Abstract
Us3 is a serine/threonine protein kinase encoded by herpes simplex virus 1 (HSV-1). Here, we report the identification of a physiological Us3 phosphorylation site on serine at position 147 (Ser-147) which regulates its protein kinase activity in vitro. Moreover, mutation of this site influences Us3 function, including correct localization of the enzyme and induction of the usual morphological changes in HSV-1-infected cells. These conclusions are based on the following observations: (i) in in vitro kinase assays, a domain of Us3 containing Ser-147 was specifically phosphorylated by Us3 and protein kinase A, while a mutant domain in which Ser-147 was replaced with alanine was not; (ii) in vitro, alanine replacement of Ser-147 (S147A) in Us3 resulted in significant impairment of the kinase activity of the purified molecule expressed in a baculovirus system; (iii) phosphorylation of Ser-147 in Us3 tagged with the monomeric fluorescent protein (FP) VenusA206K (VenusA206K-Us3) from Vero cells infected with a recombinant HSV-1 encoding VenusA206K-Us3 was specifically detected using an antibody that recognizes phosphorylated serine or threonine residues with arginine at the −3 and −2 positions; and (iv) the S147A mutation influenced some but not all Us3 functions, including the ability of the protein to localize itself properly and to induce wild-type cytopathic effects in infected cells. Our results suggest that some of the regulatory activities of Us3 in infected cells are controlled by phosphorylation at Ser-147.
The herpes simplex virus 1 (HSV-1) Us3 gene encodes a serine/threonine protein kinase with an amino acid sequence conserved in the subfamily Alphaherpesvirinae (12, 31, 48). In vitro biochemical studies characterized the consensus target sequence of an HSV-1 Us3 homologue encoded by pseudorabies virus (PRV) as RnX(S/T)YY, where n is greater than or equal to 2; X can be Arg, Ala, Val, Pro, or Ser; and Y can be any amino acid except an acidic residue (26, 27, 47). The target site specificity of HSV-1 and HSV-2 Us3 and varicella-zoster virus open reading frame 66 (ORF66) has been reported to be broadly similar to that of the PRV homologue (7, 9, 47). Based on studies showing that recombinant Us3 mutant viruses have impaired growth properties in cell cultures and virulence in mouse models (32, 52), it is concluded that HSV-1 Us3 is a positive regulator of viral replication and viral pathogenicity. At present the mechanisms by which this viral protein kinase acts in viral replication and pathogenicity remain largely undetermined, but several lines of evidence suggest possible functions of Us3, as discussed below.
(i) The Us3 protein kinase blocks apoptosis induced by replication-incompetent viruses, osmotic shock, or overexpression of proapoptotic cellular proteins (15, 18, 28, 35, 36, 42). Although the critical Us3 substrate(s) which mediates the antiapoptotic activity of the viral protein kinase has not yet been identified, evidence suggesting mechanisms by which Us3 prevent apoptosis is gradually accumulating. Recently, it has been reported that Us3 activates protein kinase A (PKA), a cellular cyclic AMP-dependent protein kinase with phosphorylation target sequences resembling those of Us3. It has also been reported that both Us3 and PKA phosphorylate the same target protein residues (1), suggesting that the former mediates antiapoptotic activity through phosphorylation of PKA substrates, by activating PKA, and/or by mimicking this cellular protein kinase. In addition, cellular apoptosis-regulating proteins, including Bad, Bid, and procaspase 3, have been implicated as being phosphorylated by Us3 (2-4, 17, 36). However, the biological significance of Us3-mediated phosphorylation of these cellular proteins in infected cells remains to be elucidated.
(ii) The Us3 protein kinase plays a role in nuclear egress of progeny nucleocapsids. In cells infected with Us3 mutant viruses, virions aggregate aberrantly within the perinuclear space in large invaginations (52). Us3 phosphorylates UL31 and UL34, both of which are crucial regulators of primary envelopment of nucleocapsids at the nuclear membrane; the catalytic activity of Us3 protein kinase is required for proper localization of UL31 and UL34 at the nuclear membrane (17, 50, 51, 54, 56). Furthermore, Us3 phosphorylates and alters the localization of lamin A/C, a fibrous meshwork lining the nucleoplasmic face of the inner nuclear membrane (34). Us3 has also been implicated in phosphorylating the inner nuclear membrane protein Emerin, which interacts with a number of nuclear components, including lamins and the DNA-binding protein BAF, and is suggested to be involved in maintaining nuclear integrity (25, 33). These observations suggest that Us3 regulates nuclear egress of nucleocapsids by mediating phosphorylation of these viral and cellular proteins. It has been reported that lack of phosphorylation of UL34 is not likely to be responsible for the aberrant nuclear membrane morphology observed in cells infected with Us3 mutant viruses, whereas the role of phosphorylation of other proteins, including UL31, lamin A/C, and Emerin, has not yet been elucidated.
(iii) The HSV-1 Us3 homologues encoded by PRV and Marek's disease virus mediate actin stress fiber breakdown in infected cells (57, 63). The cytoskeletal rearrangement mediated by the PRV Us3 homologue has been implicated as being associated with the changed morphology of infected cells and intracellular viral spread (11). Consistent with this, it has been reported that the cytopathic effects (CPEs) on cells infected with Us3 mutants of HSV-1 and HSV-2 are different from those with wild-type viruses (36, 38, 48). In addition, overexpression of HSV-2 Us3 in the absence of other viral proteins has been shown to induce actin stress fiber breakdown and to affect the cdc42/Rac pathway, which controls actin cytoskeletal organization (37). Although it remains uncertain whether HSV Us3 is in fact involved in rearrangement of actin stress fibers in infected cells as observed in PRV and Marek's disease virus infection, these observations indicate that Us3 influences the morphology of infected cells, probably by modifying the cytoskeleton.
(iv) Other than the Us3 substrates described above, ICP22, Us9, UL12, UL46, cytokeratin 17, histone deacetylase 1 (HDAC1), and HDAC2 (6, 8, 17, 30, 38, 45, 49) have been reported to be putative substrates for Us3. Although the biological significance of the phosphorylation of the putative substrates by Us3 during the viral life cycle remains to be elucidated, these observations imply an additional novel function(s) of Us3.
While the functional manifestations of Us3 have been gradually unveiled as described above, little is known regarding the mechanisms by which the protein kinase activity of Us3 protein is regulated. In cells, the activity of cellular protein kinases is tightly regulated; they are turned on or off by phosphorylation, binding of regulatory subunits, the presence of small molecules, or by virtue of their subcellular localization (55). Therefore, knowledge of the mechanisms by which protein kinases are activated or inhibited is also crucial for understanding the overall features of the enzymes as well as the functional consequences of their manipulation. In the studies presented here, we focus on phosphorylation of Us3 and its effect on regulatory activities of this protein kinase. It has been reported that Us3 is autophosphorylated in vitro (17, 47), as well as by another HSV-1-encoded protein kinase, UL13, in infected cells (18). However, a direct link between Us3 autophosphorylation and phosphorylation by UL13, and the regulatory activity of Us3, has not been demonstrated. Here, we report the identification of a physiological Us3 phosphorylation site at Ser-147, which regulates its protein kinase activity in vitro, as well as certain of its functions in infected cells.
MATERIALS AND METHODS
Cells and viruses.
Vero, rabbit skin, and Spodoptera frugiperda Sf9 cells were described previously (21, 59), as was HSV-1 wild-type strain HSV-1(F) (10, 59). Human embryonic lung fibroblast (HEL) cells were kindly provided by Shinya Watanabe and were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. A Us3 deletion mutant virus, R7041, and a recombinant virus, R7306, in which the Us3 gene has been repaired (48) were kindly provided by B. Roizman. The recombinant virus YK304 was reconstituted from pYEbac102, containing a complete HSV-1(F) sequence with the bacterial artificial chromosome (BAC) sequence inserted into the HSV intergenic region between UL3 and UL4 (59). YK304 has been shown to have a phenotype identical to that of wild-type HSV-1(F) in cell cultures and in mouse models (59).
Plasmids.
(i) A plasmid, pGEX-Us3-P1, for generating a fusion protein of glutathione S-transferase (GST) and a part of Us3, was constructed by amplifying the domains carrying Us3 codons 98 to 364 by PCR from pBS-Us3 (17) and cloning the DNA fragments into pGEX4T-1 (GE Healthcare) in frame with GST. pBS-Us3S147A, in which the serine codon at position 147 (Ser-147) of Us3 was replaced with an alanine, was generated according to the manufacturer's instructions using the QuikChange site-directed mutagenesis XL kit with complementary oligonucleotides containing the specific nucleotide substitution (Stratagene). (ii) Plasmids pMAL-Us3-P4 and pMAL-Us3-P4-S147A, for generating a fusion protein of maltose-binding protein (MBP) and a part of Us3, were constructed by amplifying the domains carrying Us3 codons 1 to 172 by PCR from pBS-Us3 or pBS-Us3S147A, respectively, and cloning the DNA fragments into pMAL-c (New England BioLabs) in frame with MBP. (iii) The transfer plasmid pAcGHLT-Us3S147A, for generating a recombinant baculovirus expressing GST fused to a mutated version of Us3 in which Ser-147 was replaced with an alanine (Us3S147A), was constructed by cloning the EcoRI-NotI fragment of pBS-Us3S147A into the EcoRI and NotI sites of pAcGHLT-A (Pharmingen) in frame with GST. (iv) The transfer plasmids pBS-VenusA206K-Us3 and pBS-VenusA206K-Us3S147A for generating a recombinant HSV-1 expressing a fusion protein of the Venus FP and Us3 or Us3S147A protein, respectively, were constructed as follows. Plasmid pBS-Venus was constructed by amplifying the Venus ORF (39) without the stop codon from Venus/pCS2 (a generous gift from A. Miyawaki) and cloning it into the EcoRI and SpeI sites of pBluescript KS(+) (Stratagene). Using pBS-Venus, pBS-VenusA206K, in which alanine at Venus residue 206 was replaced with lysine, was generated using the QuikChange site-directed mutagenesis XL kit. Because green FP (GFP) and its variants form dimers at high concentrations, the A206K mutation was used to prevent dimerization without significant alteration of FP spectral properties (66). The 1.5-kb sequence upstream of the HSV-1 Us3 start codon was amplified by PCR from pBC1013 (a generous gift from B. Roizman) (22) using the primers 5′-GCAAGCTTAGGAGGGCTCCCAGCCCTGG-3′ and 5′-GCGAATTCTCGCCGCACCGTGAGTGCCA-3′ and cloned into the pBS-VenusA206K EcoRI and HindIII sites to produce pBS-VenusA206K-1.5kb. The entire Us3, or the Us3S147A ORF without the Us3 start codon, was amplified by PCR from pBS-Us3 or pBS-Us3S147A and cloned into the pBS-VenusA206K-1.5kb SpeI and NotI sites in frame with VenusA206K to produce pBS-VenusA206K-Us3 and pBS-VenusA206K-Us3S147A, respectively. The resultant plasmid consisted of VenusA206K-Us3 or VenusA206K-Us3S147A bounded by Us3 flanking sequences.
Mutagenesis of viral genomes in E. coli and generation of recombinant HSV-1.
