (including the type II secretion system [T2SS], autotransporter, type IV secretion system, and chaperone-usher secretion system) and the Sec-independent secretion system (including the type I secretion system and the type III secretion system) (30). Sec-dependent secretion is a mode of translocation in which the proteins are secreted in two steps. Firstly, proteins produced as precursors containing an N-terminal cleavable signal sequence are transported through the inner membrane via a proteinaceous complex of the Sec translocon (6). During export, the signal peptide is cleaved by the leader peptidase and the mature proteins are released into the periplasmic space, where they undergo further modifications and folding (disulfide bond formation and subunit assembly). Secondly, the folded proteins are translocated across the outer membrane (OM). Conversely, the Sec-independent pathway leads to protein translocation directly from the cytoplasm to the extracellular environment apart from the periplasmic space (3, 15).
The T2SS, the main terminal branch of GSP (7), found in a wide range of gram-negative species, is responsible for the extracellular transport of hydrolytic enzymes, toxins, and other proteins crucial in the pathogenesis of many microorganisms (27, 30). Secretion across the OM requires 12 to 16 different proteins forming a secretion complex located mainly in the inner membrane (7). The most important structural element of T2SS is a translocation channel in the OM created by only one integral OM protein, designated GspD (19, 32). GspD protein is a ring-shaped oligomer of 12 to 14 subunits forming a central pore in the OM. The common feature of many proteins translocated by T2SS is a structure composed of β-strands in a barrel-like arrangement (27).
Gram-negative bacteria use the chaperone-usher pathway (Sec-dependent system) to secrete proteins across their OM for the formation of more than 30 different complex structures associated with virulence, such as adhesive pili or fimbriae. All bacterial OM proteins with known structures span the membrane via a series of transmembrane antiparallel β-strands arranged to form a β-barrel (28).
Urinary tract infections are a serious health problem affecting millions of people each year. The most frequent etiologic agent of urogenital infections is uropathogenic Escherichia coli, accounting for 65 to 90% of cases (18). E. coli strains bearing the Dr family of adhesins account for 40% of pyelonephritis cases in the third trimester of pregnancy, 50% of chronic diarrhea cases in children, and 20% of recurrent urinary tract infections in young women (8, 11, 13).
Genes important for the biogenesis of Dr fimbriae are encoded by a dra gene cluster (draA, draB, draC, draD, draP, and draE) (22). Unlike these genes, the structural adhesin-encoding gene, designated draE, is highly heterogeneous within the Dr family of adhesins. Dr fimbriae are homopolymeric structures built from repeating monomers of DraE fimbrial subunits (25). Assembly of DraE subunits into the fimbrial structure depends on two secretion components: the periplasmic chaperone DraB and the OM platform DraC. Fimbrial subunits cross the inner membrane in an unfolded form via the Sec GSP and then must form a stable interaction with the chaperone in the periplasm. DraB is a boomerang-shaped protein composed of two complete immunoglobulin domains. DraE subunits have incomplete immunoglobulin-like folds with the absence of the seventh (C-terminal) β-strand. The missing β-strand of DraE is donated by the G1 β-strand of the chaperone in a mechanism termed donor strand complementation. Then the chaperone-subunit complexes are targeted to the OM assembly platform DraC for fimbrial biogenesis. Interaction with the usher triggers an exchange of chaperone-subunit interactions for subunit-subunit interactions in a process termed donor strand exchange. Subsequently the chaperone-subunit complexes dissociate and the G1 β-strand of the chaperone is exchanged for the N-terminal extension of an incoming subunit into the growing fiber (25).
The final structure of DraD exhibits an immunoglobulin-like topology composed of antiparallel β-strands arranged to form a β-barrel. Its native sequence does not have the N-terminal donor strand, and therefore, DraD can be located only at the tip of the fiber (16).
The cooperation between DraE and DraD may be beneficial for E. coli invasion. The data obtained indicated that the export of DraD protein to the cell surface (in the draC mutants) does not involve the DraC OM usher protein. Additionally, the expression of DraE fimbrial protein at the cell surface does not require the expression of DraD protein, and conversely, the surface expression of DraD does not require DraE adhesin expression (35).
In this paper, we report the identification of a type II secretion pathway allowing the translocation of DraD subunits to the cell surface of uropathogenic E. coli Dr+ strains. Additionally, we show the receptor-specific adhesion of DraD invasin to HeLa cells expressing α5β1 integrin. We inactivated the secretion pathway with a “targetron” chromosomal insertion knockout constructed in the gspD gene (site-specific gene disruption by the group II intron) of a Dr− mutant of clinical E. coli isolate IH11128 (an insertional draC mutant) (12) and recombinant E. coli strains harboring the dra gene cluster and its mutants with a mutation in the draE, draD, and draC genes. DraD seems to be an ideal candidate for translocation via the T2SSs with regard to an N-terminal cleavable signal sequence in the protein precursor and an immunoglobulin-like topology of a mature protein, in which two β-sheets pack against each other. The absence of DraD surface secretion in the gspD mutants was confirmed by immunofluorescence microscopy (IFM) with purified rabbit anti-DraD (raised against DraD) antibodies and an assay of binding of E. coli strains containing the dra gene cluster to HeLa cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, enzymes, and reagents.
The overexpression was carried out in Escherichia coli BL21(λDE3) (Novagen, Nottingham, United Kingdom) (20) and the BL21(λDE3)/gspD mutant (GspD− mutant), with an inactivated gspD gene (this paper). The strains are λDE3 lysogens, which carry the gene for T7 RNA polymerase under lacUV5 promoter control.
An insertional draC mutant, E. coli DR14, of the clinical E. coli isolate IH11128 (isolated from a human with pyelonephritis) bearing Dr fimbriae was described previously (12) (Table 1).
TABLE 1.
E. coli strains and plasmids
| E. coli straina | Plasmid | Mutation | Relevant genotype
|
Source or reference | ||
|---|---|---|---|---|---|---|
| draC | draD | draE | ||||
| BL21(λDE3) | ompT lon dcm | − | − | − | 20 | |
| BL21(λDE3)/gspD | Intron GspD | − | − | − | This work | |
| pBJN406 | + | + | 21 | |||
| pBJN17 | Transposon DraE-TnPhoA | + | − | 22 | ||
| pBJN16 | Transposon DraD-TnPhoA | + | − | + | 22 | |
| pBJN417 | Transposon DraC-Tn5 | − | + | 22 | ||
| pCC90 | + | + | 5 | |||
| pCC90D54stop | Dr-D54stop | + | − | 5 | ||
| pCC90DraDmut | ΔdraDstop | + | − | + | 35 | |
| pCC90DraCmut | DraC-K15stop | − | + | 35 | ||
| pInvDsyg-C-His | − | − | 35 | |||
| pInvDsygstop | − | − | 35 | |||
| DR14 | Insertion DraC | − | + | 12 | ||
| DR14/gspD | Insertion DraC, intron GspD | − | + | This work | ||
Symbols separated by the backslash indicate gspD+/gspD E. coli strains. All E. coli GspD− mutant strains were complemented in trans with the gspD gene encoded by the pET30-gspD plasmid.
Bacterial cells were grown in Luria broth (LB) without glucose or on Luria agar plates (containing 1.5% agar) supplemented with the appropriate antibiotics (Sigma, St. Louis, MO).
Plasmid pBJN406, carrying the whole dra gene cluster, and its transposon mutants with a mutation in the draE gene (draE::TnPhoA) (pBJN17), draD gene (draD::TnPhoA) (pBJN16), or draC gene (draC::Tn5) (pBJN417) were described previously (21, 22) (Table 1).
