Abstract
Eukaryotic elongation factor 1 alpha 2 (eEF1A2) is a transforming gene product that is highly expressed in human tumors of the ovary, lung, and breast. eEF1A2 also stimulates actin remodeling, and the expression of this factor is sufficient to induce the formation of filopodia, long cellular processes composed of bundles of parallel actin filaments. Here, we find that eEF1A2 stimulates formation of filopodia by increasing the cellular abundance of cytosolic and plasma membrane-bound phosphatidylinositol-4,5 bisphosphate [PI(4,5)P2]. We have previously reported that the eEF1A2 protein binds and activates phosphatidylinositol-4 kinase III beta (PI4KIIIβ), and we find that production of eEF1A2-dependent PI(4,5)P2 and generation of filopodia require PI4KIIIβ. Furthermore, PI4KIIIβ is itself capable of activating both the production of PI(4,5)P2 and the creation of filopodia. We propose a model for extrusion of filopodia in which eEF1A2 activates PI4KIIIβ, and activated PI4KIIIβ stimulates production of PI(4,5)P2 and filopodia by increasing PI4P abundance. Our work suggests an important role for both eEF1A2 and PI4KIIIβ in the control of PI(4,5)P2 signaling and actin remodeling.
Filopodia are fingerlike projections from the plasma membrane that are the first cellular structures to reach new spaces during cell migration. Filopodia are composed of bundled actin filaments and actin-associated proteins (9, 13). Transmembrane receptors within filopodia respond to extracellular cues and guide directional movement toward chemoattractants (26). In addition, filopodia contain abundant adhesion molecules that regulate cellular attachment to growth substrates and cell-cell interactions (37). As such, filopodia regulate several key physiological processes, including cell migration, wound healing, and development. For example, filopodia are essential for neurogenesis in mice and for cell-cell adhesion during Drosophila melanogaster embryogenesis (9, 13).
We have previously described a role for eukaryotic elongation factor 1 alpha 2 (eEF1A2) in the initiation and maintenance of filopodia (1). In several types of mammalian cells, eEF1A2 expression is sufficient to stimulate formation of filopodia (1). eEF1A2 is one of two members of the eEF1A family of proteins, eEF1A1 and eEF1A2. During the elongation phase of protein synthesis, GTP-bound eEF1A proteins interact with amino-acylated tRNA and recruit them to the ribosome (18). While eEF1A1 and eEF1A2 are believed to have equivalent roles in protein translation, their tissue-specific expression patterns are each markedly different. eEF1A1 is expressed ubiquitously, whereas eEF1A2 is detectably expressed only in normal tissues of mammalian heart, brain, and skeletal muscle (24a, 24b, 28). Homozygous deletion of eEF1A2 occurs in the wasted mouse (7). These mice develop normally but suffer from neuromuscular abnormalities and immunodeficiency and die at approximately 1 month of age (35, 36). Importantly, eEF1A2 is likely to be a human oncogene, it is highly expressed, and its gene is amplified in 30 to 60% of human tumors of the breast, ovary, and lung (2, 24, 24a, 25, 40). eEF1A2 is transforming, and its expression in mammalian cells increases the cells’ in vitro growth rate, allows cells to grow in soft agar, and enhances cells’ tumorigenicity in xenograft models (2).
The mechanism by which eEF1A2 stimulates production of filopodia is currently unclear. The production of filopodia is regulated in major part by the plasma membrane abundance of phosphatidylinositol-4,5 bisphosphate [PI(4,5)P2] (9). PI(4,5)P2 cooperates with the WASP family of proteins and the Cdc42 GTPase to stimulate the ability of the Arp2-Arp3 complex to assemble actin (9). As such, proteins that control PI(4,5)P2 abundance are likely to have critical roles in controlling initiation of filopodia. Consistent with a role for eEF1A2 in phospholipid signaling, we have previously reported that eEF1A2 binds to the lipid kinase phosphatidylinositol-4 kinase III beta (PI4KIIIβ) and increases its kinase activity (21). PI4KIIIβ is a member of the PI4K family of lipid kinases that phosphorylates the D4 carbon of the inositol ring in phosphatidylinositol to yield phosphatidylinositol-4 phosphate (PI4P) (5, 17). PI4Ks are emerging as important mediators of cell physiology because PI4P is itself a regulatory phospholipid and additionally an obligate precursor for PI(4,5)P2 and PI(3,4,5)P3 (4, 29).
Here, we investigated whether eEF1A2 might activate production of filopodia through PI4KIIIβ. We find that eEF1A2 expression is sufficient to increase the cytoplasmic and plasma membrane abundance of PI(4,5)P2. This increase in plasma membrane PI(4,5)P2 is necessary for eEF1A2-induced filopodium formation. eEF1A2-mediated formation of filopodia and PI(4,5)P2 accumulation are both dependent on PI4KIIIβ. Cdc42 activation is also required for eEF1A2-induced formation of filopodia, and eEF1A2 expression is sufficient to activate Cdc42. Furthermore, we find that PI4KIIIβ expression is sufficient to induce formation of filopodia and localization of plasma membrane PI(4,5)P2. Our work is consistent with a model for filopodium production in which eEF1A2 stimulates production of filopodia through Cdc42 activation and a PI4KIIIβ-mediated increase in PI(4,5)P2 abundance. Our work also suggests an important regulatory role for PI4KIIIβ in controlling actin remodeling.
MATERIALS AND METHODS
Cell lines and vectors.
Rat2, BT549, and MCF7 cells were obtained from the American Type Culture Collection (Manassas, VA) and grown according to their instructions. MCF7 cell lines with stable eEF1A2 ablation were generated using a pSilencer Neo short interfering RNA (siRNA) expression vector (Ambion) with eEF1A2-directed siRNA, using sequences previously described (21). Bradykinin was purchased from Calbiochem (San Diego, CA). Generation of the eEF1A2 plasmid (eEF1A2-pcDNA3.1) is described in the report by Anand et al. (2). Generation of the adenoviral vectors (Ad-eEF1A2 and Ad-GFP) was described previously (1, 24). The PI4KIIIβ-expressing stable cell lines and the vector controls were generated using the pLXSN retroviral system described by Grignani et al. (12). PI4K111B, PM-FRB-CFP, mRFP-FKBP-5-ptase, and PLCδ-PH-GFP plasmids were a generous gift of T. Balla (41, 42).