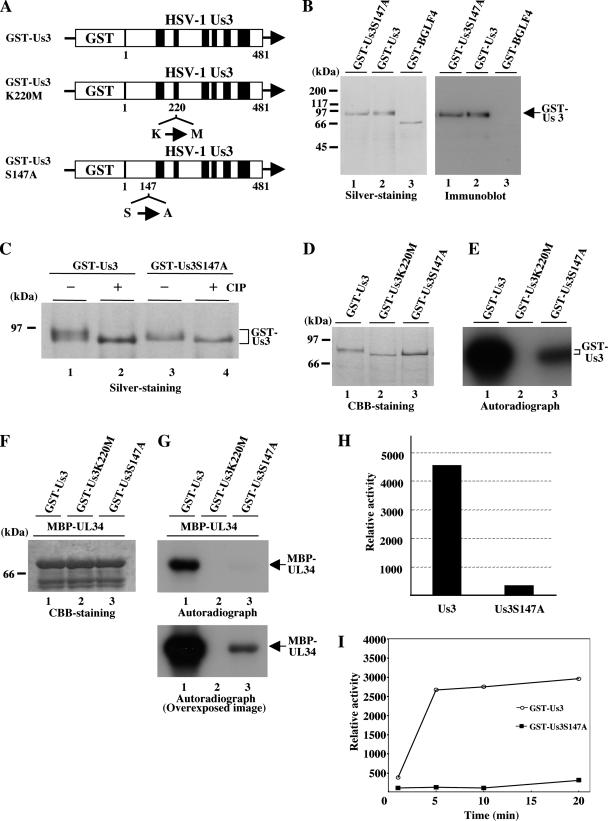
To generate a Us3 deletion mutant virus (YK502) in which a gene encoding kanamycin resistance was substituted for the domain of Us3 carrying codons 1 to 220 (Fig. 1), the one-step mutagenesis method known as ET cloning was performed as described previously (60, 67). Briefly, linear fragments containing a gene encoding kanamycin resistance and 50 bp of flanking Us3 sequences on each side generated by PCR using the primers 5′-CGGGGCCCGTCGTTCGGGGTGCTCGTTGGTTGGCACTCACGGTGCGGCGAGACAGCAAGCGAACCGGAAT-3′ and 5′-AGTCGCCTCAGCAGTCGCGCCTCGTGGCTCGTGCTCGTGTACCACCCCGCCGGAAATGTTGAATACTCATACTCTTCCTTTTTC-3′ were electroporated into YEbac202, an E. coli DH10B strain containing an HSV-1 (F)-BAC plasmid (pYEbac102) (59) and pGETrec, encoding recombinases E and T (a generous gift from P. A. Ioannou) (40). These primers were designed to replace codons 1 to 220 of Us3 with a kanamycin resistance gene. Kanamycin-resistant colonies were then screened by PCR with appropriate primers, which led to the identification of E. coli YEbac502 harboring the mutant HSV-BAC plasmid pYEbac502. The Us3 deletion mutant virus YK502 was generated by transfection of pYEbac502 into rabbit skin cells as described previously (59) and verified by Southern blotting.
FIG. 1.
Schematic diagram of genome structures of wild-type YK304 and relevant domains of the recombinant viruses. Line 1, a linear representation of the YK304 genome. The YK304 genome has bacmid (BAC) in the intergenic region between UL3 and UL4. Line 2, the HindIII(N) fragment in the genome domain carrying the Us2, Us3, Us4, and Us5 ORFs. Line 3, predicted amino acid sequence of Us3. Lines 4 to 7, 8, 9, 11, 13, and 15, schematic diagrams of the recombinant viruses YK502 to YK505, YK510, YK511, YK513, YK515, and YK517, respectively. Lines 10, 12, and 14, schematic diagrams of plasmids pYEbac512, pYEbac514, and pYEbac516, respectively.
To construct the recombinant virus YK503 encoding VenusA206K fused to a Us3 mutant protein in which alanine was substituted for Ser-147 (VenusA206K-Us3S147A) (Fig. 1), rabbit skin cells were cotransfected with the transfer plasmid pBS-VenusA206K-Us3S147A and intact YK502 viral DNA, using the calcium phosphate precipitation technique, as described previously (22). Viral DNAs were extracted from infected cells and purified on 5 to 20% potassium acetate gradients as described previously (22). At 3 days posttransfection, the transfected cells were harvested and subjected to freeze-thawing and sonication. The cell lysates were diluted and inoculated onto Vero cells, and plaques were screened for fluorescence with an inverted fluorescence microscope (Olympus IX71). Recombinants were plaque purified three times on Vero cells and verified by Southern blotting.
The recombinant virus YK505, in which the alanine replacement of Ser-147 in Us3 of YK503 had been repaired (VenusA206K-Us3-repair) (Fig. 1), was generated as follows. First, recombinant virus YK504, in which a gene encoding kanamycin resistance was substituted for VenusA206K and the domain of YK503 Us3 carrying codons 1 to 220 was constructed. To this end, circular viral DNA of YK503 isolated from infected Vero cells by the Hirt method was electroporated into E. coli DH10B (Invitrogen), and E. coli YEbac503 harboring the YK503 genome was isolated as described previously (59). ET cloning was performed in E. coli YEbac503. The Us3 deletion mutant virus YK504 (Fig. 1) was reconstituted in the same way to generate YK502 as described above. Second, rabbit skin cells were cotransfected with the transfer plasmid pBS-VenusA206K-Us3 and intact YK504 viral DNA, and the recombinant virus YK505 was generated in the same way to produce YK503, as described above.
To generate a recombinant virus (YK511) carrying a methionine replacing the lysine codon at position 220 (Lys-220) (Fig. 1), the two-step Red-mediated mutagenesis procedure (61) was performed. Briefly, E. coli GS1783 (14) containing pYEbac102 (a kind gift from G. A. Smith and N. Osterrieder) was grown in LB medium containing 20 μg/ml of chloramphenicol at 32°C to an optical density at 600 nm of 0.5 to 0.7. The E. coli culture was then transferred into a 42°C water bath and left for 15 min to induce the expression of the Red recombination system. Finally, bacteria were cooled on ice for 20 min and harvested, and electrocompetent cells were prepared as described elsewhere (59). Linear fragments containing a gene encoding the I-SceI site, kanamycin resistance, and 60 bp of Us3 sequences carrying the methionine substitution of Lys-220 were generated by PCR using the primers 5′-CAGCAGCCATCCAGATTACCCCCAACGGGTAATCGTGATGGCGGGGTGGTACACGAGCACAGGATGACGACGATAAGTAGGG-3′ and 5′-GCAGTCGCGCCTCGTGGCTCGTGCTCGTGTACCACCCCGCCATCACGATTACCCGTTGGGCAACCAATTAACCAATTCTGATTAG-3′ with pEPkan-S (61) (a kind gift from N. Osterrieder) as the template. The primers were designed to insert the I-SceI site, kanamycin resistance, and 60 bp of Us3 sequences carrying the methionine substitution of Lys-220 into the Lys-220 locus of the Us3 gene. The linear PCR-generated fragments were electroporated into the electrocompetent cells. These bacteria were then incubated at 32°C for 1.5 h and plated on LB agar plates containing 20 μg/ml of chloramphenicol and 40 μg/ml of kanamycin to select E. coli clones harboring the kanamycin resistance gene inserted into the Us3 locus. Kanamycin-resistant colonies were screened by PCR with appropriate primers, which led to the identification of YEbac510, a E. coli GS1783 strain harboring the mutant HSV-BAC plasmid pYEbac510. A Us3 deletion mutant virus (YK510) was generated by transfection of pYEbac510 into rabbit skin cells as described previously and verified by Southern blotting. Next, the kanamycin resistance gene cassette was excised by expressing the I-SceI restriction enzyme in YEbac510 through induction with arabinose, followed by induction of the Red recombination machinery by raising the temperature. Briefly, 100 μl of an overnight culture of E. coli YEbac510 cells grown in LB medium containing chloramphenicol and kanamycin was inoculated into 2 ml of LB medium containing only chloramphenicol. Bacteria were incubated at 32°C for 2 to 4 h with shaking, followed by addition of 10% (wt/vol) l-arabinose (Wako) to the culture at a 1:5 ratio, and incubated for another 1 h at 32°C. Finally, E. coli cells were incubated at 42°C for 30 min. The culture was then shaken at 32°C for another 1 to 2 h, and 100 μl of 10−1 to 10−6 dilutions of the culture was plated onto LB agar plates containing only chloramphenicol. After 24 to 48 h, individual colonies were restreaked in duplicate on chloramphenicol- or chloramphenicol-kanamycin-containing plates and grown overnight at 32°C. Chloramphenicol-resistant and kanamycin-sensitive clones were further screened by PCR with the appropriate primers, followed by nucleotide sequencing for confirmation of the desired mutation. One of the colonies (YEbac511) harboring pYEbac511 was selected and used for construction of the Us3 kinase-negative mutant virus YK511.
To generate the recombinant virus YK513 in which the methionine replacement of Lys-220 in Us3 of YK510 had been repaired (Fig. 1), the same procedure as used to generate YK511 was performed except that E. coli YEbac511 and the primers 5′-CAGCAGCCATCCAGATTACCCCCAACGGGTAATCGTGAAGGCGGGGTGGTACACGAGCACAGGATGACGACGATAAGTAGGG-3′ and 5′-GCAGTCGCGCCTCGTGGCTCGTGCTCGTGTACCACCCCGCCTTCACGATTACCCGTTGGGCAACCAATTAACCAATTCTGATTAG-3′ were used.
A recombinant virus (YK515) carrying an alanine replacement of Ser-147 in Us3 (Fig. 1) was constructed by the same procedure as used to generate YK511 except that the primers 5′-CCCGTGTGGCGCATCTCCCCCCGGTATACGACGACGCGCCCGGGATGAGATTGGGGCCACAGGATGACGACGATAAGTAGGG-3′ and 5′-CTTCCGCGGTAAATCCCGTGGCCCCAATCTCATCCCGGGCGCGTCGTCGTATACCGGGGGCAACCAATTAACCAATTCTGATTAG-3′ were used. To generate the recombinant virus YK517, in which the alanine replacement of Ser-147 in Us3 of YK515 had been repaired (Fig. 1), the same procedure as used to generate YK511 was performed except that E. coli YEbac515, which was obtained in the procedure to generate YK515 and harbored pYEbac515 carrying an alanine replacement of Ser-147 in Us3, and the primers 5′-CCCGTGTGGCGCATCTCCCCCCGGTATACGACGACGCAGCCGGGATGAGATTGGGGCCACAGGATGACGACGATAAGTAGGG-3′ and 5′-CTTCCGCGGTAAATCCCGTGGCCCCAATCTCATCCCGGCTGCGTCGTCGTATACCGGGGGCAACCAATTAACCAATTCTGATTAG-3′ were used.
Production and purification of GST and MBP fusion proteins expressed in E. coli.
GST fusion protein (GST-Us3-P1) was expressed in E. coli transformed with pGEX-Us3-P1, purified as described previously (19), and used for generation of rabbit polyclonal antibody to Us3 as described below. MBP fusion proteins (MBP-Us3-P4, MBP-Us3-P4-S147A, or MBP-UL34) were expressed in E. coli transformed with pMAL-Us3-P4, pMAL-Us3-P4-S147A, or pMAL-UL34 (17), respectively, and purified as described previously (21).
Generation of recombinant baculovirus.
Recombinant baculoviruses Bac-GST-Us3, Bac-GST-Us3K220M, and Bac-GST-BGLF4 have been described previously (17, 21). To generate Bac-GST-Us3S147A, pAcGHLT-Us3S147A was cotransfected with linearized baculovirus DNA BaculoGold (Pharmingen) into Sf9 cells using Lipofectin (Invitrogen) as described previously (21). The recombinant baculoviruses were propagated in Sf9 cells.
Purification of GST fusion proteins from baculovirus-infected cells.
GST-Us3, GST-Us3K220M, GST-Us3S147A, and GST-BGLF4 proteins were purified from Sf9 cells infected with Bac-GST-Us3, Bac-GST-Us3K220M, Bac-GST-Us3S147A, and Bac-GST-BGLF4, respectively, as described previously (21).
Antibodies.
To generate polyclonal antibody to Us3, two rabbits were immunized with purified GST-Us3-P1, following a standard protocol at MBL (Nagoya, Japan). Chicken polyclonal antibody to UL34 was kindly provided by R. Roller (51). Rabbit polyclonal antibodies to UL31 (68) and VP22 (41) were described previously. Rabbit polyclonal antibody to HDAC2 and rat monoclonal antibody to GFP were purchased from Sigma and MBL, respectively. Rabbit phospho-PKA substrate (100G7) monoclonal antibody was purchased from Cell Signaling Technology.
In vitro kinase assays.