Plasmid pCC90, corresponding to the dra operon with its promoter region and regulatory genes upstream of a draB gene deleted, and pCC90D54stop (the DraE-negative mutant), with a mutated draE gene, were provided by S. Moseley, University of Washington (Seattle) (5) (Table 1).
Plasmid pCC90DraDmut (the DraD-negative mutant), with a mutated draD gene, and pCC90DraCmut (the DraC-negative mutant), with a mutated draC gen,e were described previously (35) (Table 1).
Plasmid pET30-gspD encoding GspD protein located in the OM is from this paper.
Plasmids pInvDsyg-C-His and pInvDsygstop, encoding DraD invasin with its N-terminal signal sequence and a polyhistidine domain at the C terminus or without any fusion domain, respectively, were described previously (35).
pACD4K-C-loxP, an E. coli expression vector (with loxP sites flanking each end of the kanamycin open reading frame) was to be used in conjunction with the TargeTron GeneKnockout System (Sigma) (29). pACD4K-C is chloramphenicol resistant for general selection and propagation of the plasmid. The kanamycin marker located within the group II intron on pACD4K-C is interrupted by a td group I intron. When the group II intron is transcribed and spliced, the td group I intron is excised, activating the kanamycin resistance gene. After insertion of the group II intron into the host genome, kanamycin resistance can be used to select for mutants containing genes disrupted by the group II intron. Then the kanamycin marker can be removed via Cre-loxP-mediated recombination (1, 14).
The plasmid DNAs were isolated from E. coli cultures using the Mini-prep Plus kit (A&A Biotechnology, Gdynia, Poland). After enzymatic reactions, the DNAs were purified using the Clean-Up kit (A&A Biotechnology). Restriction enzymes were purchased from New England BioLabs (Frankfurt, Germany). The reagents for PCR were obtained from DNA-Gdańsk II s.c. (Poland), and other reagents were purchased from Sigma.
Cell lines.
HeLa cells were maintained in minimum essential medium supplemented with 10% (vol/vol) fetal bovine serum (Sigma) and penicillin-streptomycin solution (Sigma) in a 5% CO2 atmosphere at 37°C. The cell line was passaged using 0.25% (vol/vol) trypsin containing EDTA (Sigma).
Antisera.
Rabbit anti-Dr adhesin antibodies raised against purified native Dr fimbriae (24) and rabbit anti-DraD invasin antibodies raised against DraD (34) were described previously. Rabbit anti-bovine serum albumin (anti-BSA) was purchased from Sigma. Anti-rabbit immunoglobulin G (IgG) (whole-molecule) antibodies conjugated to horseradish peroxidase and tetramethylrhodamine isothiocyanate (TRITC) and anti-mouse IgG (whole-molecule) antibodies conjugated to fluorescein isothiocyanate (FITC) were purchased from Sigma. α5β1 mouse monoclonal antibodies were from Chemicon (Chemicon International Inc., Temecula, CA).
DNA analysis of gspD gene as an integral component of the T2SS.
The gspD region of the various E. coli strains was first amplified using 35 cycles of PCR (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min in a Biometra thermocycler) with primers for gspD1 (5′-ATTCGCCGTGCTCAGGTG-3′) and gspD2 (5′-GTCGGACGGATAAACACCATCAGG-3′). The 799-bp PCR products were purified from agarose gel bands by using the DNA Clean-Up kit and digested with Cfr10I restriction enzyme. Then they were sequenced using the ABI Prism 377 automatic sequencing system. Multiple sequence alignments of the gspD genes from various E. coli strains were generated using the GeneDoc and ClustalX computer programs.
TargeTron intron-mediated gspD gene disruption.
The gspD gene disruption was performed by insertion of the group II intron using the TargeTron GeneKnockout System according to the instructions of the manufacturer (Sigma) (29). The group II intron inserts via the activity of an RNA protein complex expressed from a single plasmid provided in the kit.
First, a computer algorithm was used to identify target sites in the gspD gene. Second, the computer algorithm output primer sequences designated IBS (5′-AAAAAAGCTTATAATTATCCTTAGGCGGCTAAAAAGTGCGCCCAGAT AGGGTG-3′), EBS2 (5′-TGAACGCAAGTTTCTAATTTCGATTCCGCCTCGATAGAGGAAAGTGTCT-3′), and EBS1d (5′-CAGATTGTACAAATGTGGTGATAACAGATAAGTCTAAAAACGTAACTTACCTTTCTTTGT-3′) to mutate the intron by PCR. Next, the mutated and digested PCR product was ligated into linearized pACD4K-C-loxP containing the remaining intron component and the T7 promoter to express the intron. In clinical E. coli strain DR14 the mutated intron was modified by insertion of the E. coli spc constitutive promoter amplified by PCR. For this purpose, the unique primers SPC (5′-AAAAAAGCTTCCGTTTATTTTTTCTACCCATATCCTTGAAGCGGTGTTA TAATGCCGCGCCCTCGATAAAAAGAGCTTATAATTATCCTTA-3′) (the underlined sequence contains the spc promoter sequence) and EBS1d were required. Then, the ligation mixture was transformed into E. coli BL21(λDE3) or E. coli DR14 followed by expression of a mutated intron. Finally, kanamycin-resistant colonies were screened for gene disruption by PCR. After successful selection the kanamycin resistance marker was removed via Cre-loxP-mediated recombination.
Complementation of the gspD mutant.
A 1.851-kb gspD gene was amplified by PCR from E. coli BL21(λDE3) genomic DNA using DNA Pfu Turbo polymerase and primers SPC-Sph (5′-ATAGCATGCCCGTTTATTTTTTCTACCCATATCCTTGAAGCGGTGTTATAATGCCGCGCCCTCGATACCAATAATTTTGTTTAACTTTAAAAGGAGACAGCTATGGGGCCGGGCGTACAGGGG-3′ [the underlined sequence contains the spc promoter]) and gspD-Hind (5′-TAAAGCTTTTAACGCGTTCTCCCGGCATTGAAGAACGCGCGAACTTCCG GCGGTAAGGCCTGGTTTTGCGC-3′). The PCR product, which contained the spc promoter and gspD gene sequence, was ligated into the SphI and HindIII sites of the kanamycin resistance pET30-b+ vector. The resulting construct, pET30-gspD, was transformed into E. coli BL21(λDE3)/gspD harboring the recombinant plasmids (encoding the dra gene cluster and its mutants) and E. coli DR14 with the gspD gene disruption by electroporation. The E. coli strains were cultured overnight in 5 ml of LB broth at 37°C in a rotary shaker (250 rpm). Then 250 ml of prewarmed LB medium (supplemented with the appropriate antibiotics) was inoculated with 4 ml of the overnight bacterial cultures and incubated at 37°C with vigorous agitation (300 rpm in a rotary shaker). To isolate proteins from culture supernatants, 6-h cultures were harvested by 10 min of centrifugation at 3,000 × g and 4°C and precipitated by addition of trichloracetic acid to a final concentration of 10% (wt/vol). After incubation at 4°C for 4 h, precipitated proteins were pelleted (3,000 × g at 4°C) and rinsed sequentially twice with an acetone-chloridic acid solution (200:1, vol/vol) and then with pure acetone. Pellets were suspended in Laemmli loading buffer so as to concentrate supernatant proteins 100 times. The periplasmic fractions were isolated by the osmotic shock procedure, as described by Jędrzejczak et al. (16). Then sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10 μl of each probe, 3 to 10 μg, was loaded per lane of the gel) and Western blotting of the analyzed probes were performed as described previously (35). The concentration of the proteins in the culture supernatants and periplasmic fractions was determined by densitometric analysis with an LMW Calibration Kit for sodium dodecyl sulfate electrophoresis (Amersham Biosciences) as a standard, using a VersaDoc system with Quantity One software (both from Bio-Rad).