Antibodies.
Generation of the rabbit polyclonal eEF1A2 antibody and its validation by Western blotting and immunohistochemistry were described previously (24). eEF1A2 expression was also detected using a V5-horseradish peroxidase (HRP) antibody (Invitrogen or Sigma). The PI4KIIIβ and goat anti-mouse, HRP-conjugated immunoglobulin G (IgG) antibodies were from Upstate Cell Signaling Solutions (Charlottesville, VA). Actin antibody was from Sigma (Oakville, Ontario), and HRP-conjugated anti-rabbit IgG was from Cell Signaling Technology (Danvers, MA). PI(4,5)P2 was visualized by using an anti-PI(4,5)P2 IgM antibody (Echelon Biosciences Inc., Salt Lake City, UT) and a goat anti-mouse IgM, (R)-phycoerythrin secondary antibody (Caltag Laboratories, Carlsbad, CA).
siRNA and transfections.
The sequence of the PI4KIIIβ siRNA is 5′-GGAGGUGUUGGA-GAAAGUCtt-3′. This siRNA and the negative control siRNA (catalog no. 4611) were purchased from Ambion (Austin, TX). siRNA transfections were performed using siPORT Lipid (Ambion) according to the manufacturer's instructions.
During the immunofluorescence experiments, parental Rat2 cells were triply transfected with the PLCδ-PH-GFP plasmid, with eEF1A2-pcDNA3.1, and either the negative control siRNA or the PI4KIIIβ siRNA. The eEF1A2 or vector plasmid was always at a 10-fold molar excess over that of the PLCδ-PH-GFP plasmid. When the Rat2 cells stably expressing eEF1A2 were used, the transfections were done using only the PLCδ-PH-GFP plasmid and the siRNA. For transfections, the total plasmid amount was 4 μg. When the stable cell lines were used for the siRNA experiments, cells were transfected with 3.7 μg of the PLCδ-PH-GFP plasmid and 20 nM of either the negative control siRNA or the PI4KIIIβ siRNA. To study the PI(4,5)P2 levels induced upon transient eEF1A2 or empty vector expression, Rat2 cells were transfected with the PLCδ-PH-GFP plasmid and either an eEF1A2-pcDNA3.1 or a pcDNA3.1 plasmid. For all transient eEF1A2 transfections, the eEF1A2 or vector plasmid was at a 10-fold molar excess over that of the other plasmids. All transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
Immunofluorescence.
Cells were plated in six-well plates containing coverslips. The next day, cells were fixed in 3.7% paraformaldehyde (15 min), permeabilized with 0.1% Triton-X (20 min), and blocked with 5% fetal bovine serum/phosphate-buffered saline (1 h; 37°C). Following staining, cells were mounted on slides, using fluorescence mounting medium (Dako Cytomation, Golstrup, Denmark). Actin was stained with either Alexa Fluor phalloidin 546 or Alexa Fluor 594 (Invitrogen). In experiments where eEF1A2 was analyzed, eEF1A2 was stained by using one of two methods. For the first method, we used an eEF1A2-specific rabbit polyclonal antibody (MCF7 cells, 1:100, overnight), followed by Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (1:300, 2 h, room temperature). In the second method, we used a monoclonal anti-V5 antibody (Rat2 cells, 1:1000, 1 h, room temperature), followed by Alexa Fluor 680 goat anti-mouse IgG secondary antibody (1:300, 1 h, room temperature). The PI(4,5)P2-specific antibody was used at 1:100 (overnight), followed by the goat anti-mouse IgM, (R)-phycoerythrin secondary antibody (1:500, 1 h, room temperature). All images were acquired using an Olympus FluoView FV1000 laser scanning confocal microscope. PI(4,5)P2 fluorescence was quantified with Olympus software (FV1000, version 01.04a; Center Valley, PA) and analyzed using GraphPad Prism version 4 software (San Diego, CA).
Phosphoinositide labeling.
The medium of near-confluent Rat2 cell cultures was removed and replaced with phosphate-free Dulbecco's modified Eagle's medium (Invitrogen) and labeled inorganic phosphate (40 μCi/ml; GE Healthcare, Piscataway, NJ) for 4 h. Following the incubation, cells were pelleted, and phospholipids were extracted with 40 μl of 1:1 methanol-water, 10 μl of salt-saturated NaCl, 2 μl of glacial acetic acid, and 40 μl of chloroform. The sample was vortexed vigorously and frozen at −20°C. Samples were thawed at room temperature, 40-μl aliquots of the organic phase were spotted onto precoated thin-layer chromatography (TLC) plates, and the plates were placed in the appropriate solvent system. The precoating protocol and details of the solvent system were described previously (21). The phosphatidylinositol-stained cells were visualized with storage phosphor screens (GE Healthcare), which were placed on top of the plates for a 24-h period. For the adenoviral experiments, Rat2 cells were incubated with the Ad-eEF1A2 or Ad-GFP virus (multiplicity of infection of 500) when they were 50 to 60% confluent for an overnight period. The next day, medium was replaced with phosphate-free medium, and the above-described procedure was followed.
In vitro PI4KIIIβ lipid kinase assay.
Total cellular protein (20 μl) from PI4KIIIβ or vector stable cells was added to 35 μl of kinase buffer (1 mM EDTA, 30 mM HEPES [pH 7.4], 100 mM NaCl, 2 mM MgCl2, and 0.2% Triton X-100) and 5 μl of 10 mM ATP containing 10 μCi of [32P]ATP. Following a 20-min incubation, the reactions were stopped by the addition of 60 μl of 1 N HCl. Phospholipids were then extracted by adding 160 μl of chloroform-methanol (1:2 [vol/vol]). After samples were vortexed briefly, they were centrifuged for 10 min at 10,000 × g. Aliquots of the organic phase were spotted onto precoated TLC plates, and the plates were placed in the same solvent system as that mentioned previously. Once complete, the plates were placed in a cassette and covered with a phosphor screen (GE Healthcare) for a 24-h period. The screen was then analyzed with a Storm 860 phosphorimager unit (GE Healthcare).