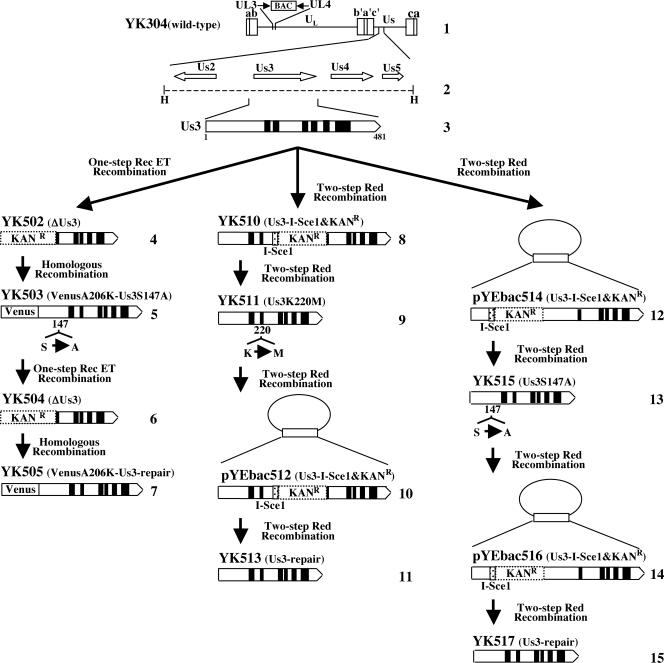
MBP fusion proteins were captured on amylose beads (New England BioLabs) and used as substrates in in vitro kinase assays with 0.34 μg (3.7 pmol) of purified GST-Us3 or GST-Us3K220M, as described previously (17). MBP fusion proteins were also used as substrates in in vitro kinase assays with 0.14 μg (3.7 pmol) (Fig. 2C and D) or 0.45 μg (11.9 pmol) (Fig. 2A, B, E, and F) of purified PKA, purchased from Invitrogen. In vitro kinase assays with PKA were performed as described previously (21) except that specific PKA reaction buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2) was used. The relative amount of radioactivity in substrates phosphorylated by GST-Us3 or GST-Us3S147A was quantified with the aid of Dolphin Doc and the software Dolphin-1D (Wealtec).
FIG. 2.
(A) Purified MBP-Us3-P4 (lane 1) and MBP-Us3-P4-S147A (lane 2) incubated in kinase buffer containing [γ-32P]ATP and purified PKA (lanes 1 and 2), separated on a denaturing gel, and stained with CBB. (B) Autoradiograph of the gel in panel A. (C) Purified MBP-Us3-P4 incubated in kinase buffer containing [γ-32P]ATP and purified PKA in the absence (lane 1) or in the presence (lane 2) of the PKA inhibitor (PKI), separated on a denaturing gel, and stained with CBB. (D) Autoradiograph of the gel in panel C. (E) Purified MBP-Us3-P4 incubated in kinase buffer containing [γ-32P]ATP and purified PKA and then either mock treated (lane 1) or treated with λ-PPase (lane 2), separated on a denaturing gel, and stained with CBB. (F) Autoradiograph of the gel in panel E. (G) Purified MBP-Us3-P4 incubated in kinase buffer containing [γ-32P]ATP and purified GST-Us3 (lanes 1 and 2) or GST-Us3K220M (lane 3) in the absence (lanes 1 and 3) or in the presence (lane 2) of the PKA inhibitor, separated on a denaturing gel, and stained with CBB. (H) Autoradiograph of the gel in panel G.
Immunoblotting and immunofluorescence.
Immunoblotting was performed as described previously (22) except that polyvinylidene difluoride membranes (Millipore) were used with the phospho-PKA substrate antibody. Indirect immunofluorescence assays were performed as described previously (16), except that anti-chicken immunoglobulin G (IgG) conjugated to Alexa Fluor 546 was used as the secondary antibody, and samples were examined with the Zeiss LSM5 laser scanning microscope.
Immunoprecipitation.
Vero cells were infected with either YK503 or YK505 at a multiplicity of infection (MOI) of 3 PFU. Infected cells were harvested at 18 h postinfection and lysed in NP-40 buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1.0% Nonidet P-40 [NP-40]) containing a protease inhibitor cocktail (Sigma). Supernatants obtained after centrifugation of the cell lysates were precleared by incubation with protein A-Sepharose beads (GE Healthcare) at 4°C for 30 min. After a brief centrifugation, supernatants were passed through 0.22-μm-pore-size filters and then reacted at 4°C for 1.5 h with 6 μl of agarose-conjugated anti-GFP (MBL), i.e., anti-GFP monoclonal antibody RQ2 coupled to agarose beads. To visualize the agarose bead pellet more easily, additional protein A-Sepharose beads were added to the supernatants. Immunoprecipitates were collected by a brief centrifugation, washed extensively with NP-40 buffer, and analyzed by immunoblotting with anti-phospho-PKA substrate (RRXS/T) antibody or anti-Us3 antibody.
Immune complex kinase assays.
Vero cells were infected with either YK304, YK511, or YK515 at an MOI of 3. Infected cells were harvested at 18 h postinfection and lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate) containing a protease inhibitor cocktail (Sigma). Supernatants obtained after centrifugation of the lysates were precleared by incubation with protein A-Sepharose beads at 4°C for 30 min and then reacted with the anti-Us3 antibody at 4°C for 1.5 h. Protein A-Sepharose beads were added to the supernatants and the reaction continued for another 2 h. Immunoprecipitates were collected by a brief centrifugation and washed twice with high-salt buffer (1 M NaCl, 10 mM Tris-HCl [pH 8.0], 0.2% NP-40), once with low-salt buffer (0.1 M NaCl, 10 mM Tris-HCl [pH 8.0], 0.2% NP-40), six times with radioimmunoprecipitation assay buffer, and finally four times with Us3 kinase buffer (50 mM Tris-HCl [pH 9.0], 20 mM MgCl2, 0.1% NP-40, and 1 mM dithiothreitol). Thereafter, they were divided into two aliquots. Immunoprecipitates in one aliquot were analyzed by in vitro kinase assays. For these assays, Us3 kinase buffer containing 10 μM ATP and 10 μCi [γ-32P]ATP was added to the mixture of protein A-Sepharose beads containing immunoprecipitated Us3 protein kinase and amylose beads containing purified MBP-UL34 and reacted at 30°C for 30 min. After incubation, the samples were washed twice with TNE buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, and 1 mM EDTA) and separated by electrophoresis in denaturing gels. After electrophoresis, the separated proteins were transferred from the gels to nitrocellulose membranes (Bio-Rad) and exposed to X-ray film. Immunoprecipitates in the other aliquot were analyzed by immunoblotting with anti-Us3 antibody.
Phosphatase treatment.
After in vitro kinase assays, the MBP fusion proteins or the immunoprecipitates were subjected to phosphatase treatment as described previously except that 200 U λ phosphatase (λ-PPase) (New England BioLabs) was used in these experiments (17). In other studies, the GST-Us3 fusion proteins were treated with 20 U alkaline phosphatase (CIP; New England BioLabs) as described previously (17).
Chemical treatments.
The PKA inhibitor 6-22 amide was purchased from Calbiochem and was used at a final concentration of 52 nM.
Purification of virions.
Virions were purified as described previously (60).
RESULTS
Identification of the in vitro autophosphorylation site of Us3 as Ser-147.
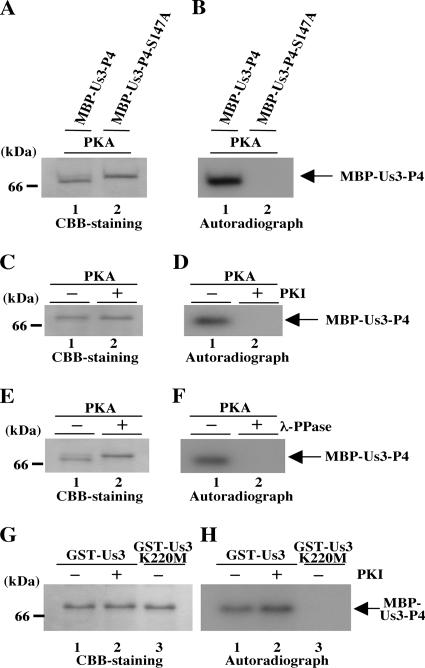
As a first step toward investigation of the regulation of Us3 by phosphorylation, we attempted to identify the in vitro autophosphorylation site(s) of Us3. Based on the consensus Us3 phosphorylation site sequence, we identified a putative autophosphorylation site at Us3 codons 144 to 149 (RRRSRD) (Fig. 3A). To confirm that Ser-147 of Us3 is in fact the autophosphorylation site in vitro, we generated and purified chimeric proteins consisting of MBP fused to peptides encoded by Us3 codons 1 to 172 (MBP-Us3-P4) and its mutant MBP-Us3-P4-S147A in which alanine was substituted for Ser-147 (Fig. 3A). The MBP fusion proteins were captured on amylose beads and used as substrates in in vitro kinase assays with purified wild-type GST-Us3 or the kinase-negative mutant GST-Us3K220M (Fig. 4A). As shown in Fig. 3C, MBP-Us3-P4 was labeled with [γ-32P]ATP in kinase assays using GST-Us3 (Fig. 3C, lane 1), while MBP-Us3-P4-S147A was not (Fig. 3C, lane 2). When the kinase-negative mutant GST-Us3K220M was used, none of the MBP fusion proteins were labeled (Fig. 3C, lanes 3 and 4). To confirm that MBP-Us3-P4 labeling by GST-Us3 was due to phosphorylation, the labeled MBP-Us3-P4 was treated with λ-PPase. As shown in Fig. 3E, MBP-Us3-P4 labeling by GST-Us3 was eliminated by phosphatase treatment, indicating that MBP-Us3-P4 was labeled by phosphorylation. The presence of each MBP fusion protein and the radiolabeled MBP-Us3-P4 band was verified by Coomassie brilliant blue (CBB) staining (Fig. 3B and D). These results indicate that Ser-147 of Us3 is one of the in vitro autophosphorylation sites.
FIG. 3.
(A) Schematic diagram of the genome structures of wild-type virus HSV-1(F) and the location of the Us3 gene. Line 1, linear representation of the HSV-1(F) genome. The unique sequences are represented as unique long (UL) and short (Us) domains, and the terminal repeats flanking them are shown as open rectangles with the designation above each repeat. Line 2, structure of the genome domain containing the Us2, Us3, and Us4 ORFs. Line 3, structure of the Us3 ORF. The shaded areas represent subdomains I to VI, which are conserved in eukaryotic protein kinases. Line 4, the domains of the Us3 gene encoding Us3 residues 98 to 364, used in these studies to generate GST-Us3-P1 fusion proteins which were used for generation of the rabbit polyclonal antibody to Us3. Lane 5, the domains of the Us3 gene encoding Us3 residues 1 to 172, used in these studies to generate MBP-Us3-P4 fusion protein. Line 6, amino acid sequence of Us3 residues 135 to 160. Sites with the consensus sequence for phosphorylation by Us3 itself are underlined. Line 7, domains of the Us3 gene encoding Us3 residues 1 to 172 carrying the S147A mutation used in these studies to generate MBP-Us3-P4-S147A fusion proteins. (B) Purified MBP-Us3-P4 (lanes 1 and 3) and MBP-Us3-P4-S147A (lanes 2 and 4) incubated in kinase buffer containing [γ-32P]ATP and purified GST-Us3 (lanes 1 and 2) or GST-Us3K220M (lanes 3 and 4), separated on a denaturing gel, and stained with CBB. Molecular masses are shown on the left. (C) Autoradiograph of the gel in panel B. (D) Purified MBP-Us3-P4 incubated in kinase buffer containing [γ-32P]ATP and purified GST-Us3 and then either mock treated (lane 1) or treated with λ-PPase (lane 2), separated on a denaturing gel, and stained with CBB. (E) Autoradiograph of the gel in panel D.
FIG. 4.