IFM of surface-exposed DraE and DraD subunits.
Bacterial cultures grown on Luria agar plates at 37°C for 24 h were harvested and washed gently in phosphate-buffered saline (PBS). Bacterial suspensions (100 μl; 105 to 106 cells/ml) were incubated with 50 μl of a 1:500 dilution (anti-fimbria Dr or anti-DraD) in PBS of the primary antibodies at room temperature for 1 h. The reaction mixtures were then washed three times with PBS containing 10% (vol/vol) glycerol and incubated with 50 μl of a 1:50 dilution (anti-rabbit IgG FITC or TRITC conjugate according to DraE and DraD proteins, respectively) in PBS of secondary antibodies at room temperature for 1 h. The reaction mixtures were then washed again three times with PBS containing 10% (vol/vol) glycerol. Bacterial suspensions (10 μl) were loaded on glass slides and observed with an immunofluorescence microscope (Olympus BX-60).
Assay of cell binding between HeLa cells and E. coli strains expressing native Dr fimbriae and DraD invasin.
HeLa cells were split into six-well plates with glass coverslips and grown in the appropriate medium for 24 h. E. coli strains (grown overnight on LB medium) harboring the dra gene cluster and its mutants suspended in PBS (adjusted to a final optical density at 600 nm of 0.4, 5 × 104 CFU/ml) were added to culture plates containing HeLa cells grown on glass coverslips and incubated for 2 h at 37°C. The cells were washed three times with PBS, fixed with 70% methanol, and stained with 10% Giemsa stain. Finally, the glass coverslips were examined for bound bacteria by using an Olympus BX-60 immunofluorescence microscope (40 cells associated with bacteria were examined by light microscopy). The amount of bacteria added to HeLa cells minus the amount in the washing fractions represents the amount left on glass coverslips. The experiment was performed in duplicate.
Expression and purification of DraD with and without His tag fusion domain.
The constructs DraD-C-His6 and native DraD were expressed for periplasmic localization using pET-30 Ek/Lic expression vector in the E. coli strain BL21(λDE3) (Novagen) (20). DraD-C-His6 and native DraD were isolated from the periplasmic space by the osmotic shock procedure, as described by Jędrzejczak et al. (16). Then, DraD-C-His6 was loaded onto a nickel-nitrilotriacetic acid affinity resin (Novagen) and eluted with 200 mM imidazole, 0.3 M NaCl, 30 mM Tris, pH 7.6 (16). After overnight dialysis against PBS, DraD-C-His6 was loaded onto a Superdex 200 column (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom). Native DraD isolated from periplasm and dialyzed against PBS was purified by size-exclusion chromatography using a Superdex 200 column. Finally, the fractions containing DraD-C-His6 and native DraD were collected and concentrated on a Centricon YM-3 instrument (Amicon) to 2 mg/ml.
α5β1 receptor-specific cell adhesion assay.
Purified DraD-C-His6 and native DraD proteins (2 mg/ml) were applied as a coating by passive adsorption onto 1-μm polystyrene microspheres (Polysciences Inc., Warrington, PA) according to the manufacturer's instructions (Polysciences Inc.) (26). Subconfluent HeLa cell monolayers grown on glass coverslips were washed twice with PBS and subsequently incubated for 1 h with a polystyrene bead suspension diluted 1:1,000 in minimum essential medium with 10% fetal bovine serum (107 beads/ml). The cells were washed with PBS and fixed for 10 min with 4% formaldehyde. Extracellular beads were then labeled by incubation of cells for 1 h with rabbit anti-DraD antibodies (diluted 1:100 in PBS). Control beads were coated with BSA (Sigma) and incubated for 1 h with rabbit anti-BSA antibodies (diluted 1:100 in PBS). The particles were washed three times and then incubated with TRITC-labeled anti-rabbit antibodies (diluted 1:100 in PBS). For the antibody blockade adhesion assay, cell monolayers were incubated with α5β1 mouse monoclonal antibodies (diluted 1:100) for 1 h and then coated beads were added for an additional 1 h. The amount of coated beads added to HeLa cells minus the amount in the washing fractions represents the amount left on glass coverslips. Finally, the glass coverslips were examined using an Olympus BX-60 immunofluorescence microscope (counting was performed on 50 cells associated with beads). The experiments were performed in duplicate.
RESULTS
gspD gene sequence analysis and disruption.
A substantial element of the T2SS is a ring-shaped GspD oligomer composed of 12 to 14 subunits forming a central pore in the OM. In our work, the GspD protein became a molecular target for further studies concentrated mainly on the determination of a DraD alternative surface secretion system. So far, two forms of GspD proteins encoded by E. coli H10407 (GenBank accession no. AF426313) (31) and K-12 (GenBank accession no. NP_417784.4) (4) strains have been published.
Multiple sequence alignments of gspD genes derived from the prototypic human enterotoxigenic E. coli strain H10407 and E. coli K-12 were generated using the GeneDoc and ClustalX computer programs. E. coli K-12, the most widely used laboratory strain, does not secrete endogenous proteins under standard growth conditions (silencing of the gsp gene transcription by H-NS), although it contains gsp genes that are homologous to those encoding other secretons (4, 9, 33). The nucleotide sequences of the analyzed gspD genes were 50.7% identical. The GspD products encoded by two forms of the gspD genes showed 53.85% amino acid sequence similarity. The analyses performed revealed a highly conserved region of the gspD gene (used for further analyses) present in both of the E. coli strains studied.
The identification of the gspD gene of the putative T2SS in laboratory strain E. coli BL21(λDE3) and clinical E. coli strains IH11128 and DR14 was determined by PCR amplification and restriction analysis of the products obtained. The amplification was done with primers (designated gspD1 and gspD2) complementary to the highly conserved region of the gspD gene from E. coli H10407 and K-12 strains. Specific PCR products flanking a 799-bp fragment of the gspD gene were obtained (data not shown). The results achieved confirmed the existence of the putative T2SS in the analyzed bacterial strains.
For further studies, E. coli DR14 with an insertional draC mutation (12), which affected the translocation of DraE fimbrial subunits but did not disrupt export of DraD to the bacterial cell surface (35), and laboratory E. coli BL21(λDE3) transformed with pBJN406 (or pCC90) and its mutants with mutations within dra genes were used. Earlier investigations based on a recombinant E. coli K-12 strain harboring the pBJN406 plasmid showed invasiveness, a virulence mechanism leading to chronic pyelonephritis, similar to that of E. coli clinical strain IH11128 (13). Taking into consideration the results obtained, we can assume that the T2SS will also be required for the secretion of DraD invasin by the clinical E. coli strain.
The gspD gene disruption from E. coli DR14 and E. coli BL21(λDE3) was made by insertion of a group II intron. The knockout of the selected bacterial gene was based on the TargeTron GeneKnockout System (Sigma) (29). Recent advances in group II intron research enabled retargeting introns to insert efficiently into virtually all desired DNA fragments (10, 23, 36).
Immunofluorescence examination of DraE and DraD surface expression.