Generation of eEF1A2 mutants.
eEF1A2 mutant proteins were generated by using a PCR-mediated deletion protocol (14). Briefly, we used the eEF1A2pLXSN retroviral plasmid (1) as the template (50 ng) and Pfu Turbo DNA polymerase (Stratagene) with the appropriate primers (0.2 μM each), 1× Pfu buffer, and 0.2 mM of each deoxynucleoside triphosphate. The 5′-phosphorylated primers used were as follows: 5′-TCGGTGAAGGACATCGGTAAGCCTATCCCTAACCCTCTC-3′ (Δ321 to 464, forward); 5′-AGGGATAGGCTTACCGATGTCCTTCACCGACACGTTCTT-3′ (Δ321 to 464, reverse); 5′-ACAGAGCCGGCCTACGGTAAGCCTATCCCTAACCCTCTC-3′ (Δ163 to 464, forward); 5′-AGGGATAGGCTTACCGTAGGCCGGCTCTGTGGAGTCCAT-3′ (Δ163 to 464, reverse); 5′-GCGCCGGAATTCATGCGGCGGGGCAACGTGTGTGGGGAC-3′ (Δ1 to 320, forward); and 5′-CACGTTGCCCCGCCGCATGAATTCCGGCGCCTAGAGAAG-3′ (Δ1 to 320, reverse). The PCR was carried out as follows: denaturation at 95°C for 45 s, annealing at 43°C (Δ163 to 464 and Δ321 to 464) or 55°C (Δ1 to 320) for 45 s, and elongation at 72°C for 14 min (2 min/kb). Following the PCR, the 50-μl reaction product was DpnI digested for 1 to 2 h at 37°C and heat inactivated for 20 min at 80°C. One to two microliters of the treated reaction mixture were then transformed into competent cells. Rat2 cells stably expressing the mutants were generated as described previously (1).
Coimmunoprecipitation.
For coimmunoprecipitation, cells were grown to 80 to 95% confluence in 100-mm cell culture plates. Cells were lysed by sonication on ice in detergent-free buffer (137 mM NaCl, 8 mM KH2PO4 [pH 7.5], 2.7 mM KCl, 2.5 mM EDTA, 1% aprotinin, 1 mg/ml leupeptin, 50 mM NaF, 1 mM Na3VO4, 10 μg/ml pepstatin, 1 mM phenylmethylsulfonyl fluoride) and centrifuged at 10,000 × g for 20 min to remove membranes. Supernatants were collected, and protein levels were quantified by using a Bradford assay (Bio-Rad) according to the manufacturer's instructions. Total protein (250 μg) was precleared with protein A-agarose (Amersham Biosciences) for 1 h at 4°C. Following this, 2 to 4 μg of PI4KIIIβ antibody (rabbit polyclonal; Upstate Cell Signaling Solutions), anti-V5 agarose affinity gel (Sigma) or anti-FLAG affinity gel (Sigma) was added and incubated overnight at 4°C. Beads were washed three times in phosphate-buffered saline, centrifuged, and boiled for 5 min in sample buffer, and the supernatant was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The antibodies used to detect the proteins were mouse anti-PI4KIIIβ antibody (BD Biosciences), anti-V5-HRP (Invitrogen), and anti-FLAG-HRP (Sigma).
RESULTS
eEF1A2 regulates formation of filopodia.
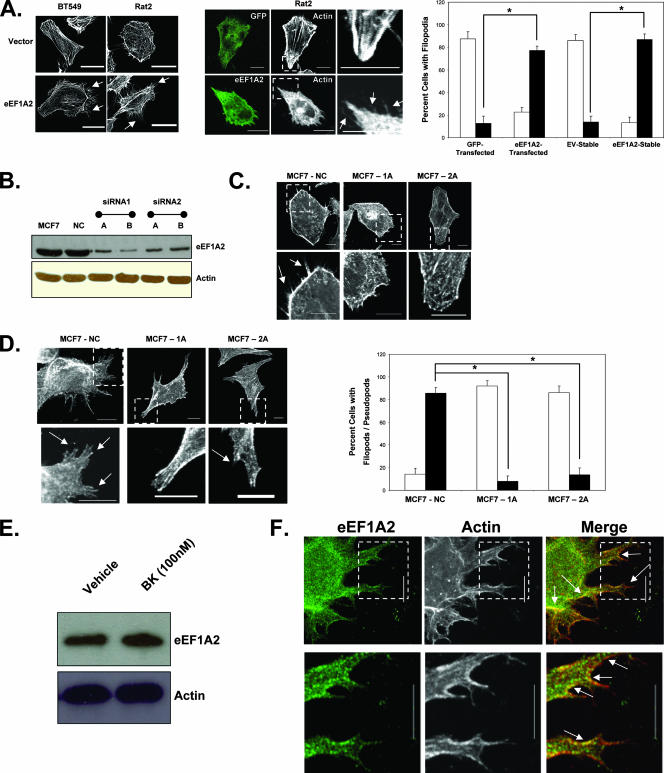
We have previously reported that eEF1A2 expression induces formation of filopodia in rodent and mammalian cell lines (1). For example, stable expression of eEF1A2 in the Rat2 fibroblast line and the BT549 breast carcinoma cell line visibly increases the number and size of filopodia (Fig. 1A). This also occurs in Rat2 cells transiently transfected with eEF1A2 (Fig. 1A). In general, eEF1A2 expression causes a significant, eight- to ninefold, increase in the number of cells with filopodia greater than 3 μm in length relative to that of empty vector controls (P < 0.05, Student's t test). To further investigate a role for eEF1A2 in production of filopodia, we studied the MCF7 human breast cancer cell line. These cells express abundant eEF1A2 message and protein (24). We generated MCF7 variants, in which endogenous eEF1A2 expression had been stably reduced by using RNA interference (Fig. 1B). While abundant filopodia are observed for control cells, their size and number are visibly reduced upon eEF1A2 ablation (Fig. 1C). To quantify the reduction in filopodia, the eEF1A2-deficient cells and control cell lines were incubated with bradykinin, a soluble stimulator of filopodium formation (23). After cells were incubated with 100 nM bradykinin, there was a significant, ninefold, decrease in the number of eEF1A2-ablated MCF7 cells with filopodia and pseudopodia relative to those of control MCF7 cells (Fig. 1D). Bradykinin has no effect on eEF1A2 protein levels (Fig. 1E). Thus, eEF1A2 likely has a physiological role in formation of filopodia. For the most part, the eEF1A2 protein is diffusely cytoplasmic but can be found within filopodia and also in the bases and edges of pseudopods, colocalizing there with actin (Fig. 1F).