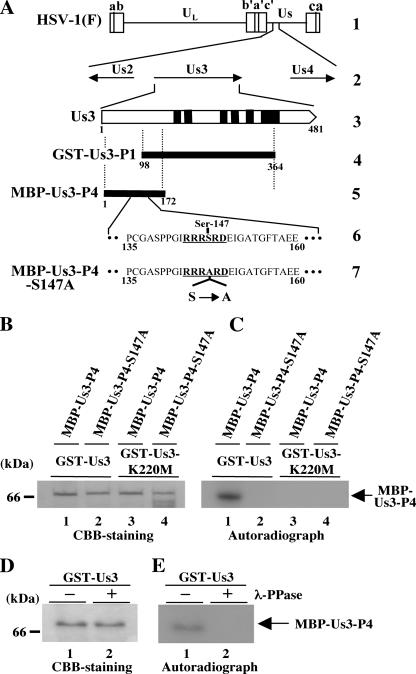
(A) Schematic diagram of the predicted amino acid sequences of GST-Us3 and its mutants GST-Us3K220M and GST-Us3S147A. The K220M and S147A mutations are also indicated. (B) A silver-stained gel (left panel) and an immunoblot (right panel) of purified GST-Us3S147A, GST-Us3, and GST-BGLF4 from Sf9 cells infected with recombinant baculovirus Bac-GST-Us3S147A (lanes 1), Bac-GST-Us3 (lanes 2), and Bac-GST-BGLF4 (lanes 3). Proteins were separated on a denaturing gel and subjected to silver staining (left panel) or transferred onto a nitrocellulose membrane and reacted with antibody to Us3 (right panel). Molecular masses are shown on the left. (C) A silver-stained gel of purified GST-Us3 and GST-Us3S147A. The purified GST-Us3 (lanes 1 and 2) and GST-Us3S147A (lanes 3 and 4) were mock treated (lanes 1 and 3) or treated with CIP (lanes 2 and 4). (D) Purified GST-Us3 (lane 1), GST-Us3K220M (lane 2), and GST-Us3S147A (lane 3) were incubated in kinase buffer containing [γ-32P]ATP, separated on a denaturing gel, and subjected to silver staining. (E) Autoradiograph of the gel in panel D. (F) Purified MBP-UL34 was incubated in kinase buffer containing [γ-32P]ATP and purified GST-Us3 (lane 1), GST-Us3K220M (lane 2), or GST-Us3S147A (lane 3), separated on a denaturing gel, and stained with CBB. (G) Autoradiograph of the gel in panel F. An overexposed image is also shown (lower panel). (H) Quantification of the relative amount of radioactivity in 32P-radiolabeled MBP-UL34 phosphorylated by GST-Us3 or GST-Us3S147A. (I) Quantification of the relative amount of radioactivity in 32P-radiolabeled MBP-UL34 phosphorylated by GST-Us3 or GST-Us3S147A in various time periods of in vitro kinase assay reactions. Experiments were carried out as described for panels F to H except that in vitro kinase reactions were performed for various time periods as indicated.
PKA also phosphorylates Ser-147 of Us3 in vitro.
An earlier report indicated that Us3 and PKA phosphorylate the same site of their target protein in vitro (1). These observations led us to test whether PKA also phosphorylates Ser-147 of Us3 in vitro. As shown in Fig. 2B, MBP-Us3-P4 was labeled with [γ-32P]ATP in kinase assays using purified PKA (Fig. 2B, lane 1), while MBP-Us3-P4-S147A was not (Fig. 2B, lane 2). That this labeling was due to PKA was implied by the observation that MBP-Us3-P4 was not labeled in the presence of a specific inhibitor of PKA, 6-22 amide (Fig. 2D). Thus, MBP-Us3-P4 labeling by PKA was eliminated by phosphatase treatment (Fig. 2F), indicating that MBP-Us3-P4 was labeled by phosphorylation. The presence of each MBP fusion protein and the radiolabeled MBP-Us3-P4 band was verified by CBB staining (Fig. 2A, C, and E). These results indicate that PKA phosphorylates Ser-147 of Us3 in vitro.
The observation that Us3 and PKA phosphorylate the same site of the target protein in our in vitro kinase assays raised the slim possibility that contamination with insect PKA during the purification of GST-Us3 proteins might have been responsible for the protein kinase activity detected using purified GST-Us3. To exclude this possibility, purified GST-Us3 was treated with the specific PKA inhibitor 6-22 amide, followed by addition of the substrate (MBP-Us3-P4). The results (Fig. 2H) indicate that 6-22 amide at a concentration sufficient to completely inhibit PKA (Fig. 2D) had no effect on the activity of GST-Us3. These observations indicate that Us3 and PKA specifically phosphorylate the same site, Ser-147, of Us3 in vitro. We note that Us3 and PKA also phosphorylate the same sites of UL34 (Thr-195 and Ser-198) (17, 56) in vitro (data not shown).
Ser-147 regulates Us3 enzymatic activity in vitro.
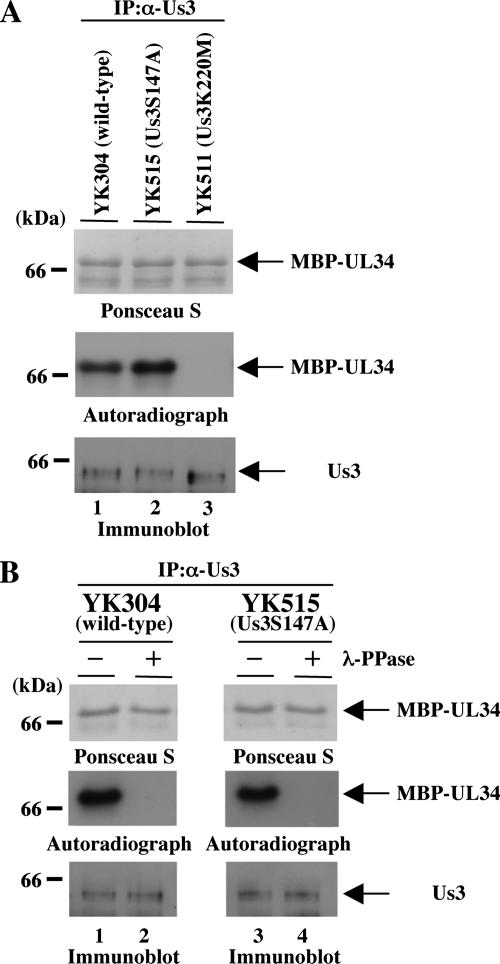
To investigate the role(s) of the identified phosphorylation site of Us3 (Ser-147) for its optimal protein kinase activity in vitro, we expressed and purified GST fused to a mutant Us3 in which alanine is substituted for Ser-147 (GST-Us3S147A) (Fig. 4A) in a baculovirus expression system. As shown in Fig. 4B, the purified GST-Us3S147A contained a major purified band with an Mr of approximately 90,000 as detected by silver staining (left panel) and reacted with antiserum to Us3 (right panel), as observed with purified GST-Us3. Interestingly, GST-Us3 proteins in the slower-migrating band were much more abundant than GST-Us3S147A proteins (Fig. 4C, lanes 1 and 3). That the difference in the electrophoretic mobility between GST-Us3 and GST-Us3S147A was due to phosphorylation was implied by the observation that after phosphatase treatment, GST-Us3 proteins migrated as fast as GST-Us3S147A proteins in a denaturing gel (Fig. 4C, lanes 2 and 4). These results indicate that the phosphorylation status of GST-Us3 expressed in baculovirus is different from that of GST-Us3S147A. Moreover, the purified GST-Us3S147A proteins treated with phosphatase exhibited a faster electrophoretic mobility than the untreated enzyme (Fig. 4C), indicating that sites other than Ser-147 in Us3 are phosphorylated. Consistently, GST-Us3S147A still autophosphorylated itself in in vitro kinase assays (Fig. 4D and E).
Next, we tested purified GST-Us3S147A in in vitro kinase assays using MBP-UL34 as a substrate. We had previously reported that GST-Us3 specifically phosphorylates MBP-UL34 in vitro (17). As shown in Fig. 4F and G, however, phosphorylation of MBP-UL34 in the presence of GST-Us3S147A was barely detectable after the normal exposure time (1 h) in the in vitro kinase assay. Nonetheless, longer exposure (24 h) revealed phosphorylation of MBP-UL34 in the presence of GST-Us3S147A but not GST-Us3K220M. Phosphorylation of MBP-UL34 mediated by GST-Us3S147A was reduced 12.6-fold compared to that mediated by wild-type GST-Us3 (Fig. 4G and H). The difference in kinase activity between GST-Us3 and GST-Us3S147A was confirmed by kinetic assays shown in Fig. 4I. These results indicate that Ser-147 of Us3 has a strong influence on the optimal kinase activity of this enzyme in vitro but is not essential for its catalytic activity.
Generation of a recombinant virus expressing VenusA206K-Us3S147A and the repaired virus.
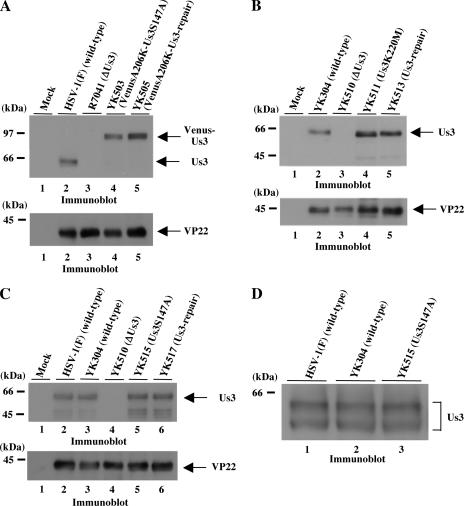
Next, we investigated whether Ser-147 of Us3 is phosphorylated in HSV-1-infected cells. To this end, we attempted to detect Ser-147 phosphorylation of Us3 immunoprecipitated by specific antibody from wild-type HSV-1(F)-infected cells, using the rabbit anti-phospho-PKA substrate monoclonal antibody 100G7. The latter antibody detects proteins containing a phosphorylated serine or threonine residue with arginine at positions −3 and −2 (RRXS or RRXT). In the Us3 polypeptide, the antibody theoretically recognizes only phosphorylated Ser-147. In preliminary experiments, however, we determined that phosphorylation of Us3 immunoprecipitated by rabbit polyclonal antibody from lysates of HSV-1(F)-infected cells was difficult to detect with the anti-phospho-PKA substrate antibody (data not shown). Because the anti-Us3 and the anti-phospho-PKA substrate antibodies were both from rabbits, the secondary antibody against the anti-phospho-PKA antibody in immunoblotting also detected the heavy and light chains of the anti-Us3 antibodies. This resulted in such a high background in the immunoblots that we were unable to discern phosphorylated Us3 protein with the anti-phospho-PKA substrate antibody. To resolve this problem, we generated a recombinant virus, YK503, expressing VenusA206K-tagged Us3 proteins carrying the S147A mutation, and YK505, expressing VenusA206K-tagged Us3, in which the alanine replacement of Ser-147 in YK503 had been repaired. Us3 proteins fused to VenusA206K could then be immunoprecipitated with commercial anti-GFP rat monoclonal antibody. This antibody reacts with the secondary antibody for the detection of the anti-phospho-PKA monoclonal rabbit antibody to a much lesser extent than does the anti-Us3 polyclonal rabbit antibody. Furthermore, VenusA206K-Us3 (92 kDa) migrated much more slowly than nontagged Us3 (65 kDa) in denaturing gels. Accordingly, detection of VenusA206K-Us3 with the anti-phospho-PKA substrate antibody was much less affected by cross-reaction of the secondary antibody with the heavy chains of the anti-GFP antibody (50 kDa) in immunoblotting. With this system, we could now easily detect Ser-147 phosphorylation of immunoprecipitated VenusA206K-Us3 using the anti-phospho-PKA antibody. The strategy for construction of YK503 and YK505 is summarized in Fig. 1. The genotypes of YK502, YK503, YK504, and YK505 were confirmed by Southern blotting and sequencing (data not shown). Expression of the predicted fusion proteins was confirmed by infecting Vero cells with each of the recombinant viruses at an MOI of 3 for 18 h and then assaying the infected cells for the appropriate fusion proteins by immunoblotting with the anti-Us3 antibody. As shown in Fig. 5A, the anti-Us3 antibody reacted with wild-type Us3 (lane 2), VenusA206K-Us3S147A (lane 4), and VenusA206K-Us3-repair (lane 5) but not with lysates from cells infected with the Us3 deletion mutant virus (lane 3). The expression levels of VenusA206K-Us3S147A mutant protein in Vero cells were similar to those of the proteins from wild-type HSV-1(F) and the recombinant viruses YK304 and YK503, encoding nontagged and tagged wild-type Us3, respectively. These results indicate that neither tagging VenusA206K with Us3 nor the S147A mutation in Us3 has any influence on the accumulation of Us3 proteins in infected cells.