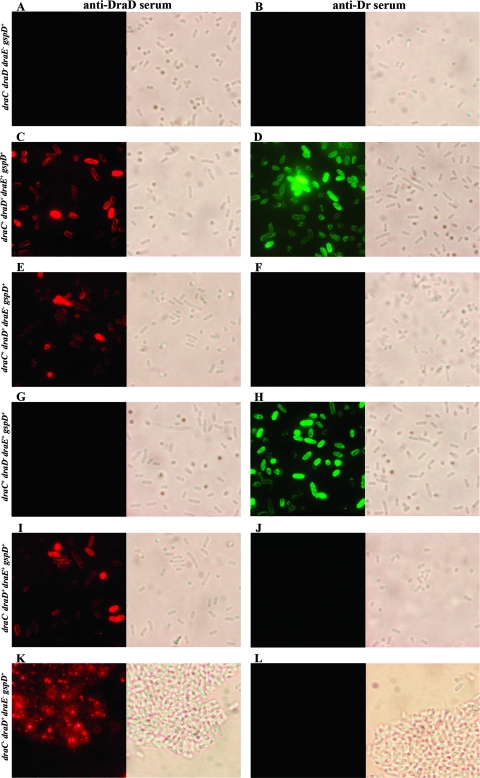
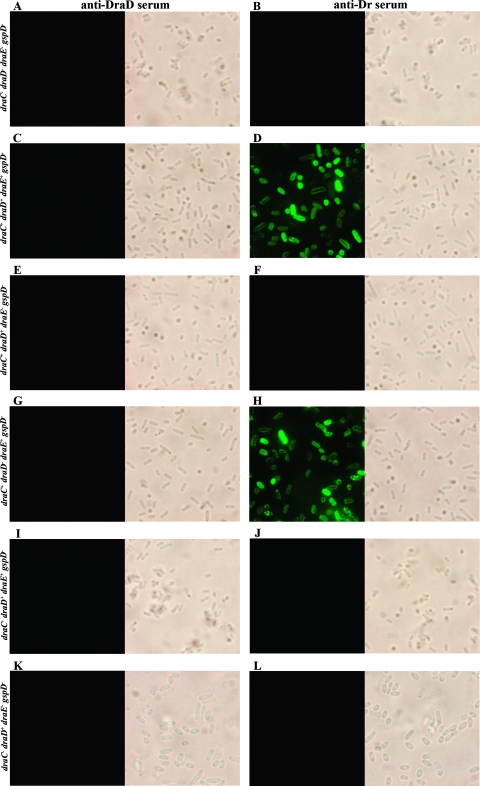
A summary of IFM results of DraE and DraD surface expression for E. coli strains, with (Fig. 1) and without (Fig. 2) the gspD gene knockout, harboring the recombinant plasmids is given in Table 2. E. coli BL21(λDE3) with the gspD gene disruption was used as a negative control for DraD surface secretion across the OM via T2SS (Fig. 2A). Therefore, immunofluorescence staining of the E. coli GspD− mutant strain transformed with pBJN406 encoding the entire dra gene cluster (Table 2) and pCC90 with the dra gene cluster without a regulatory region (Fig. 2C) and pCC90D54stop (Fig. 2E) and pBJN17 (Table 2) with inactivated draE or pCC90DraCmut (Fig. 2I) and pBJN417 (Table 2) with the inactivated draC gene did not show any DraD surface presentation. IFM of E. coli BL21(λDE3)-pInvDsyg-C-His (Table 2) and BL21(λDE3)-pInvDsygstop (Fig. 2K) with the gspD gene knockout (GspD−) also revealed the lack of DraD at the bacterial cell surface. The same results were obtained for the Dr− mutant, E. coli DR14, with an insertional inactivation of gspD (Fig. 3E). Conversely, IFM with anti-DraD and anti-rabbit TRITC-labeled antibodies detected the presence of DraD invasin on the cell surfaces of the clinical E. coli isolate IH11128 bearing Dr fimbriae (Fig. 3A) and its insertional draC mutant DR14 (Fig. 3C) and E. coli BL21(λDE3) GspD+ harboring all above-analyzed plasmids (Table 2). DraE surface exposition detected with anti-Dr and anti-rabbit FITC-labeled antibodies was independent of GspD (Fig. 2D and H). The results achieved corroborated the data obtained previously that showed that DraD expression at the bacterial cell surface is independent of DraE fimbrial subunit production and does not require contact with the OM DraC channel by which the polymerization process of Dr fimbriae occurs (35). To our knowledge, this is the first report which identified a secretion system of the DraD protein on the bacterial surface by the GspD OM channel, the main component of the type II secretion pathway.
FIG. 1.
Surface display of DraD and DraE examined by IFM of E. coli strains with a functional gspD gene (GspD+ strains). Bacteria were incubated with primary antiserum (anti-DraD [A, C, E, G, I, and K] or anti-Dr [B, D, F, H, J, and L]), washed, incubated with secondary antiserum (conjugated with TRITC [A, C, E, G, I, and K] or FITC [B, D, F, H, J, and L]), washed, and subjected to microscopy. (A and B) E. coli BL21(λDE3); (C and D) E. coli BL21(λDE3)-pCC90; (E and F) E. coli BL21(λDE3)-pCC90D54stop; (G and H) E. coli BL21(λDE3)-pCC90DraDmut; (I and J) E. coli BL21(λDE3)-pCC90DraCmut; (K and L) E. coli BL21(λDE3)-pInvDsygstop. Magnification, ×10,000 (Olympus BX-60 microscope).
FIG. 2.
Surface display of DraD and DraE examined by IFM of E. coli strains with a disrupted gspD gene (GspD− strains). Bacteria were incubated with primary antiserum (anti-DraD [A, C, E, G, I, and K] or anti-Dr [B, D, F, H, J, and L]), washed, incubated with secondary antiserum (conjugated with TRITC [A, C, E, G, I, and K] or FITC [B, D, F, H, J, and L]), washed, and subjected to microscopy. (A and B) E. coli BL21(λDE3)/gspD; (C and D) E. coli BL21(λDE3)/gspD-pCC90; (E and F) E. coli BL21(λDE3)/gspD-pCC90D54stop; (G and H) IFM of E. coli BL21(λDE3)/gspD-pCC90DraDmut; (I and J) E. coli BL21(λDE3)/gspD-pCC90DraCmut; (K and L) E. coli BL21(λDE3)/gspD-pInvDsygstop. Magnification, ×10,000 (Olympus BX-60 microscope).
TABLE 2.
Surface expression of a DraD invasin and a DraE adhesin in the tested recombinant E. coli strains with and without the gspD gene knockout
| E. coli straina | Relevant genotype
|
Surface staining of:
|
|||
|---|---|---|---|---|---|
| draC | draD | draE | DraD invasin | DraE adhesin | |
| BL21(λDE3)/gspD | − | − | − | −/− | −/− |
| BL21(λDE3)-pBJN406/gspD | + | + | + | +/− | +/+ |
| BL21(λDE3)-pBJN17/gspD | + | + | − | +/− | −/− |
| BL21(λDE3)-pBJN16/gspD | + | − | + | −/− | +/+ |
| BL21(λDE3)-pBJN417/gspD | − | + | + | +/− | −/− |
| BL21(λDE3)-pCC90/gspD | + | + | + | +/− | +/+ |
| BL21(λDE3)-pCC90D54stop/gspD | + | + | − | +/− | −/− |
| BL21(λDE3)-pCC90DraDmut/gspD | + | − | + | −/− | +/+ |
| BL21(λDE3)-pCC90DraCmut/gspD | − | + | + | +/− | −/− |
| pInvDsyg-C-His | − | + | − | +/− | −/− |
| pInvDsygstop | − | + | − | +/− | −/− |
| DR14/gspD | − | + | + | +/− | −/− |
Symbols separated by the backslash indicate gspD+/gspD E. coli strains. All E. coli GspD− mutant strains were complemented in trans with the gspD gene encoded by the pET30-gspD plasmid.