FIG. 1.
eEF1A2 regulates formation of filopodia. (A) Left panel, BT549 and Rat2 cells stably expressing eEF1A2 have more filopodia (arrows) than do the empty vector control cells. Middle panel, transient transfection of eEF1A2 in Rat2 cells increases the number of filopodia (arrows) relative to those of GFP-transfected controls. Right panel, quantification of the number of cells with filopodia (n = 138, 152, 131 and 157, for GFP-transfected, eEF1A2-transfected, stable-EV, and stable-eEF1A2 cells, respectively). Filled columns indicate cells with at least 10 filopodia greater than or equal to 3 μm in length. Open columns indicate cells with any number of filopodia less than 3 μm in length. Significance (P < 0.05, Student's t test) is indicated by an asterisk. (B) Expression of eEF1A2 protein in MCF7 variants with stable downregulation of eEF1A2 by RNAi. NC, negative control. (C) MCF7 stable cell lines with control RNAi (NC) cells show abundant filopodia (arrows), while those showing constitutive eEF1A2 downregulation (1A and 2A) show little or none. (D) Left panel, MCF7 cells grown in the presence of 100 nM bradykinin. eEF1A2-deficient 1A and 2A cells show fewer filopodia and pseudopodia (arrows) than control cells. Right panel, quantification of filopodia/pseudopodia (n = 148, 165, and 154 for MCF7-NC, MCF7-1A and MCF7-2A cells, respectively). Filled columns indicate cells with at least 10 pseudopodia/filopodia greater than or equal to 5 μm in length. Open columns indicate cells with any number of filopodia/pseudopodia less than 5 μm in length. (E) eEF1A2 protein levels in MCF7 cells are not detectably altered upon bradykinin treatment. (F) MCF7 cells grown in the presence of bradykinin were stained for eEF1A2 and actin. eEF1A2 is found in filopodia and pseudopodia, as well as along their bases (arrows). Scale bars represent 10 μm.
eEF1A2 regulates development of filopodia through Cdc42 activation.
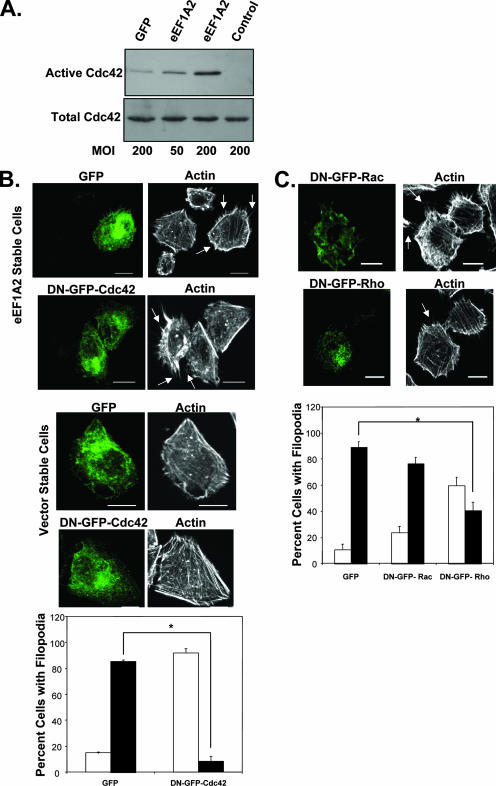
One of the key proteins that regulates formation of filopodia is the Rho family GTPase Cdc42 (9). Bradykinin, for example, stimulates filopodia through Cdc42 activation (23). eEF1A2 expression is sufficient to increase Cdc42 activity (Fig. 2A). In four separate experiments, eEF1A2 expression increased Cdc42 activity two- to eightfold relative to that of green fluorescent protein (GFP) controls. To investigate whether Cdc42 played a role in eEF1A2-dependent filopodium production, we inhibited Cdc42 activity in eEF1A2-expressing Rat2 cells by using dominant-negative GFP-tagged Cdc42 (DN-GFP-Cdc42) (39). Untransfected eEF1A2-expressing cells or those transfected with a GFP control plasmid showed many filopodia, but those expressing DN-GFP-Cdc42 showed a visibly reduced number of filopodia (Fig. 2B). Moreover, the filopodia that remained were reduced in length relative to those of controls. Overall, there was a significant, eightfold, decrease (P < 0.05, Student's t test) in the number of eEF1A2/DN-GFP-Cdc42-expressing cells with filopodia greater than 3 μm in length relative to those of eEF1A2/GFP-expressing control cells (Fig. 2B). DN-GFP-Cdc42 expression had no visible effect on the overall shape or cytoplasmic actin architecture in the vector control cell lines. To confirm the specificity of these effects for dominant-negative Cdc42, we repeated the experiments using dominant-negative Rac (DN-GFP-Rac) or a dominant-negative Rho (DN-GFP-Rho). Cells expressing DN-GFP-Rac showed no significant decrease in the size or length of filopodia relative to those of the GFP-expressing controls (Fig. 2C). However, there was a nearly twofold increase in the number of DN-GFP-Rho-expressing cells with filopodia greater than 3 μm in length compared to those of GFP controls (Fig. 2C). This is still fourfold more filopodia than in cells expressing DN-GFP-Cdc42 (Fig. 2B), suggesting that either eEF1A2 may regulate development of filopodia through a mechanism that is partially Rho dependent or that DN-Rho is interfering with Cdc42 function. Taken together, however, our findings indicate that eEF1A2 regulates filopodia primarily through Cdc42.