FIG. 5.
(A) Immunoblots of electrophoretically separated lysates of Vero cells mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), R7041 (ΔUs3) (lane 3), YK503 (VenusA206K-Us3S147A) (lane 4), or YK505 (VenusA206K-Us3-repair) (lane 5) at an MOI of 3. The infected Vero cells were harvested at 18 h postinfection and subjected to immunoblotting with polyclonal antibody to Us3 (upper panel) or VP22 (lower panel). (B) Immunoblots of electrophoretically separated lysates of Vero cells mock infected (lane 1) or infected with YK304 (wild type) (lane 2), YK510 (ΔUs3) (lane 3), YK511 (Us3K220M) (lane 4), or YK513 (repair) (lane 5) at an MOI of 3. The infected Vero cells were harvested at 18 h postinfection and immunoblotted with polyclonal antibody to Us3 (upper panel) or VP22 (lower panel). (C) Immunoblots of electrophoretically separated lysates of Vero cells mock infected (lane 1) or infected with HSV-1(F) (wild-type) (lane 2), YK304 (wild type) (lane 3), YK510 (ΔUs3) (lane 4), YK515 (Us3S147A) (lane 5), or YK517 (repair) (lane 6) at an MOI of 3. The infected Vero cells were harvested at 18 h postinfection and immunoblotted with polyclonal antibody to Us3 (upper panel) or VP22 (lower panel). (D) Immunoblot of electrophoretically separated lysates of Vero cells infected with HSV-1(F) (wild type) (lane 1), YK304 (wild type) (lane 2), or YK515 (Us3S147A) (lane 3). The infected Vero cells were harvested at 18 h postinfection and immunoblotted with polyclonal antibody to Us3.
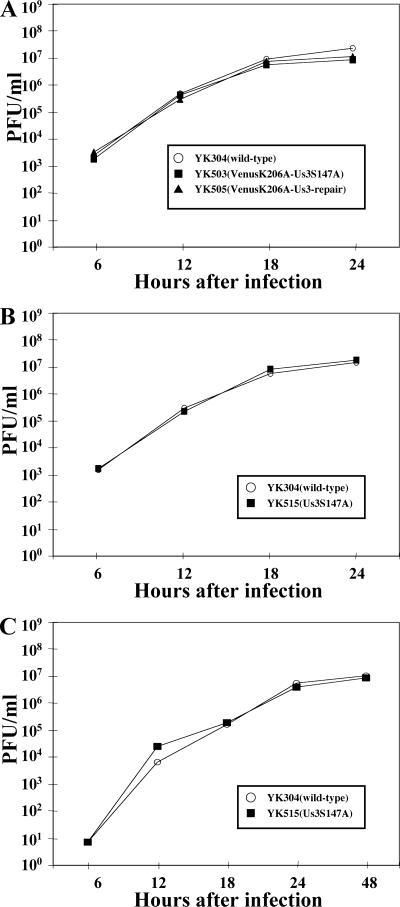
Next, Vero cells were infected with wild-type YK304, YK503 (VenusA206K-Us3S147A), or YK505 (VenusA206K-Us3-repair) at an MOI of 3, and the total virus yield from the cell culture supernatants and the infected cells was harvested at the indicated time points (Fig. 6A). The titers of each sample were determined by standard plaque assays on Vero cells. As shown in Fig. 6A, the growth curve of YK503 was essentially identical to that of YK505, indicating that Ser-147 and its phosphorylation are not critical for optimal viral replication in Vero cells. The FP-tagged recombinant viruses (YK503 and YK505) displayed growth characteristics similar to those of wild-type YK304 but produced a slightly (two- to threefold) lower progeny yield than YK304 (Fig. 6A). Because the growth of the FP-tagged recombinant virus (YK505) was not appreciably affected by fusing the Us3 protein with VenusA206K and because the other phenotypes of YK505 (VenusA206K-Us3-repair) tested in the present studies were identical to those of wild-type YK304 (Fig. 5; see Fig. 9 and 10), it is likely that tagging VenusA206K with Us3 has little effect on the functions of Us3 in infected cells.
FIG. 6.
Growth curves for recombinant viruses. (A) Vero cells were infected at an MOI of 3 with wild-type YK304 or FP-tagged recombinant virus YK503 (VenusA206KUs3S147A) or YK505 (VenusA206KUs3-repair). (B) Vero cells were infected at an MOI of 3 with wild-type YK304 or recombinant virus YK515 (Us3S147A). (C) Vero cells were infected at an MOI of 0.01 with wild-type YK304 or recombinant virus YK515 (Us3S147A). Total virus from the cell culture supernatants and the infected cells was harvested at the indicated times and assayed on Vero cells.
FIG. 9.
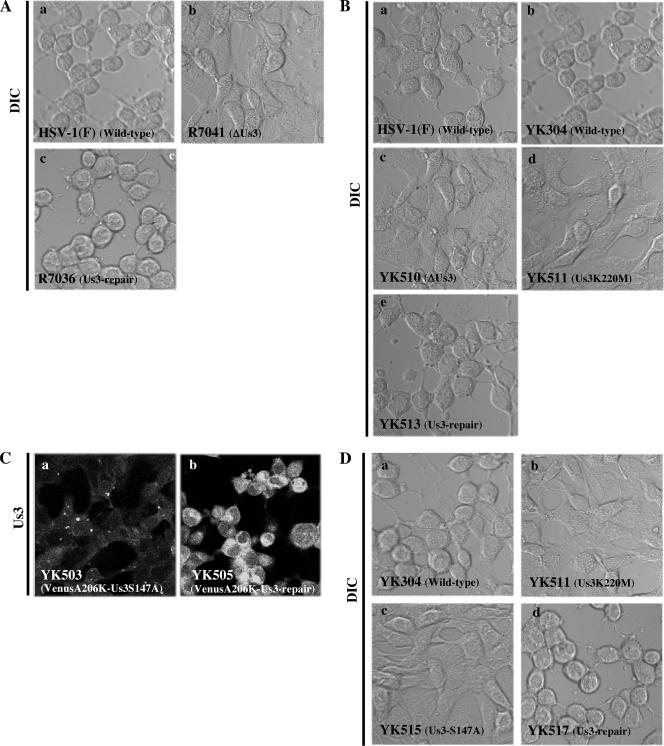
Digital confocal microscope images of CPEs in Vero cells infected at an MOI of 10 with (A) wild-type HSV-1(F) (a), R7041 (ΔUs3) (b), or R7306 (repair) (c); (B) wild-type HSV-1(F) (a), wild-type YK304 (b), YK510 (ΔUs3) (c), YK511 (Us3K220M) (d), or YK513 (repair) (e); (C) YK503 (VenusA206K-Us3S147A) (a) or YK505 (VenusA206K-Us3-repair) (b); and (D) wild-type YK304 (a), YK511 (Us3K220M) (b) YK515 (Us3S147A) (c), or YK517 (repair) (d). The live cells were examined at 24 h postinfection by confocal microscopy. Differential interference contrast (DIC) (A, B, and D) and fluorescence (C) images are shown.
FIG. 10.
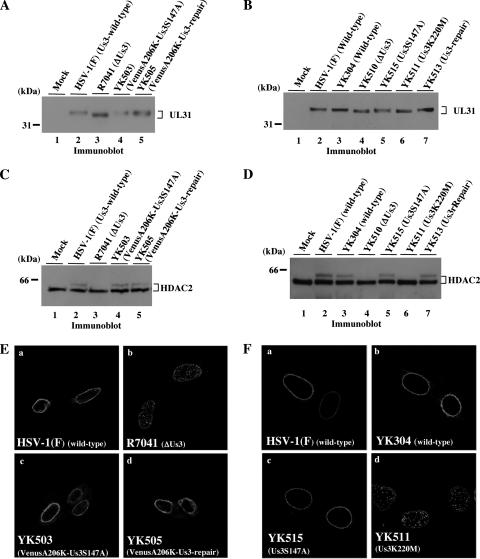
(A) Immunoblot of electrophoretically separated lysates from Vero cells mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), R7041 (ΔUs3) (lane 3) YK503 (VenusA206K-Us3S147A) (lane 4), or YK505 (VenusA206K-Us3-repair) (lane 5) at an MOI of 3. Infected cells were harvested at 18 h postinfection and immunoblotted with anti-UL31 antibody. (B) Immunoblot of electrophoretically separated lysates from Vero cells mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), wild-type YK304 (lane 3), YK510 (ΔUs3) (lane 4), YK515 (Us3S147A) (lane 5), YK511 (Us3K220M) (lane 6), or YK513 (repair) (lane 7) at an MOI of 3. Infected cells were harvested at 18 h postinfection and immunoblotted with anti-UL31 antibody. (C) Immunoblot of electrophoretically separated lysates from Vero cells mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), R7041 (ΔUs3) (lane 3), YK503 (VenusA206K-Us3S147A) (lane 4), or YK505 (VenusA206K-Us3-repair) (lane 5) at an MOI of 3. Infected cells were harvested at 18 h postinfection and immunoblotted with anti-HDAC2 antibody. (D) Immunoblot of electrophoretically separated lysates from Vero cells mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), wild-type YK304 (lane 3), YK510 (ΔUs3) (lane 4), YK515 (Us3S147A) (lane 5), YK511 (Us3K220M) (lane 6), or YK513 (repair) (lane 7) at an MOI of 3. Infected cells were harvested at 18 h postinfection and immunoblotted with anti-HDAC2. (E) Digital confocal microscope images showing localization of UL34 in Vero cells infected with wild-type HSV-1(F) (a), R7041 (ΔUs3) (b), YK503 (VenusA206K-Us3S147A) (c), or YK505 (VenusA206K-Us3-repair) (d). After 15 h, infected cells were fixed, permeabilized, and immunostained with chicken polyclonal antibody to UL34 detected with Alexa-546 conjugated anti-chicken IgG antibody. (F) Digital confocal microscope images showing localization of UL34 in Vero cells infected with wild-type HSV-1(F) (a), wild-type YK304 (b), YK515 (Us3S147A) (c), or YK511 (Us3K220M) (d). After 15 h, infected cells were fixed, permeabilized, and immunostained with chicken polyclonal antibody to UL34 detected with Alexa-546 conjugated anti-chicken IgG antibody.
Ser-147 of Us3 is phosphorylated in infected cells.
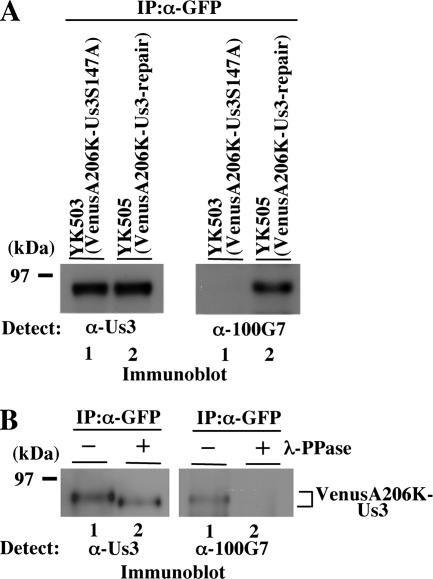
To investigate whether Ser-147 of Us3 was phosphorylated in infected cells, Vero cells were infected with YK503 expressing VenusA206K-Us3S147A or with YK505 expressing VenusA206K-Us3-repair at an MOI of 3, harvested at 18 h postinfection, solubilized, immunoprecipitated with the anti-GFP antibody, and finally analyzed by immunoblotting with the anti-phospho-PKA substrate antibody or anti-Us3 antibody. As shown in Fig. 7A, the anti-phospho-PKA substrate antibody reacted with VenusA206K-Us3-repair purified from YK505-infected Vero cells but not with VenusA206K-Us3S147A purified from YK503-infected Vero cells. The expression levels of total Us3 proteins in Vero cells infected with YK505 were equivalent to those in cells infected with YK503. To confirm that the VenusA206K-Us3-repair product detected by the anti-phospho-PKA substrate antibody was due to phosphorylation, material purified by immunoprecipitation was treated with λ-PPase. As shown in Fig. 7B, the reactivity of VenusA206K-Us3-repair with the anti-phospho-PKA substrate antibody was eliminated by phosphatase treatment, indicating that the antibody did indeed react with phosphorylated VenusA206K-Us3-repair. These results indicate that the anti-phospho-PKA substrate antibody specifically recognized the phosphorylated Ser-147 of Us3 and that this residue is phosphorylated in infected cells.