FIG. 3.
Surface display of DraD and DraE examined by IFM of clinical E. coli isolates. Bacteria were incubated with primary antiserum (anti-DraD [A, C, and E] or anti-Dr [B, D, and F]), washed, incubated with secondary antiserum (conjugated with TRITC [A, C, and E] or FITC [B, D, and F]), washed, and subjected to microscopy. (A and B) E. coli IH11128; (C and D) E. coli DR14; (E and F) E. coli DR14 with a gspD gene knockout (GspD− strain). Magnification, ×10,000 (Olympus BX-60 microscope).
Binding patterns of E. coli Dr+ strains and α5β1 receptor-specific cell adhesion of DraD invasin.
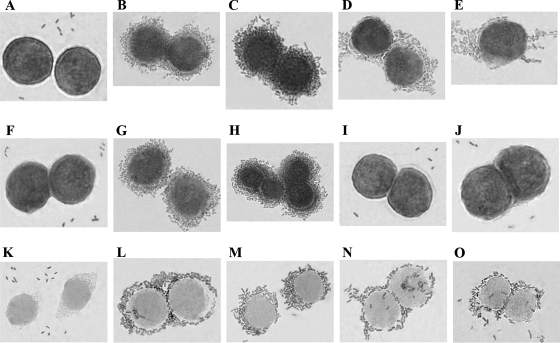
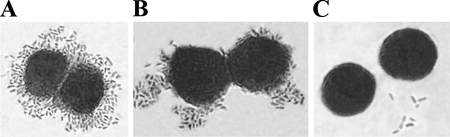
A summary of binding phenotypes of the analyzed recombinant E. coli strains with and without the gspD gene disruption is presented in Table 3. To examine the adherence patterns of the E. coli strains (with and without the gspD gene disruption) harboring the dra gene cluster and its mutants to decay-accelerating factor (DAF) and α5β1 integrin receptors (according to DraE and DraD proteins, respectively), a HeLa cell line was used. E. coli strains (grown overnight on LB medium) were suspended in PBS (adjusted to a final optical density at 600 nm of 0.4) and added to culture plates containing HeLa cells grown on glass coverslips. Bound bacterial cells were stained with Giemsa stain. Binding of DraD invasin to HeLa cells was not observed in the E. coli BL21(λDE3) strain with the disrupted gspD gene (GspD−) and transformed with pCC90 (Fig. 4G) or pBJN406 plasmids (Table 3). The same results were obtained for E. coli BL21(λDE3) GspD− mutant strains harboring pCC90D54stop (Fig. 4I) and pBJN17 (Table 3) or pCC90DraCmut (Fig. 4J) and pBJN417 (Table 3) plasmids with the inactivated draE and draC gene, respectively. As a positive control for DraD binding to HeLa cells, the clinical E. coli isolates (IH11128 and DR14 with and without Dr fimbria bioassembly, respectively) (Fig. 5A and B) and E. coli BL21(λDE3)-pCC90 (Fig. 4B) (average number of associated bacteria was 200 ± 4) and BL21(λDE3)-pBJN406 (Table 3), BL21(λDE3)-pCC90D54stop (Fig. 4D) and BL21(λDE3)-pBJN17 (with the functional OM DraC channel) (Table 3) and BL21(λDE3)-pCC90DraCmut (Fig. 4E) and BL21(λDE3)-pBJN417 (with the inactivated chaperone-usher secretion pathway) (Table 3) strains with no disrupted gspD gene were used. Binding of Dr fimbriae to DAF was independent of the GspD OM channel. Additionally, the binding patterns of DraE-negative mutants, E. coli BL21(λDE3)-pCC90D54stop (Fig. 4D) (average number of associated bacteria was 94 ± 6) and BL21(λDE3)-pCC90DraCmut (Fig. 4E) (average number of associated bacteria was 90 ± 4), were different from that of a DraD-negative mutant, E. coli BL21(λDE3)-pCC90DraDmut (Fig. 4C) (average number of associated bacteria was 150 ± 5). The surface expression of DraD protein without Dr fimbriae induced aggregation of bacterial cells often adjacent in certain parts of HeLa cells (aggregative adhesion) (Fig. 4D and E and 5B). The presence of Dr fimbriae with or without DraD as a capping fimbrial domain was related to strict adhesion of bacteria surrounding the whole surface of HeLa cells (Fig. 4B and C and 5A). The results obtained were in agreement with electron microscopic studies of polystyrene beads coated with periplasmic DraD-C-His6 which also revealed aggregation in a concentration-dependent manner. DraD-coated beads entered HeLa cells in the form of conglomerates containing three to five beads (35). To confirm the binding specificities of DraD for α5β1 integrin receptor, we tested the ability of native DraD and DraD-C-His6 to promote association with HeLa cells in the presence and absence of monoclonal α5β1 integrin antibodies. Polystyrene beads were coupled with DraD and DraD-C-His6 by passive adsorption. DraD-coated beads adhered to the HeLa cells only in the absence of α5β1 monoclonal antibodies (about 24 ± 5 beads were bound). The same results were obtained for native DraD (Fig. 6D) as well as for DraD-C-His6 with the polyhistidine domain which did not influence the interactions of DraD with α5β1 cellular receptor (data not shown). An antibody blockade involving anti-α5β1 integrin abolished association of both forms of DraD with the HeLa cells (Fig. 6B). The examinations performed allowed corroboration of specificity of DraD-mediated bacterial binding for the α5β1 integrin receptor. These investigations also indicated that the gspD gene disruption affects the interactions between the DraD invasin and α5β1 integrin, which was connected with the lack of DraD at the bacterial cell surfaces. This research confirmed the role of the GspD OM channel (the crucial component of the T2SS) in DraD surface translocation across the OM also.
TABLE 3.
Binding phenotype of the analyzed recombinant E. coli strains with and without the gspD gene disruption
| E. coli straina | Relevant genotype
|
Binding phenotype
|
|||
|---|---|---|---|---|---|
| draC | draD | draE | DraD invasinb,c | DraE adhesinc | |
| BL21(λDE3)/gspD | − | − | − | −/− | −/− |
| BL21(λDE3)-pBJN406/gspD | + | + | + | SA+/− | SA+/+SA |
| BL21(λDE3)-pBJN17/gspD | + | + | − | AA+/− | −/− |
| BL21(λDE3)-pBJN16/gspD | + | − | + | −/− | SA+/+SA |
| BL21(λDE3)-pBJN417/gspD | − | + | + | AA+/− | −/− |
| BL21(λDE3)-pCC90/gspD | + | + | + | SA+/− | SA+/+SA |
| BL21(λDE3)-pCC90D54stop/gspD | + | + | − | AA+/− | −/− |
| BL21(λDE3)-pCC90DraDmut/gspD | + | − | + | −/− | SA+/+SA |
| BL21(λDE3)-pCC90DraCmut/gspD | − | + | + | AA+/− | −/− |
| DR14/gspD | − | + | + | AA+/− | −/− |
Symbols separated by the backslash indicate gspD+/gspD E. coli strains. All E. coli GspD− mutant strains were complemented in trans with the gspD gene encoded by the pET30-gspD plasmid.