FIG. 2.
eEF1A2 regulates development of filopodia through Cdc42. (A) Cdc42 activation in cells transduced with a GFP adenovirus or an eEF1A2 adenovirus. Multiplicity of infection (MOI) is as indicated. Cdc42 activity, as measured by glutathione S-transferase (GST)-Pak1 precipitation, is increased in cells transduced with the eEF1A2 adenovirus in a dose-dependent manner. Total Cdc42 protein is derived from a whole-cell lysate of infected cells. The control is GST-rhotekin precipitation. (B) Expression of a dominant-negative GFP-Cdc42 (DN-GFP-Cdc42) in eEF1A2-expressing cells inhibits development of filopodia (arrows). Expression of DN-GFP-Cdc42 has no noticeable effect on the overall shape or actin architecture in vector control cells. Scale bars represent 10 μm. Bottom panel, quantification of filopodia after Cdc42 inhibition (n = 137 and 144 for GFP and DN-GFP-Cdc42 cells, respectively). Filled columns indicate cells with at least 10 filopodia greater than or equal to 3 μm in length. Open columns indicate cells with any number of filopodia less than 3 μm in length. Significance (P < 0.05, Student's t test) is indicated by an asterisk. (C) eEF1A2-expressing Rat2 cells transfected with a dominant-negative Rho (DN-GFP-Rho) or Rac (DN-GFP-Rac) and stained for actin. DN-GFP-Rac-expressing cells show filopodia similar to those of untransfected cells, but those expressing DN-GFP-Rho show a decrease. Bottom panel, quantification of filopodia after Rac or Rho inhibition (n = 125, 166, and 176 for GFP, DN-GFP-Rac, and DN-GFP-Rho cells, respectively). Filled columns indicate cells with at least 10 filopodia greater than or equal to 3 μm in length. Open columns indicate cells with any number of filopodia less than 3 μm in length. Significance (P < 0.05, Student's t test) is indicated by an asterisk. Scale bars represent 10 μm.
eEF1A2 stimulates PI(4,5)P2 accumulation and membrane localization.
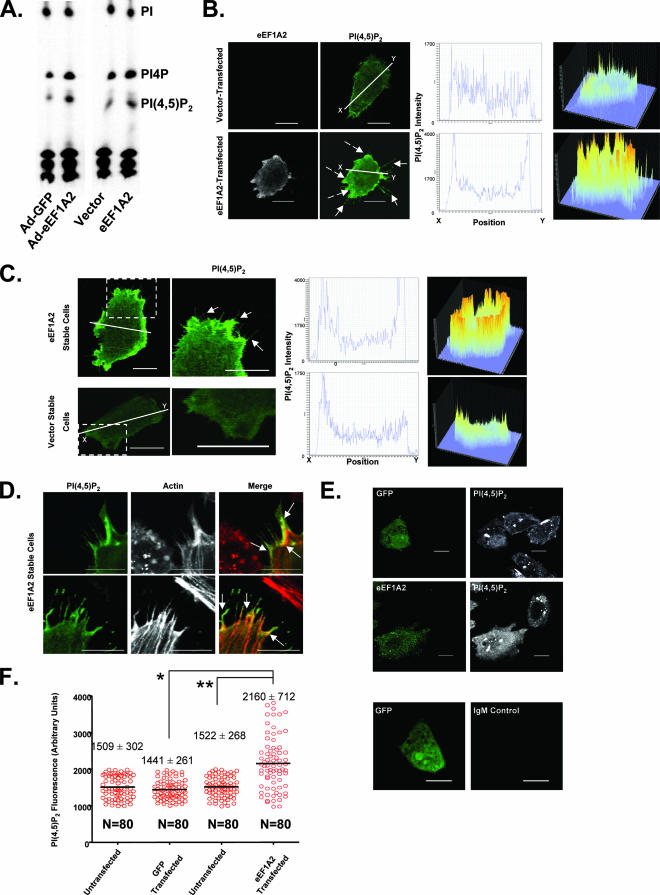
A major pathway for the production of filopodia is controlled through Cdc42 and the abundance of PI(4,5)P2 (9). Active Cdc42 and PI(4,5)P2 cooperate to activate actin nucleation by the Arp2/3 complex (19). We have previously found that eEF1A2 can activate PI4KIIIβ, leading to an increase in the intracellular abundance of PI4P (21), a precursor for PI(4,5)P2. We reasoned that eEF1A2 might be inducing filopodia through an increase in cellular PI(4,5)P2 abundance. To test this idea, we first used TLC to measure total PI(4,5)P2 levels in Rat2 cells transduced with an eEF1A2 adenovirus and in Rat2 cells stably expressing eEF1A2. We observed an increase in PI(4,5)P2 levels upon eEF1A2 expression relative to that of controls (2.1-fold ± 0.4-fold in adenovirally transduced cells and 1.5-fold ± 0.2-fold in the eEF1A2 stable cell line) (Fig. 3A). Because Cdc42 cooperates with membrane-bound PI(4,5)P2 during filopodium generation, we next investigated PI(4,5)P2 localization in eEF1A2-expressing cells. To this end, cells were transfected with a fluorescent PI(4,5)P2 reporter constructed from the fusion of the PI(4,5)P2-binding pleckstrin homology (PH) domain of phospholipase C delta (PLCδ) and GFP as described previously (41). PLCδ-PH-GFP binds to membrane-bound PI(4,5)P2. When Rat2 cells were cotransfected with eEF1A2 and PLCδ-PH-GFP, PI(4,5)P2 was observed to be prominent at the cell membrane (Fig. 3B). The staining in eEF1A2-expressing cells was visibly thicker and more intense than that found in control cells transfected with an empty vector and the reporter. Furthermore, the ratio of the PI(4,5)P2 staining in the membrane relative to that in the cytosol was greater in eEF1A2-expressing cells than in vector controls. A similar increase in membrane PI(4,5)P2 staining was observed for cell lines stably expressing eEF1A2 (Fig. 3C). PI(4,5)P2 can also be visualized within filopodium-like structures and at their bases (Fig. 3D). To further extend these studies, we used a PI(4,5)P2 antibody to quantify cytosolic PI(4,5)P2. This antibody does not detect PI(4,5)P2 at the cell membrane. We transiently transfected Rat2 cells with either GFP or eEF1A2 and then stained fixed cells with this antibody (Fig. 3E). Cells transfected with eEF1A2 showed visibly more PI(4,5)P2 staining than untransfected cells or those transfected with GFP. We quantified PI(4,5)P2 fluorescence intensity in individual eEF1A2-transfected and -untransfected cells, as well as in GFP-transfected and -untransfected cells (Fig. 3F). PI(4,5)P2 fluorescence intensity is significantly higher (P < 0.0001, Mann-Whitney U test) in eEF1A2-expressing cells (2,160 ± 712 fluorescence units) than in either untransfected (1,522 ± 268 fluorescence units) or GFP-transfected cells (1,441 ± 261 fluorescence units). Thus, eEF1A2 expression increases the abundance of both plasma membrane-bound and cytoplasmic PI(4,5)P2.