FIG. 7.
(A) Immunoblots of electrophoretically separated Us3 immunoprecipitates from Vero cells infected with YK503 (VenusA206K-Us3S147A) (lane 1) or YK505 (VenusA206K-Us3-repair) (lane 2) at an MOI of 3. The infected Vero cells were harvested at 18 h postinfection and immunoprecipitated with the anti-GFP antibody. The immunoprecipitates were separated on a denaturing gel, transferred to a polyvinylidene difluoride membrane, and analyzed by immunoblotting with anti-Us3 antibody (left panel) or anti-phospho-PKA substrate antibody (right panel). (B) Immunoprecipitates prepared as for panel A were either mock treated (lanes 1) or treated with λ-PPase (lanes 2), separated on a denaturing gel, transferred to a polyvinylidene difluoride membrane, and analyzed by immunoblotting with anti-Us3 antibody (left panel) or anti-phospho-PKA substrate antibody (right panel).
The S147A mutation in Us3 affects enzyme localization in infected cells.
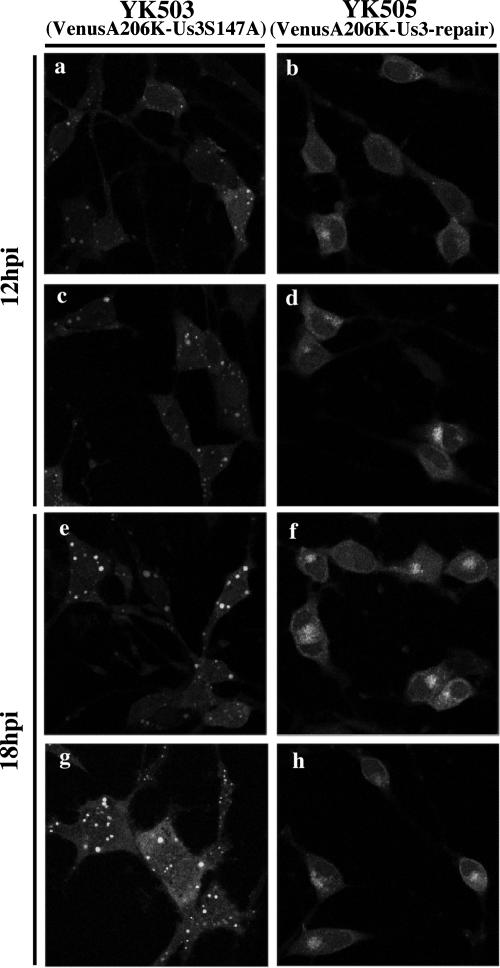
An earlier publication by Ryckman and Roller reported that in Vero cells infected with a recombinant HSV-1 encoding enzymatically inactive Us3, the mutant Us3 protein was aberrantly localized to large, punctate structures distributed throughout the cells, indicating that the protein kinase activity of Us3 regulates its localization (56). This observation, together with our results that Ser-147 of Us3 is required for optimal kinase activity of Us3 in vitro, led us to examine the localization of the Us3S147A mutant in infected cells more closely. However, Reynolds et al. had noted that the anti-Us3 antibody used in earlier studies had a consistent background fluorescence in the cytoplasm of cells infected with HSV-1 Us3 null mutants in immunofluorescence assays (52), making it difficult to draw conclusions about the exact localization of Us3. Consistent with this, the anti-Us3 antibody generated in the present study also showed such background fluorescence in some cell lines infected with the Us3 deletion mutant virus (R7041) (data not shown). Therefore, we investigated the effect of the S147A mutation in Us3 on localization of this molecule in infected cells using recombinant viruses (YK503 and YK505) encoding FP-tagged Us3. Because Us3 was tagged with FP in YK503 and YK505, the specific subcellular localization of Us3 proteins in live infected cells could easily be investigated without immunofluorescence assays relying on anti-Us3 antibodies. Vero cells were mock infected or infected with YK503 (VenusA206K-Us3S147A) or YK505 (VenusA206K-Us3-repair) at an MOI of 10 for 12 or 18 h and then examined by confocal microscopy. As shown in Fig. 8, the VenusA206K-Us3-repair protein was detected mainly in the cytoplasm of YK505-infected Vero cells, and as infection progressed, it accumulated preferentially at the juxtanuclear regions (Fig. 8b, d, f, and h). In contrast, when Vero cells were infected with YK503, Us3 localization differed significantly from that in YK505-infected Vero cells. In Vero cells infected with YK503, the VenusA206K-Us3S147A protein was aberrantly localized to large, punctate structures in the cytoplasm, the appearance of which was strongly reminiscent of those reported earlier by Ryckman and Roller (56). The number of these punctate structures increased as infection progressed (Fig. 8a, c, e, and g). These results indicate that Ser-147 of Us3 is necessary for the correct localization of the enzyme in infected cells.
FIG. 8.
Digital confocal microscope images showing localization of VenusA206K-Us3S147A and VenusA206K-Us3-repair proteins in live Vero cells infected with YK503 or YK505, respectively. Vero cells were infected with YK503 (VenusA206K-Us3S147A) (a, c, e, and g) or YK505 (VenusA206K-Us3-repair) (b, d, f, and h) at an MOI of 10, and live cells were examined at 12 h (a to d) or 18 h (e to h) postinfection by confocal microscopy.
The S147A mutation in Us3 affects the ability of the protein to induce wild-type CPEs in infected cells.
It has been reported that the CPEs of Us3 deletion mutant viruses of HSV-1 and HSV-2 are clearly different from those of wild-type viruses (36, 38, 48), indicating that Us3 influences the morphology of infected cells. To confirm this, Vero cells were infected with wild-type HSV-1(F), R7041 (ΔUs3), and R7306 (repair) at an MOI of 10 for 24 h, and then CPEs of infected cells were observed by confocal microscopy. Consistent with the previous reports, CPEs of R7041 (ΔUs3) were apparently different from those of wild-type HSV-1(F) or R7306 (repair) (Fig. 9A). Infection of Vero cells with wild-type HSV-1(F) or R7306 (repair) efficiently induced cell rounding, while R7041 (ΔUs3) did so only partially. Next, we examined whether the ability of Us3 to induce wild-type CPEs is associated with catalytic activity of Us3, because this issue has not been addressed. To this end, we generated three Us3 mutant viruses, i.e., YK510 (ΔUs3), YK511 (Us3K220M), and their repaired version, YK513 (Fig. 1). YK510 is a Us3 null mutant, whereas YK511 encodes the enzymatically inactive Us3 mutant Us3K220M (17), in which the lysine codon at position 220 is replaced by methionine. The genotypes of YK510, YK511, and YK513 were confirmed by Southern blotting and sequencing (data not shown). Expression of the predicted proteins was confirmed by infecting Vero cells with each of the recombinant viruses and assaying the infected cells for the appropriate proteins by immunoblotting (Fig. 5B). As shown in Fig. 9B, CPEs in Vero cells infected with YK511 (Us3K220M) were similar to those in cells infected with YK510 (ΔUs3), with both viruses only partially inducing cell rounding. In contrast, wild-type HSV-1(F), YK304, and YK513 (repair) efficiently induced cell rounding. These results indicate that the catalytic function of the Us3 protein is necessary for induction of wild-type CPEs in infected cells.
Next, we examined whether the S147A mutation in Us3 affects the ability of the protein to induce wild-type CPEs. As shown in Fig. 9C, CPEs in Vero cells infected with YK503 (VenusA206K-Us3S147A) resembled those in cells infected with the Us3 mutant viruses YK510 (ΔUs3), R7041 (ΔUs3), and YK511 (Us3K220M) (Fig. 9A and B). In contrast, YK505 (VenusA206K-Us3-repair) induced cell rounding efficiently in Vero cells, as observed with the wild-type viruses HSV-1(F) and YK304 (Fig. 9A and B).
We also generated an additional Us3 mutant virus (YK515) in which Ser-147 of nontagged Us3 was replaced with alanine (Fig. 1). The repaired version was also created (YK517) (Fig. 1). The genotypes of YK515 and YK517 were confirmed by Southern blotting and sequencing (data not shown). We then examined the effect of the S147A mutation in nontagged Us3 on the ability of the protein to induce wild-type CPEs. As shown in Fig. 9D, the results obtained with YK515 (Us3S147A) and YK517 (repair) were consistent with those obtained with FP-tagged viruses (Fig. 9C). These results indicate that Ser-147 of Us3 is required for induction of wild-type CPEs in infected cells. We also examined growth of YK515 in Vero cells at an MOI of 3 or 0.01 and the level of expression of Us3S147A protein in YK515-infected Vero cells. The results (Fig. 5C and 6B and C) confirmed the conclusions drawn using FP-tagged recombinant viruses (YK503 and YK505), namely, that Ser-147 and its phosphorylation are not required for either accumulation of Us3 protein (Fig. 5A) or optimal viral replication in Vero cells (Fig. 6 A). Similar growth properties of YK515 were observed in resting HEL cells at MOIs of 3 and 0.01, and plaques produced by both YK515 and wild-type YK304 were of similar size in Vero and resting HEL cells (data not shown). Furthermore, the efficiency of packaging of Us3S147A mutant proteins into YK515 virions was similar to that of packaging of wild-type Us3 proteins into wild-type virions (data not shown). We note that the pattern of elecrophoretic mobilities of the Us3S147A mutant from Vero cells infected with YK515 could not be differentiated from that of wild-type Us3 from cells infected with HSV-1(F) or YK304 (Fig. 5D), unlike the case with Us3 proteins expressed in a baculovirus expression system (Fig. 4C).
The S147A mutation in Us3 does not affect the ability of Us3 to induce posttranslational modification of UL31 and HDAC2 and to regulate localization of UL34 in infected cells.
In this part of the work, we investigated whether the S147A mutation in Us3 affects the other reported functions of Us3. It was demonstrated earlier that Us3 is required for induction of posttranslational modification of UL31 and HDAC2 and for proper localization of UL34 in infected cells (17, 45, 51). Here, in the first series of experiments, Vero or rabbit skin cells were mock infected or infected with wild-type HSV-1(F), wild-type YK304, R7041 (ΔUs3), YK510 (ΔUs3), YK511 (Us3K220M), YK513 (repair), YK503 (VenusA206K-Us3S147A), YK505 (VenusA206K-Us3-repair), or YK515 (Us3S147A) at an MOI of 3 for 18 h and then immunoblotted with anti-UL31 or anti-HDAC2 antibody. As shown in Fig. 10A to D, UL31 and HDAC2 proteins in cells infected with recombinant viruses carrying the S147A mutation in Us3 (YK503 and YK515) were posttranslationally modified at a level similar to those in cells infected with wild-type or recombinant viruses encoding wild-type Us3 [HSV-1(F), YK304, YK505 or YK513]. In contrast, such modifications of these proteins were not observed in Vero cells infected with Us3-null virus (R7041) or a recombinant virus expressing enzymatically inactive Us3 (YK511). These results indicate that Ser-147 and phosphorylation of Ser-147 in Us3 are not necessary for the protein to induce posttranslational modification of UL31 and HDAC2 in infected cells. In addition, the finding that, like the Us3-null virus, YK511 carrying the K220M Us3 mutation was also defective in inducing posttranslational modification of UL31 and HDAC2 indicates that the catalytic function of Us3 is required for the modification of the target proteins.