AA, aggregative adhesion of DraD.
SA, strict adhesion of Dr fimbrial structures (with or without DraD tip subunit) surrounding the HeLa cells.
FIG. 4.
DAF and α5β1 integrin binding properties of DraE and DraD examined by light microscopy of E. coli strains stained with Giemsa stain. Adherence of E. coli strains to HeLa cells. (A to E) Microscopic examination of E. coli BL21(λDE3) (negative control of cellular adhesion) (A), E. coli BL21(λDE3)-pCC90 (positive control of cellular adhesion) (B), E. coli BL21(λDE3)-pCC90DraDmut (C), E. coli BL21(λDE3)-pCC90D54stop (D), and E. coli BL21(λDE3)-pCC90DraCmut (E). (F to J) Micrographs of E. coli BL21(λDE3) with the gspD gene knockout (GspD− strain) transformed with the same plasmids as shown in panels A, B, C, D, and E, respectively. (K to O) Micrographs of E. coli BL21(λDE3) GspD− mutant strain complemented with plasmid-encoded GspD and harboring the same plasmids as shown in panels A, B, C, D, and E, respectively. Forty cells associated with bacteria were examined by light microscopy. Magnification, ×10,000 (Olympus BX-60 microscope).
FIG. 5.
DAF and α5β1 integrin binding properties of DraE and DraD examined by light microscopy of clinical E. coli isolates stained with Giemsa stain. Adherence of E. coli strains to HeLa cells. HeLa cells were incubated with E. coli strains, fixed, stained with Giemsa stain, and subjected to microscopy. ME of E. coli IH11128 (positive control of DraE and DraD adhesion) (A), E. coli DR14 (positive control of DraD adhesion) (B), and E. coli DR14 with a gspD gene knockout (GspD− strain) (negative control for DraE and DraD adhesion) (C). Forty cells associated with bacteria were analyzed by light microscopy. Magnification, ×10,000 (Olympus BX-60 microscope).
FIG. 6.
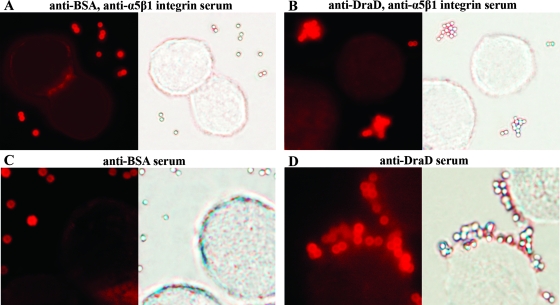
Specificity of DraD-mediated binding for the α5β1 integrin receptor examined by IFM. Visualization of interactions between DraD-coated beads and HeLa cells in the presence or absence of the monoclonal α5β1 integrin antibodies. HeLa cells were incubated with polystyrene beads coated with BSA (as a negative control) or the native DraD, fixed, and subjected to microscopy. Extracellular polystyrene beads were labeled with rabbit anti-BSA or anti-DraD and visualized by the red fluorescence of TRITC-labeled anti-rabbit antibodies. IFM of polystyrene beads coated with BSA (negative control) (A and C) and DraD (B and D) incubated with HeLa cells in the presence (A and B) or absence (C and D) of monoclonal α5β1 integrin antibodies, respectively. Counting was performed on 50 cells associated with beads. The beads associated with other cells were too aggregated; thus, counting was not possible. Magnification, ×10,000 (Olympus BX-60 microscope).
Complementation experiments.
The DraD surface localization defect in the GspD mutant strains was complemented with plasmid pET30-gspD-encoded GspD. The expression of gspD restored detection of surface-exposed DraD, which was shown by IFM with anti-DraD serum. The E. coli BL21(λDE3)/gspD-pET30-gspD strain was used as a negative control for DraD surface secretion across the OM via T2SS (Fig. 7A). Immunofluorescence staining of the E. coli GspD− mutant strain complemented in trans with the gspD gene and transformed with pCC90 (Fig. 7B) and pCC90D54stop (Fig. 7C) or pCC90DraCmut (Fig. 7E) showed DraD surface presentation. After complementation, the gspD mutant of E. coli DR14 also retained the ability to expose DraD at the bacterial cell surface (Fig. 7F). Additionally, HeLa cell monolayers were coinfected with E. coli BL21(λDE3)/gspD strains harboring the recombinant plasmids pET30-gspD and pCC90 (Fig. 4L) harboring the dra gene cluster and its mutants in draD (pCC90DraDmut) (Fig. 4M), draE (pCC90D54stop) (Fig. 4N), or draC (pCC90DraCmut) (Fig. 4O), respectively. Complementation of GspD− mutant cells with a wild-type copy of gspD in trans on plasmid pET30-gspD also restored adherence of the DraD surface-exposed protein, similar to E. coli strains with the functional type II secretory pathway. To further verify the results obtained, a subcellular fraction experiment was performed. The complementation with GspD OM protein shifted the location of DraD from periplasmic space to the cell surface exterior, which was confirmed by Western blotting with anti-DraD serum (Fig. 8B).
FIG. 7.
Surface display of DraD examined by IFM of E. coli gspD mutants (GspD− strains) complemented with plasmid-encoded GspD. Bacteria were incubated with primary antiserum (anti-DraD), washed, incubated with secondary antiserum (conjugated with TRITC), washed, and subjected to microscopy. (A) E. coli BL21(λDE3)/gspD-pET30-gspD; (B) E. coli BL21(λDE3)/gspD-(pET30-gspD, pCC90); (C) E. coli BL21(λDE3)/gspD-(pET30-gspD, pCC90D54stop); (D) E. coli BL21(λDE3)/gspD-(pET30-gspD, pCC90DraDmut); (E) IFM of E. coli BL21(λDE3)/gspD-(pET30-gspD, pCC90DraCmut); (F) E. coli DR14/gspD-pET30-gspD. Magnification, ×10,000 (Olympus BX-60 microscope).
FIG. 8.
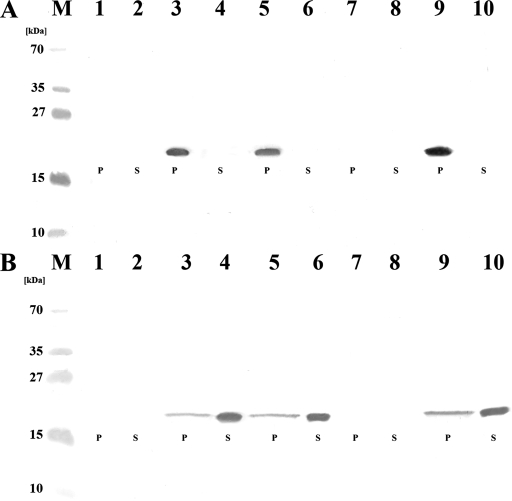
Western blot analysis of E. coli periplasmic fractions and broth cultures with antisera against DraD. Visualization of DraD expressed by E. coli strains with a disrupted (A) and complemented (B) gspD gene with a rabbit anti-DraD serum. All preparations were isolated from the same amount of bacteria (optical density at 600 nm, 0.5). A rabbit anti-DraD serum was used as the primary serum, and a peroxidase-coupled anti-rabbit serum was employed as the secondary serum. (A) Lane M, PageRuler Prestained Protein Ladder Plus (Fermentas) for 250, 130, 100, 70, 55, 35, 27, 15, and 10 kDa; lanes 1 and 2, periplasmic (P) and supernatant (S) fractions of E. coli BL21(λDE3)/gspD; lanes 3 and 4, periplasmic (P) and supernatant (S) fractions of E. coli BL21(λDE3)/gspD-pCC90; lanes 5 and 6, periplasmic (P) and supernatant (S) fractions of E. coli BL21(λDE3)/gspD-pCC90D54stop; lanes 7 and 8, periplasmic (P) and supernatant (S) fractions of E. coli BL21(λDE3)/gspD-pCC90DraDmut; lanes 9 and 10, periplasmic (P) and supernatant (S) fractions of E. coli BL21(λDE3)/gspD-pCC90DraCmut. (B) Lane M, PageRuler Prestained Protein Ladder (Fermentas) for 250, 130, 100, 70, 55, 35, 27, 15, and 10 kDa; lanes 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 contain periplasmic (P) and supernatant (S) fractions of E. coli BL21(λDE3)/gspD (lanes 1 and 2) complemented with pET30-gspD and harboring pCC90 (lanes 3 and 4), pCC90D54stop (lanes 5 and 6), pCC90DraDmut (lanes 7 and 8), or pCC90DraCmut (lanes 9 and 10) plasmids.