FIG. 3.
eEF1A2 stimulates overall accumulation and membrane localization of PI(4,5)P2. (A) TLC of labeled phosphatidylinositol from Rat2 cells transduced with either a GFP-adenovirus (Ad-GFP) or an eEF1A2-adenovirus (Ad-eEF1A2) or from vector only or eEF1A2-expressing stable cells. The TLC shown is a representative sample of three independent experiments. (B) Rat2 cells, transiently transfected with eEF1A2, show greater membrane-localized PI(4,5)P2 fluorescence (dashed arrows) than vector-transfected cells. The first and second columns (at left) show eEF1A2 and PI(4,5)P2 staining, respectively. The third column (middle) shows a cross-section of PI(4,5)P2 fluorescence intensity along the white line indicated in the second column. The fourth column (at right) shows a three-dimensional (3D) representation of PI(4,5)P2 staining in the entire cell. (C) eEF1A2-expressing Rat2 stable cells show greater membrane-bound PI(4,5)P2 than vector stable cells. First and second columns (at left) show PI(4,5)P2 staining. The third column (middle) shows a cross-section of PI(4,5)P2 fluorescence intensity along the line indicated in the first column. The fourth column (at right) shows a 3D representation of the PI(4,5)P2 staining pattern of cells seen in the first column. Filopodium-like structures are visible (solid arrows). (D) Colocalization of PI(4,5)P2 and actin in eEF1A2-expressing Rat2 cells. Some colocalization is observed along the bases of the filopodia, as well as along the filopodia (arrows). (E) Rat2 cells transfected with either eEF1A2 or GFP, stained for eEF1A2 (green) or GFP (green) and for PI(4,5)P2 (white). eEF1A2-transfected cells show more intense PI(4,5)P2 staining than either GFP-transfected cells or untransfected control cells. The bottom two panels show a GFP-transfected cell stained with the IgM secondary antibody alone. (F) Quantification of the PI(4,5)P2 fluorescence in the GFP control-transfected cells or the eEF1A2-transfected cells. Red circles represent fluorescence intensity in individual cells, with the means and standard deviations for each transfection condition indicated. eEF1A2-transfected cells have significantly more PI(4,5)P2 fluorescence than controls (* and **, P < 0.0001, Mann-Whitney test). Scale bars represent 10 μm.
eEF1A2-mediated formation of filopodia is dependent on PI(4,5)P2.
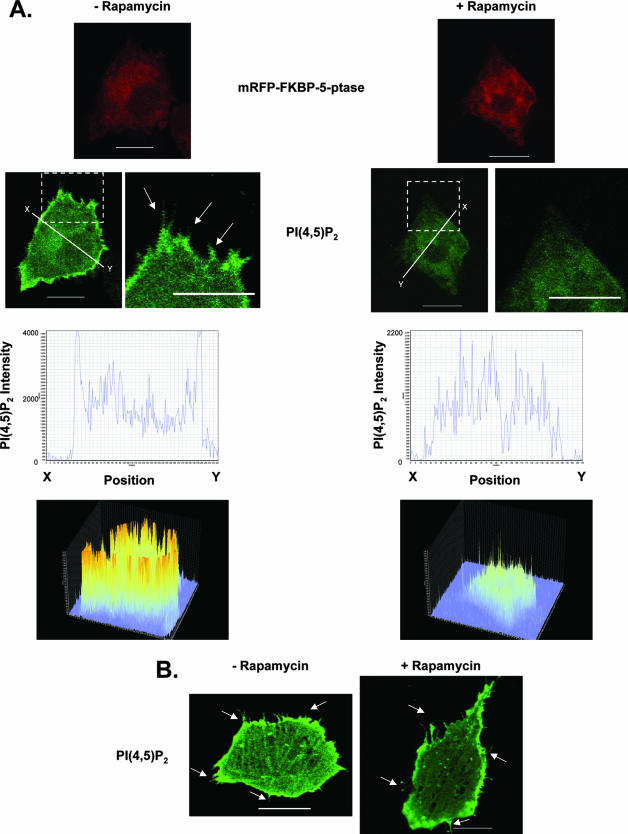
To determine whether the eEF1A2-mediated increase in PI(4,5)P2 levels was necessary for production of filopodia, we used a rapamycin-inducible system to decrease membrane-bound PI(4,5)P2. This system uses two plasmids, PM-FRB-CFP and mRFP-FKBP-5-ptase (42). The mRFP-FKBP-5-ptase contains the phosphoinositide-5 phosphatase domain of the inositol polyphosphate 5-phosphatase enzyme fused to the rapamycin-binding FKBP12 protein and the red fluorescent protein (mRFP). The PM-FRB-CFP is a fusion of the plasma membrane-bound FRB domain of mTOR (containing the palmitoylation sequence of the human GAP43 protein) and cyan fluorescent protein (CFP). Upon addition of rapamycin, mRFP-FKBP-5-ptase translocates to the membrane, where it heterodimerizes with the PM-FRB-CFP protein via the FRB domain. Once it is localized to the plasma membrane, the phosphatase in mRFP-FKBP-5-ptase depletes plasma membrane PI(4,5)P2 by converting it to PI4P. We transfected Rat2 cells stably expressing eEF1A2 with these plasmids (Fig. 4A). In the presence of rapamycin, the eEF1A2-expressing cells transfected with PM-FRB-CFP and mRFP-FKBP-5-ptase show visibly decreased plasma membrane-bound PI(4,5)P2 and a concomitant reduction in the number and length of filopodia relative to those of cells without rapamycin. Rapamycin treatment of eEF1A2-expressing cells without PM-FRB-CFP and mRFP-FKBP-5-ptase was unable to elicit changes in the appearance of filopodia or PI(4,5)P2 membrane localization in eEF1A2-expressing cells (Fig. 4B). Therefore, the induction of filopodia by eEF1A2 is dependent on plasma membrane PI(4,5)P2.