In the second series of experiments, Vero cells were infected with wild-type HSV-1(F), wild-type YK304, R7041 (ΔUs3), YK511 (Us3K220M), YK503 (VenusA206K-Us3S147A), YK505 (VenusA206K-Us3-repair), or YK515 (Us3S147A) at an MOI of 10, fixed at 15 h postinfection, and processed for immunofluorescence with anti-UL34 antibody. As shown in Fig. 10E and F, the UL34 protein localized mainly at the nuclear envelope with a uniform distribution in cells infected with HSV-1(F) (wild type), YK304 (wild type), YK503 (VenusA206K-Us3S147A), YK505 (VenusA206K-Us3-repair), or YK515 (Us3S147A), while it was mislocalized to punctate structures as reported previously (51, 56) in cells infected with the Us3 mutant virus R7041 (ΔUs3) or YK511 (Us3K220M). These results indicate that Ser-147 and phosphorylation of Ser-147 are not required for proper localization of UL34 in infected cells.
The S147A mutation in Us3 does not affect the total Us3 protein kinase activity in infected cells.
Finally, we investigated protein kinase activity of Us3 carrying the S147A mutation in infected cells. Vero cells infected with wild-type YK304, YK515 (Us3S147A), or YK511 (Us3K220M) at an MOI of 3 were harvested at 18 h postinfection, solubilized, and immunoprecipitated with the anti-Us3 antibody. The immunoprecipitates were then used in in vitro kinase assays with purified MBP-UL34 as a substrate. To reduce the possibility that the anti-Us3 antibody might coprecipitate a contaminating kinase(s), the precipitated material containing Us3 protein kinase was washed with a high-salt buffer containing 1 M NaCl prior to the assays. As shown in Fig. 11A, MBP-UL34 was labeled with [γ-32P]ATP in the presence of Us3S147A mutant protein immunoprecipitated from YK515-infected cells at a level similar to that in the presence of wild-type Us3 protein immunoprecipitated from wild-type YK304-infected cells. In contrast, MBP-UL34 was not labeled in the presence of the kinase-negative mutant Us3K220M protein immunoprecipitated from YK511-infected cells. The labeling of Us3 proteins was due to phosphorylation, as determined by studies showing that it was eliminated by phosphatase treatment (Fig. 11B). The presence of MBP-UL34 protein and the radiolabeled MBP-UL34 band was verified by Ponceau S staining. Similar results were also observed with the FP-tagged recombinant viruses YK503 (VenusA206K-Us3S147A) and YK505 (VenusA206K-Us3-repair) (data not shown). These results indicate that the total protein kinase activity of the Us3S147A mutant in infected cells is similar to that of wild-type Us3.
FIG. 11.
Autoradiographic images of MBP-UL34 subjected to in vitro kinase assay with Us3 immunoprecipitates. (A) Vero cells were infected with wild-type YK304 (lane 1), YK515 (Us3S147A) (lane 2), or YK511 (Us3K220M) (lane 3) at an MOI of 3, harvested at 18 h postinfection, and immunoprecipitated with antibody to Us3. The immunoprecipitates were divided into two aliquots. One aliquot was incubated in kinase buffer containing [γ-32P]ATP and MBP-UL34, separated on a denaturing gel, transferred to a nitrocellulose membrane, stained with Ponceau S (upper panel), and analyzed by autoradiography (middle panel). The other was separated on a denaturing gel, transferred to a nitrocellulose membrane, and subjected to immunoblotting with the anti-Us3 antibody (lower panel). (B) MBP-UL34 incubated with the immunoprecipitates prepared for panel A was either mock treated (lane 1) or treated with λ-PPase (lane 2), separated on a denaturing gel, transferred to a nitrocellulose membrane, stained with Ponceau-S (upper panel), and analyzed by autoradiography (middle panel). Immunoprecipitates prepared for panel A were also separated on a denaturing gel, transferred to a nitrocellulose membrane, and subjected to immunoblotting with anti-Us3 antibody (lower panel).
DISCUSSION
HSV-1 possesses at least three protein kinases encoded by the UL13, Us3, and UL39 genes. The amino acid sequence of Us3 is conserved in the Alphaherpesvirinae subfamily, while UL13 is conserved throughout all herpesvirus subfamilies (5, 31, 58). In Alphaherpesvirinae, only HSV-1 and HSV-2 have a third viral protein kinase, RR1, encoded by the UL39 gene, in addition to Us3 and UL13 (20). Thus, every herpesvirus encodes at least one viral protein kinase, and these have been reported to play multiple roles in viral replication by regulation of viral gene expression (49), apoptosis (28, 43), encapsidation (64), nuclear egress of nucleocapsid (13, 24, 52), and replication of viral genomes (64). While downstream events of herpesviral protein kinases have been extensively investigated, there is a dearth of information on any herpesviral protein kinases regarding how their activity is regulated. The question we have posed in the studies reported here is whether the activity of the Us3 protein kinase is regulated by phosphorylation. The salient features of these studies are as follows.
(i) Ser-147 of Us3 is phosphorylated in vitro and in infected cells. In the studies presented here, bioinformatics analysis predicted a putative autophosphorylation site of Us3 at codons 144 to 149 (RRRSRD), and in vitro kinase assays confirmed that Us3 phosphorylates Ser-147 of Us3. Consistent with the previous report that the activity of Us3 kinase functionally overlaps that of PKA in vitro and in infected cells (1), we have also demonstrated that PKA phosphorylated Ser-147 of Us3 in vitro. It is known that some protein kinases are somewhat promiscuous in vitro if allowed sufficient reaction time and provided with enough substrate. Therefore, in vitro phosphorylation, although a necessary characteristic, is not sufficient for defining a new phosphorylation site; thus, the site identified in vitro must be shown to be a physiological phosphorylation site in cells in vivo. To this end, we used an anti-phospho-PKA substrate antibody which recognizes phosphorylated Ser or Thr residues in the context of the sequence RRXS or RRXT. Phospho-substrate antibodies have been extremely useful for identifying and characterizing phosphorylation sites of new substrates over the past several years (29). In the studies presented here, we have demonstrated that the anti-phospho-PKA substrate antibody specifically reacted with the phosphorylated Ser-147 of Us3 purified by immunoprecipitation from infected cells. This indicates that Ser-147 of Us3 is phosphorylated in infected cells and therefore fulfills the requirements for a physiological phosphorylation site. At present, the protein kinase(s) responsible for the phosphorylation of Ser-147 of Us3 in infected cells is unknown, although in vitro assays suggest that Us3 itself and PKA are involved. As mentioned above, in vitro phosphorylation often does not reflect in vivo phosphorylation, and the sequence flanking Ser-147 of Us3 also matches the consensus phosphorylation sites for other protein kinases, including a set of protein kinases in the AGC protein kinase family, such as protein kinase C, protein kinase G, and p21 activation kinase 2 (23, 62). Further studies will be needed to clarify which protein kinase(s) is responsible for the phosphorylation of Us3 Ser-147 in infected cells.
(ii) The S147A mutation attenuated the activity of Us3 in vitro and affected some but not all Us3 functions in infected cells. It is well established that activity of cellular protein kinases is controlled by phosphorylation of the enzymes (55). Consistent with this, we have demonstrated that the protein kinase activity of the GST-Us3S147A mutant generated in a baculovirus expression system was much lower than that of wild-type GST-Us3 in vitro (Fig. 4), indicating that optimal activity of Us3 is regulated by Ser-147. Furthermore, we have shown that Us3 from recombinant viruses carrying the S147A mutation in Us3 failed to localize correctly (Fig. 8) and did not alter the morphology of cells like the wild-type virus (Fig. 9), two activities both requiring the catalytic activity of Us3 (Fig. 9) (56). Together with the observation that Ser-147 is the physiological phosphorylation site of Us3, this makes it reasonable to conclude that phosphorylation of Us3 Ser-147 is the event that regulates the optimal kinase activity of this enzyme and the manifestations of its catalytic activity in infected cells. However, we cannot formally exclude the slim possibility that the phenotypes observed with recombinant viruses carrying the S147A mutation are due to steric hindrance of Us3 caused by the amino acid replacement, rather than prevention of phosphorylation on Ser-147 of Us3.
(iii) HSV-1 Us3 has been reported to encode two transcriptional units directing the synthesis of the Us3 and Us3.5 proteins (44, 53). The Us3.5 protein is a truncated form of Us3 protein and is expressed in wild-type HSV-1-infected cells at a much lower level than Us3 protein (46). Like Us3 protein kinase, Us3.5 mediates posttranslational modification of HDAC1, HDAC2, the PKA regulatory II subunit, and UL31, suggesting that Us3.5 protein is a protein kinase (44). In contrast, these proteins differ with respect to their functions in blocking apoptosis and in nuclear egress of progeny nucleocapsids (44). Since the physiological phosphorylation site (Ser-147) in Us3 identified in this study is present in Us3.5 protein, it is possible that phosphorylation of Ser-147 also controls regulatory activities of Us3.5. Further studies, such as studies of the effects of Ser-147 mutation in Us3.5 on its protein kinase activity in vitro and its functions in infected cells, are of interest and are under way in this laboratory.
(iv) It appears that Ser-147 in Us3 is conserved among HSV-1 strains, whereas the residue is not among other alpaherpesvirus Us3 homologues and even the HSV-2 Us3 protein lacks a comparable serine residue (Fig. 12). This suggests that the phosphorylation of Us3 at Ser-147 and its relevance in viral replication are specific to HSV-1 and are not applicable to Us3 homologues of other alphaherpesviruses. However, we note that the amino acid residue in Us3 of HSV-2 strain HG52 corresponding to HSV-1 Us3 Ser-147 is asparatic acid (Asp-147) (Fig. 12), and Asp-147 in Us3 appears to be conserved among HSV-2 strains because Asp-147 is also present in Us3 of HSV-2 strain G (data not shown). It is known that acidic amino acids such as Asp or glutamic acid sometimes mimic the negative charges exerted by phosphorylation (65). Therefore, it would be interesting to investigate whether Asp-147 in HSV-2 Us3 plays roles in its proper localization and induction of the wild-type morphological change in HSV-2-infected cells.
FIG. 12.
Sequence alignment of the Us3 protein homologues of HSV-1(F), HSV-1 strain 17, and HSV-2 strain HG52. In the last line, residues conserved among three of the sequences are shown as asterisks.
(v) The total protein kinase activity of Us3 purified by immunoprecipitation from cells infected with the recombinant virus YK503 or YK515 carrying the S147A mutation in Us3 was similar to that from cells infected with recombinant viruses expressing wild-type Us3 (YK505 and YK304). These observations appeared to contradict the result that Ser-147 is critical for optimal protein kinase activity of Us3 in vitro. One hypothesis to account for this discrepancy is that only a small fraction of Us3 molecules are phosphorylated at Ser-147 at any one time, and another is that phosphorylation of the majority does take place but is transient in HSV-1-infected cells. Thus, phosphorylation of Us3 on Ser-147 is presumably tightly regulated in HSV-1-infected cells and occurs transiently or only in a limited subcellular domain(s). Therefore, the majority of Us3 in HSV-1-infected cells would be unphosphorylated on Ser-147 at any one time, resulting in little difference in the total protein kinase activity between wild-type Us3 from cells infected with wild-type viruses and Us3S147A mutants from cells infected with YK503 or YK515. This hypothesis is supported by the finding that only some of the Us3 functions examined here, including regulation of localization of Us3 itself and the morphology of infected cells, were affected by the S147A mutation, while others, including regulation of localization of UL34 and modification of UL31 and HDAC2, were not. Probably overexpression of large amounts of GST-Us3 in the baculovirus expression system in insect cells resulted in a different phosphorylation status of Us3 proteins than for the majority of Us3 expressed in mammalian cells infected with HSV-1. Consistent with this possibility, the phosphorylation status of GST-Us3 expressed in the baculovirus expression system was clearly different from that of GST-Us3S147A (Fig. 4C), while the phosphorylation status of Us3 from Vero cells infected with the wild-type viruses, detected in a denaturing gel, could not be differentiated from that of Us3S147A in Vero cells infected with YK515 (Fig. 5D). Thus, GST-Us3 may be efficiently phosphorylated at Ser-147 in insect cells infected with the recombinant baculovirus, resulting in significant differences in protein kinase activity between GST-Us3 and GST-Us3S147A. Further studies, such as time-space tracking of phosphorylation of Us3 on Ser-147 in HSV-1-infected cells by using antibodies that specifically recognize phosphorylated Ser-147 of Us3 and identification of the protein kinase(s) responsible for this phosphorylation, will be needed and are in progress in this laboratory. Such information will provide further insights into the important issue of how Us3 functions are regulated.