DISCUSSION
DraD protein encoded by a dra gene cluster of E. coli Dr+ strains may be expressed in two forms, fimbria associated and fimbria nonassociated (prone to interaction with the N-terminal extension of the DraE protein located on the fimbrial tip) (35). The ability of the surface-exposed form of DraD to interact with a donor strand of the top DraE fimbrial subunit is indirectly confirmed by the structural studies. The acceptor cleft of the DraD is relatively firm, and as a result of complementation by a donor strand its conformation is practically not changed (the cleft remains open to solvent). Such structure of acceptor cleft in DraD is probably able to reverse interaction with a donor peptide based on stiff complementation. Certainly the above-mentioned model must be confirmed.
Until now, very little was known about a translocation system of the DraD invasin. In this study, we demonstrated for the first time an identification of the T2SS in E. coli Dr+ strains (22) which leads to DraD secretion at the bacterial cell surface. Inactivation of this pathway by mutagenesis of the gspD gene completely impeded DraD secretion.
The widespread and conserved T2SS among gram-negative pathogens is used to export the largest number of virulence determinants. The secretion pathway requires the functions of 12 to 15 proteins that are believed to form a complex that is able to recognize fully folded proteins and translocate them across the OM. Among the proteins of an active secreton, GspD is presumed to form a gated OM channel (27). The DraD synthesized as a protein precursor with an N-terminal cleavable signal sequence and showing the final structure as a modified immunoglobulin fold composed of antiparallel β-strands (16) appears to be ideal for secretion via the type II secretion pathway.
In clinical E. coli Dr+ isolates, the DraE protein forms extended surface-exposed fimbrial structures. These structures are composed of DraE, which acts as both an adhesin (the main virulence determinant in E. coli Dr+ strains) and a structural fimbrial subunit, and DraD capping the Dr fiber (2). The interaction of the DraB chaperone-DraE adhesin with the DraC usher protein (a highly conserved chaperone-usher pathway) is absolutely required for initiation of Dr fimbria biogenesis. The growing fiber moves through a pore in the OM formed by the DraC and adopts its final quaternary structure (25). The latest research revealed that the assembly of Dr fimbriae did not require DraD. In the appropriate DraE and DraD mutants, the expression of one protein at the bacterial cell surface did not require the production of the other. Additionally, the lack of DraD immunoidentification in the isolated Dr fimbrial fractions suggested that a majority of Dr fimbrial structures were made up of only the DraE subunits, and most of the DraD may form surface-located independent structures of Dr fimbriae (35).
Available data also showed that the export of DraD at the surface of bacteria in E. coli DraC insertional mutants [E. coli DR14 and E. coli BL21(λDE3)-pCC90DraCmut] does not require a functional DraC OM channel (35). Therefore, we inactivated the T2SS by chromosomal insertion knockout in the gspD gene by site-specific insertion of the group II intron of E. coli clinical strain DR14 (12) and recombinant E. coli strains harboring the dra gene cluster and mutations in the draE, draD, and draC genes. We predict that the same events described here would occur in the E. coli clinical isolate IH11128. DraD secretion was restored when the gspD mutant was complemented in trans with a copy of the wild-type gspD gene. Giemsa staining of HeLa cells associated with bacteria confirmed that DraD was exposed at the bacterial cell surface. However, DraD was also able to detach from the bacterial surface to the culture supernatant, which was determined by Western blot analysis with anti-DraD serum.
Immunofluorescence examinations with anti-DraD and anti-rabbit TRITC-labeled antibodies in E. coli strains with the gspD gene disruption (GspD− mutants) transformed with pCC90, pCC90D54stop (DraE mutant), pCC90DraCmut (DraC mutant), pInvDsyg-C-His, or pInvDsygstop did not detect the presence of DraD at the bacterial cell surface. For comparison, parallel experiments were conducted with the transposon-insertion mutations inactivating the draE and draC genes. We also obtained the same results for a Dr− mutant, E. coli DR14, with insertional inactivation of gspD. Conversely, immunofluorescence labeling of the E. coli clinical strains IH11128 and DR14 and E. coli BL21(λDE3) (without gspD gene knockout) harboring all above-mentioned plasmids showed DraD surface expression. The gspD gene disruption also did not influence the Dr fimbria biogenesis via the chaperone-usher pathway. The surface secretion defect of DraD in E. coli GspD− mutant strains was restored by expression of a plasmid encoding GspD. Taken together, these data indicate that the GspD OM channel, the main component of the type II secretion pathway, is essential for DraD surface translocation.
For researching the specificity of binding of E. coli strains (with or without the gspD gene inactivation) containing the dra gene cluster and its negative mutants to DAF (for DraE) and α5β1 integrin (for DraD) receptors, a HeLa cell line was used. The bacterial cells adhering to cellular receptors of HeLa cells were visualized by Giemsa staining. The binding specificity of DraD for α5β1 integrin receptor was also confirmed by incubation of DraD-coated beads with HeLa cells in the presence and absence of α5β1 mouse monoclonal antibodies. Binding of Dr fimbriae to DAF was independent of the GspD OM channel. The adherence of DraD to HeLa cells was not identified in E. coli GspD− mutant strains harboring pCC90, pCC90D54stop, pCC90DraCmut, or pCC90DraDmut (a negative control for DraD binding). The same results were observed for DraE, DraC, and DraD transposon mutants. We could distinguish adhesion mediated by DraD from the Dr fimbrial structures (alone or with DraD as a tip subunit) because the binding patterns of DraE-negative mutants [E. coli BL21(λDE3)-pCC90D54stop and BL21(λDE3)-pCC90DraCmut] were different from that of a DraD-negative mutant [E. coli BL21(λDE3)-pCC90DraDmut]. The lack of Dr fimbriae at the surface of E. coli strains induced aggregation of bacterial cells expressing only the DraD protein. Using this mechanism, the bacterial cells most likely tried to compensate for the absence of the main virulence factor, Dr fimbriae, and to intensify the invasion process.
For the first time, our results demonstrated a mechanism for DraD secretion by E. coli strains containing the dra gene cluster. The gspD gene inactivation entirely abolished DraD export at the cell surface. Our findings are very important for resolving a molecular basis of pathogenesis of infections caused by E. coli Dr+ strains. The confirmed independent DraE and DraD surface expression can influence the intracellular survival of bacterial cells, which will be the next step to explore in our studies. Additionally the ability of DraD to be released from the fimbrial structures may be a molecular basis for establishing chronic and recurrent urinary tract infections.