FIG. 4.
eEF1A2-mediated formation of filopodia is dependent on PI(4,5)P2. Rat2 cells stably expressing eEF1A2 were transfected with mRFP-FKBP-5-ptase, PM-FRB-CFP, and PLCδ-PH-GFP. (A) Addition of rapamycin (+Rapamycin) depletes the levels of membrane-bound PI(4,5)P2 and reduces production of filopodia (arrows). (B) Addition of rapamycin, without mRFP-FKBP-5-ptase and PM-FRB-CFP, has no effect on eEF1A2-dependent PI(4,5)P2 accumulation on the plasma membrane or on development of filopodia (arrows). Scale bars represent 10 μm.
eEF1A2-mediated formation of filopodia is dependent on eEF1A2 interaction with PI4KIIIβ.
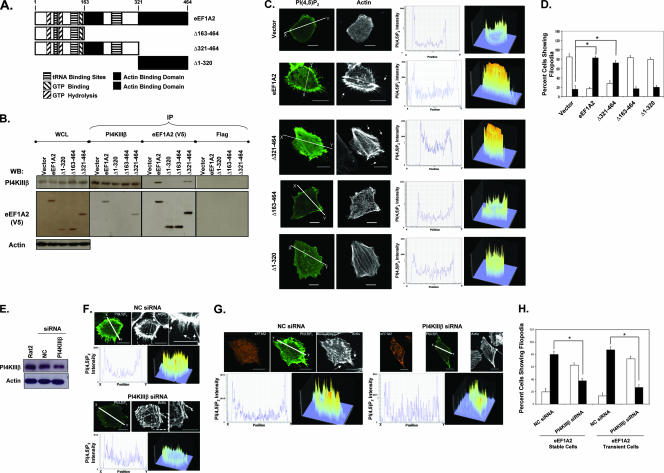
Thus far, we have determined that eEF1A2 can increase PI(4,5)P2 production and that membrane PI(4,5)P2 is required for eEF1A2-mediated formation of filopodia. Because we have previously shown that eEF1A2 can increase the cellular pool of PI4P by binding to and activating PI4KIIIβ (21), we next investigated the role of PI4KIIIβ in eEF1A2-dependent PI(4,5)P2 generation and production of filopodia. We first designed eEF1A2 variants that did not interact with PI4KIIIβ. Based on its homology with yeast and Dictyostelium eEF1A, the eEF1A2 protein contains domains for GTP binding, GTP hydrolysis, tRNA binding, and two putative actin binding domains (Fig. 5A). We generated mutants lacking amino acids 163 to 464, 321 to 464, and 1 to 320 (Δ163 to 464, Δ321 to 464, and Δ1 to 320) and tagged them with a C-terminal V5 epitope. We generated polyclonal cell lines expressing each of these proteins (protein expression levels in the polyclonal cell lines are shown in Fig. 5B). To determine which of these mutants interact with PI4KIIIβ, we performed coimmunoprecipitation assays with each of these cell lines. As shown in Fig. 5B, both the full-length eEF1A2 and the Δ321-to-464 mutant bind to PI4KIIIβ, but the Δ163-to-464 and Δ1-to-320 mutants do not. Thus, a putative PI4KIIIβ interaction domain is likely contained within eEF1A2 residues 163 to 320.
FIG. 5.
eEF1A2-PI4KIIIβ interaction is necessary for eEF1A2-mediated development of filopodia. (A) Schematic domain map of eEF1A2 and the generated mutants. (B) Polyclonal cell lines expressing eEF1A2 or mutants were used for coprecipitation analysis with PI4KIIIβ. Whole-cell lysate (WCL) lanes indicate expression of PI4KIIIβ and eEF1A2 in the cell lines used. eEF1A2 is V5 tagged, and actin is used as a loading control. Reciprocal PI4KIIIβ and eEF1A2 coimmunoprecipitation proteins were detected by immunoprecipitation (IP) with the indicated antibody, followed by the indicated Western blotting antibody. No precipitation of either protein was detected using the Flag antibody control. The WCL lane contains 40 μg of total cellular protein, and each IP was performed using 250 μg of protein lysate. (C) eEF1A2 mutants that cannot bind PI4KIIIβ do not show increased membrane-bound PI(4,5)P2 staining or any formation of filopodia. The first and second columns (at left) show PI(4,5)P2 and actin staining, respectively. The third column (middle) shows a cross-section of PI(4,5)P2 fluorescence intensity along the white line indicated in the first column. The fourth column (at right) shows a three-dimensional representation of PI(4,5)P2 staining in the entire cell. (D) Quantification of filopodia (n = 142, 158, 132, 133, and 135 for vector, eEF1A2, Δ163-to-464, Δ1-to-320, and Δ321-to-464 stable cells, respectively). Filled columns indicate cells with at least 10 filopodia greater than or equal to 3 μm in length. Open columns indicate cells with any number of filopodia less than 3 μm in length. Significance (P < 0.05, Student's t test) is indicated by an asterisk. (E) Western blots showing downregulation of PI4KIIIβ in Rat2 cells using siRNA. (F) Downregulation of PI4KIIIβ in eEF1A2-expressing Rat2 stable cells leads to a decrease in membrane-bound PI(4,5)P2 and a decrease in the number and length of filopodia. (G) Data shown are the same as in panel F, except Rat2 cells are transiently expressing eEF1A2. (H) Quantification of filopodia (n = 141, 157, 122, and 127 for negative control [NC; stable], PI4KIIIβ [stable], NC [transient], and PI4KIIIβ [transient], respectively). Filled columns indicate cells with at least 10 filopodia greater than or equal to 3 μm in length. Open columns indicate cells with any number of filopodia less than 3 μm in length. Significance (P < 0.05, Student's t test) is indicated by an asterisk. Scale bars represent 10 μm.