Acknowledgments
We thank B. Roizman for R7041, R7306, and pBC1013; R. Roller for chicken polyclonal antibody to UL34; A. Miyawaki for Venus/pCS2; P. A. Ioannou for pGETrec; G. A. Smith for E. coli GS1783; N. Osterrieder for E. coli GS1783 containing pYEbac102 and pEPkan-S; and S. Watanabe for HELs. We thank S. Koyama for excellent technical assistance.
This study was supported in part by Grants for Scientific Research and Grants for Scientific Research in Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan. A.K. and R.A. were supported by a Research Fellowship of the Japanese Society for the Promotion of Science for Young Scientists.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Benetti, L., and B. Roizman. 2004. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. USA 1019411-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benetti, L., and B. Roizman. 2006. Protein kinase B/Akt is present in activated form throughout the entire replicative cycle of deltaU(S)3 mutant virus but only at early times after infection with wild-type herpes simplex virus 1. J. Virol. 803341-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartier, A., E. Broberg, T. Komai, M. Henriksson, and M. G. Masucci. 2003. The herpes simplex virus-1 Us3 protein kinase blocks CD8T cell lysis by preventing the cleavage of Bid by granzyme B. Cell Death Differ. 101320-1328. [DOI] [PubMed] [Google Scholar]
- 4.Cartier, A., T. Komai, and M. G. Masucci. 2003. The Us3 protein kinase of herpes simplex virus 1 blocks apoptosis and induces phosporylation of the Bcl-2 family member Bad. Exp. Cell Res. 291242-250. [DOI] [PubMed] [Google Scholar]
- 5.Chee, M. S., G. L. Lawrence, and B. G. Barrell. 1989. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J. Gen. Virol. 701151-1160. [DOI] [PubMed] [Google Scholar]
- 6.Daikoku, T., R. Kurachi, T. Tsurumi, and Y. Nishiyama. 1994. Identification of a target protein of US3 protein kinase of herpes simplex virus type 2. J. Gen. Virol. 752065-2068. [DOI] [PubMed] [Google Scholar]
- 7.Daikoku, T., Y. Yamashita, T. Tsurumi, K. Maeno, and Y. Nishiyama. 1993. Purification and biochemical characterization of the protein kinase encoded by the US3 gene of herpes simplex virus type 2. Virology 197685-694. [DOI] [PubMed] [Google Scholar]
- 8.Daikoku, T., Y. Yamashita, T. Tsurumi, and Y. Nishiyama. 1995. The US3 protein kinase of herpes simplex virus type 2 is associated with phosphorylation of the UL12 alkaline nuclease in vitro. Arch. Virol. 1401637-1644. [DOI] [PubMed] [Google Scholar]
- 9.Eisfeld, A. J., S. E. Turse, S. A. Jackson, E. C. Lerner, and P. R. Kinchington. 2006. Phosphorylation of the varicella-zoster virus (VZV) major transcriptional regulatory protein IE62 by the VZV open reading frame 66 protein kinase. J. Virol. 801710-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2357-364. [DOI] [PubMed] [Google Scholar]
- 11.Favoreel, H. W., G. Van Minnebruggen, D. Adriaensen, and H. J. Nauwynck. 2005. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an alphaherpesvirus are associated with enhanced spread. Proc. Natl. Acad. Sci. USA 1028990-8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frame, M. C., F. C. Purves, D. J. McGeoch, H. S. Marsden, and D. P. Leader. 1987. Identification of the herpes simplex virus protein kinase as the product of viral gene US3. J. Gen. Virol. 682699-2704. [DOI] [PubMed] [Google Scholar]
- 13.Gershburg, E., S. Raffa, M. R. Torrisi, and J. S. Pagano. 2007. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 815407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarosinski, K. W., N. G. Margulis, J. P. Kamil, S. J. Spatz, V. K. Nair, and N. Osterrieder. 2007. Horizontal transmission of Marek's disease virus requires US2, the UL13 protein kinase, and gC. J. Virol. 8110575-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jerome, K. R., R. Fox, Z. Chen, A. E. Sears, H. Lee, and L. Corey. 1999. Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3. J. Virol. 738950-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanamori, M., S. Watanabe, R. Honma, M. Kuroda, S. Imai, K. Takada, N. Yamamoto, Y. Nishiyama, and Y. Kawaguchi. 2004. Epstein-Barr virus nuclear antigen leader protein induces expression of thymus- and activation-regulated chemokine in B cells. J. Virol. 783984-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, A., M. Yamamoto, T. Ohno, H. Kodaira, Y. Nishiyama, and Y. Kawaguchi. 2005. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J. Virol. 799325-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 801476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 711019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi, Y., and K. Kato. 2003. Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev. Med. Virol. 13331-340. [DOI] [PubMed] [Google Scholar]
- 21.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1δ. J. Virol. 772359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 717328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennelly, P. J., and E. G. Krebs. 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 26615555-15558. [PubMed] [Google Scholar]
- 24.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leach, N., S. L. Bjerke, D. K. Christensen, J. M. Bouchard, F. Mou, R. Park, J. Baines, T. Haraguchi, and R. J. Roller. 2007. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 8110792-10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leader, D. P. 1993. Viral protein kinases and protein phosphatases. Pharmacol. Ther. 59343-389. [DOI] [PubMed] [Google Scholar]
- 27.Leader, D. P., A. D. Deana, F. Marchiori, F. C. Purves, and L. A. Pinna. 1991. Further definition of the substrate specificity of the alpha-herpesvirus protein kinase and comparison with protein kinases A and C. Biochim. Biophys. Acta 1091426-431. [DOI] [PubMed] [Google Scholar]
- 28.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 947891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manning, B. D., and L. C. Cantley. 2007. AKT/PKB signaling: navigating downstream. Cell 1291261-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuzaki, A., Y. Yamauchi, A. Kato, F. Goshima, Y. Kawaguchi, T. Yoshikawa, and Y. Nishiyama. 2005. US3 protein kinase of herpes simplex virus type 2 is required for the stability of the UL46-encoded tegument protein and its association with virus particles. J. Gen. Virol. 861979-1985. [DOI] [PubMed] [Google Scholar]
- 31.McGeoch, D. J., and A. J. Davison. 1986. Alphaherpesviruses possess a gene homologous to the protein kinase gene family of eukaryotes and retroviruses. Nucleic Acids Res. 141765-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meignier, B., R. Longnecker, P. Mavromara-Nazos, A. E. Sears, and B. Roizman. 1988. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology 162251-254. [DOI] [PubMed] [Google Scholar]
- 33.Morris, J. B., H. Hofemeister, and P. O'Hare. 2007. Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 814429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mou, F., T. Forest, and J. D. Baines. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 816459-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munger, J., A. V. Chee, and B. Roizman. 2001. The U(S)3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 755491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munger, J., and B. Roizman. 2001. The US3 protein kinase of herpes simplex virus 1 mediates the posttranslational modification of BAD and prevents BAD-induced programmed cell death in the absence of other viral proteins. Proc. Natl. Acad. Sci. USA 9810410-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murata, T., F. Goshima, T. Daikoku, H. Takakuwa, and Y. Nishiyama. 2000. Expression of herpes simplex virus type 2 US3 affects the Cdc42/Rac pathway and attenuates c-Jun N-terminal kinase activation. Genes Cells 51017-1027. [DOI] [PubMed] [Google Scholar]
- 38.Murata, T., F. Goshima, Y. Nishizawa, T. Daikoku, H. Takakuwa, K. Ohtsuka, T. Yoshikawa, and Y. Nishiyama. 2002. Phosphorylation of cytokeratin 17 by herpes simplex virus type 2 US3 protein kinase. Microbiol. Immunol. 46707-719. [DOI] [PubMed] [Google Scholar]
- 39.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2087-90. [DOI] [PubMed] [Google Scholar]
- 40.Narayanan, K., R. Williamson, Y. Zhang, A. F. Stewart, and P. A. Ioannou. 1999. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 6442-447. [DOI] [PubMed] [Google Scholar]
- 41.Nozawa, N., Y. Kawaguchi, M. Tanaka, A. Kato, A. Kato, H. Kimura, and Y. Nishiyama. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J. Virol. 796947-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogg, P. D., P. J. McDonell, B. J. Ryckman, C. M. Knudson, and R. J. Roller. 2004. The HSV-1 Us3 protein kinase is sufficient to block apoptosis induced by overexpression of a variety of Bcl-2 family members. Virology 319212-224. [DOI] [PubMed] [Google Scholar]
- 43.Perkins, D., E. F. Pereira, M. Gober, P. J. Yarowsky, and L. Aurelian. 2002. The herpes simplex virus type 2 R1 protein kinase (ICP10 PK) blocks apoptosis in hippocampal neurons, involving activation of the MEK/MAPK survival pathway. J. Virol. 761435-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poon, A. P., L. Benetti, and B. Roizman. 2006. U(S)3 and U(S)3.5 protein kinases of herpes simplex virus 1 differ with respect to their functions in blocking apoptosis and in virion maturation and egress. J. Virol. 803752-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon, A. P., Y. Liang, and B. Roizman. 2003. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J. Virol. 7712671-12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poon, A. P., and B. Roizman. 2005. Herpes simplex virus 1 ICP22 regulates the accumulation of a shorter mRNA and of a truncated US3 protein kinase that exhibits altered functions. J. Virol. 798470-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purves, F. C., A. D. Deana, F. Marchiori, D. P. Leader, and L. A. Pinna. 1986. The substrate specificity of the protein kinase induced in cells infected with herpesviruses: studies with synthetic substrates [corrected] indicate structural requirements distinct from other protein kinases. Biochim. Biophys. Acta 889208-215. [DOI] [PubMed] [Google Scholar]
- 48.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 612896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 906701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 655757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rixon, F. J., and D. J. McGeoch. 1985. Detailed analysis of the mRNAs mapping in the short unique region of herpes simplex virus type 1. Nucleic Acids Res. 13953-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roux, P. P., and J. Blenis. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol Rev. 68320-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schumacher, D., B. K. Tischer, S. Trapp, and N. Osterrieder. 2005. The protein encoded by the US3 orthologue of Marek's disease virus is required for efficient de-envelopment of perinuclear virions and involved in actin stress fiber breakdown. J. Virol. 793987-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith, R. F., and T. F. Smith. 1989. Identification of new protein kinase-related genes in three herpesviruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J. Virol. 63450-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 771382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka, M., Y. Nishiyama, T. Sata, and Y. Kawaguchi. 2005. The role of protein kinase activity expressed by the UL13 gene of herpes simplex virus 1: the activity is not essential for optimal expression of UL41 and ICP0. Virology 341301-312. [DOI] [PubMed] [Google Scholar]
- 61.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 62.Tuazon, P. T., W. C. Spanos, E. L. Gump, C. A. Monnig, and J. A. Traugh. 1997. Determinants for substrate phosphorylation by p21-activated protein kinase (gamma-PAK). Biochemistry 3616059-16064. [DOI] [PubMed] [Google Scholar]
- 63.Van Minnebruggen, G., H. W. Favoreel, L. Jacobs, and H. J. Nauwynck. 2003. Pseudorabies virus US3 protein kinase mediates actin stress fiber breakdown. J. Virol. 779074-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 981895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yokoyama, A., M. Tanaka, G. Matsuda, K. Kato, M. Kanamori, H. Kawasaki, H. Hirano, I. Kitabayashi, M. Ohki, K. Hirai, and Y. Kawaguchi. 2001. Identification of major phosphorylation sites of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP): ability of EBNA-LP to induce latent membrane protein 1 cooperatively with EBNA-2 is regulated by phosphorylation. J. Virol. 755119-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296913-916. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20123-128. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, H. Y., H. Yamada, Y. M. Jiang, M. Yamada, and Y. Nishiyama. 1999. Intracellular localization of the UL31 protein of herpes simplex virus type 2. Arch. Virol. 1441923-1935. [DOI] [PubMed] [Google Scholar]