Acknowledgments
This work was supported by the Polish State Committee for Scientific Research, grant N401 156 32/3040 to B.Z.-P.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Abremski, K., B. Frommer, and R. H. Hoess. 1986. Linking-number changes in the DNA substrate during Cre-mediated loxP site-specific recombination. J. Mol. Biol. 19217-26. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, K. L., J. Billington, D. Pettigrew, E. Cota, P. Simpson, P. Roversi, H. A. Chen, P. Urvil, L. du Merle, P. N. Barlow, M. E. Medof, R. A. Smith, B. Nowicki, C. Le Bougenec, S. M. Lea, and S. Matthews. 2004. An atomic resolution model for assembly, architecture, and function of the Dr adhesins. Mol. Cell 15647-657. [DOI] [PubMed] [Google Scholar]
- 3.Binet, R., S. Letoffe, J. M. Ghigo, P. Delepelaire, and C. Wandersman. 1997. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene 1927-11. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Carnoy, C., and S. L. Moseley. 1997. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol. Microbiol. 23365-379. [DOI] [PubMed] [Google Scholar]
- 6.de Keyzer, J., C. van der Does, and A. J. Driessen. 2003. The bacterial translocase: a dynamic protein channel complex. Cell. Mol. Life Sci. 602034-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filloux, A. 2004. The underlying mechanism of type II protein secretion. Biochim. Biophys. Acta 1694163-179. [DOI] [PubMed] [Google Scholar]
- 8.Foxman, B., L. X. Zhang, P. Tallman, K. Palin, C. Rode, C. Bloch, B. Gillespie, and C. F. Marrs. 1995. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of 2nd infection. J. Infect. Dis. 1721536-1541. [DOI] [PubMed] [Google Scholar]
- 9.Francetic, O., D. Belin, C. Badut, and A. P. Pugsley. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 196697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazier, C. L., J. San Filippo, A. M. Lambowitz, and D. A. Mills. 2003. Genetic manipulation of Lactococcus lactis by using targeted group II introns: generation of stable insertions without selection. Appl. Environ. Microbiol. 691121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Germani, Y., E. Begaud, P. Duval, and C. Le Bouguenec. 1996. Prevalence of enteropathogenic, enteroaggregative, and diffusely adherent Escherichia coli among isolates from children with diarrhea in New Caledonia. J. Infect. Dis. 1741124-1126. [DOI] [PubMed] [Google Scholar]
- 12.Goluszko, P., S. L. Moseley, L. D. Truong, A. Kaul, J. R. Williford, R. Selvarangan, S. Nowicki, and B. Nowicki. 1997. Development of experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H-bearing Dr fimbriae. J. Clin. Investig. 991662-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goluszko, P., V. Popov, R. Selvarangan, S. Nowicki, T. Pham, and B. J. Nowicki. 1997. Dr fimbriae operon of uropathogenic Escherichia coli mediate microtubule-dependent invasion to the HeLa epithelial cell line. J. Infect. Dis. 176158-167. [DOI] [PubMed] [Google Scholar]
- 14.Hartung, M., and B. Kisters-Woike. 1998. Cre mutants with altered DNA binding properties. J. Biol. Chem. 27322884-22891. [DOI] [PubMed] [Google Scholar]
- 15.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jędrzejczak, R., Z. Dauter, M. Dauter, R. Piątek, B. Zalewska, M. Mróz, K. Bury, B. J. Nowicki, and J. Kur. 2006. Structure of DraD invasin from uropathogenic Escherichia coli: a dimer with swapped beta-tails. Acta Crystallogr. D Biol. Crystallogr. 62157-164. [DOI] [PubMed] [Google Scholar]
- 17.Kostakioti, M., C. L. Newman, D. G. Thanassi, and C. Stathopoulos. 2005. Mechanisms of protein export across the bacterial outer membrane. J. Bacteriol. 1874306-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millar, L. K., and S. M. Cox. 1997. Urinary tract infections complicating pregnancy. Infect. Dis. Clin. N. Am. 1113-26. [DOI] [PubMed] [Google Scholar]
- 19.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 968173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novagen. 2002-2003. Competent cells overview. Strain descriptions. Novagen, Nottingham, United Kingdom.
- 21.Nowicki, B., J. P. Barrish, T. Korhonen, R. A. Hull, and S. I. Hull. 1987. Molecular cloning of the Escherichia coli O75X adhesin. Infect. Immun. 553168-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowicki, B., C. Svanborg-Eden, R. Hull, and S. Hull. 1989. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect. Immun. 57446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perutka, J., W. Wang, D. Goerlitz, and A. M. Lambowitz. 2004. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J. Mol. Biol. 336421-439. [DOI] [PubMed] [Google Scholar]
- 24.Pham, T. Q., P. Goluszko, V. Popov, S. Nowicki, and B. Nowicki. 1997. Molecular cloning and characterization of Dr-II, a nonfimbrial adhesin-I-like adhesin isolated from gestational pyelonephritis-associated Escherichia coli that binds to decay-accelerating factor. Infect. Immun. 654309-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piątek, R., B. Zalewska, O. Kolaj, M. Ferens, B. Nowicki, and J. Kur. 2005. Molecular aspects of biogenesis of Escherichia coli Dr fimbriae: characterization of DraB-DraE complexes. Infect. Immun. 73135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polysciences Inc. 2006-2007. Protocol for adsorbing proteins on polystyrene microparticles: guide and convenience. Polysciences Inc., Warrington, PA.
- 27.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40271-283. [DOI] [PubMed] [Google Scholar]
- 28.Saulino, E. T., E. Bullitt, and S. J. Hultgren. 2000. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc. Natl. Acad. Sci. USA 979240-9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigma. 2005. TargeTron GeneKnockout System: user guide. Sigma, St. Louis, MO.
- 30.Stathopoulos, C., D. R. Hendrixon, D. G. Thanassi, S. J. Hultgren, J. W. St. Geme III, and R. Curtiss III. 2000. Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 21061-1072. [DOI] [PubMed] [Google Scholar]
- 31.Tauschek, M., R. J. Gorrell, R. A. Strugnell, and R. M. Robins-Browne. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA 997066-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thanassi, D. G. 2002. Ushers and secretins: channels for the secretion of folded proteins across the bacterial outer membrane. J. Mol. Microbiol. Biotechnol. 411-20. [PubMed] [Google Scholar]
- 33.Whitchurch, C. B., and J. S. Mattick. 1994. Escherichia coli contains a set of genes homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene 1509-15. [DOI] [PubMed] [Google Scholar]
- 34.Zalewska, B., R. Piątek, H. Cieśliński, B. J. Nowicki, and J. Kur. 2001. Cloning, expression, and purification of the uropathogenic Escherichia coli invasin DraD. Protein Expr. Purif. 23476-482. [DOI] [PubMed] [Google Scholar]
- 35.Zalewska, B., R. Piątek, K. Bury, A. Samet, B. J. Nowicki, S. Nowicki, and J. Kur. 2005. A surface-exposed DraD protein of uropathogenic Escherichia coli bearing Dr fimbriae may be expressed and secreted independently from DraC usher and DraE adhesin. Microbiology 1512477-2486. [DOI] [PubMed] [Google Scholar]
- 36.Zhong, J., M. Karberg, and A. M. Lambowitz. 2003. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 311656-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]