We next investigated whether these eEF1A2 mutants were competent for increasing PI(4,5)P2 or producing filopodia. As shown in Fig. 5C, both the full-length eEF1A2 and the Δ321-to-464 mutant activate production of filopodia and membrane PI(4,5)P2 accumulation, but the Δ163-to-464 and Δ1-to-320 mutants do not. Overall, the wild-type eEF1A2-expressing cells and the Δ321-to-464 mutant-expressing cells had a significant, ca. fourfold, increase (P < 0.05, Student's t test) in the number of cells showing filopodia greater than 3 μm in length (Fig. 5D). The Δ163-to-464 and Δ1-to-320 mutant-expressing cells were no different from vector controls with respect to filopodium appearance (Fig. 5D). Thus, eEF1A2 proteins that do not interact with PI4KIIIβ do not activate PI(4,5)P2 production or generate filopodia.
To further study the requirement for PI4KIIIβ interaction in eEF1A2-medated formation of filopodia, we used siRNA to decrease PI4KIIIβ protein levels in Rat2 cells (Fig. 5E). The siRNA decreased PI4KIIIβ protein levels by ∼60%. We transfected the eEF1A2-expressing Rat2 stable cell lines with PLCδ-PH-GFP to visualize PI(4,5)P2 production and either PI4KIIIβ or negative control siRNA (Fig. 5F). Cells with the PI4KIIIβ siRNA show a marked decrease in the intensity of PI(4,5)P2 fluorescence relative to that of the control siRNA-treated cells. Importantly, the number of cells with filopodia was similarly decreased. This attenuation was not absolute, however, likely because some PI4KIIIβ was still present in the cells. To confirm these results, we repeated this experiment with Rat2 cells cotransfected with the wild-type eEF1A2 and the siRNAs (Fig. 5G). As with the eEF1A2 stable cell lines, eEF1A2-expressing cells transfected with PLCδ-PH-GFP and the PI4KIIIβ siRNA had decreased filopodia compared to those with the control siRNA. In general, eEF1A2-expressing cells with ablated PI4KIIIβ protein levels had a significant, two- to threefold, decrease (P < 0.05, Student's t test) in the number of cells with filopodia greater than 3 μm in length (Fig. 5H).
PI4KIIIβ is sufficient for the formation of filopodia.
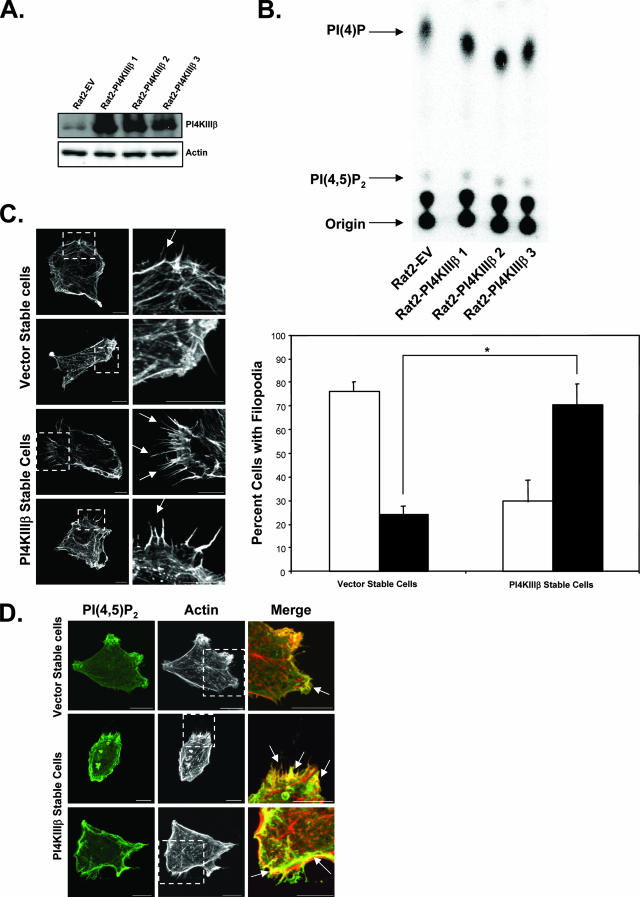
The data thus far suggest that PI4KIIIβ is necessary for eEF1A2-induced development of filopodia. We thus decided to determine whether PI4KIIIβ alone was sufficient to activate both the generation of PI(4,5)P2 and the production of filopodia. We generated Rat2 cell lines that stably overexpressed PI4KIIIβ (Fig. 6A). These cells have an ∼1.5-fold increase in PI4KIIIβ lipid kinase activity (Fig. 6B). Like eEF1A2-expressing cells, there is a significant, ca. fourfold, increase (P < 0.05, Student's t test) in the number of PI4KIIIβ-expressing cells with filopodia greater than 3 μm in length (Fig. 6C), as well as an increase in membrane-bound PI(4,5)P2 staining compared to that of vector controls (Fig. 6D). The PI(4,5)P2 also shows some degree of colocalization with the actin in the filopodia (Fig. 6D). PI4KIIIβ has previously been reported to be enriched in the trans-Golgi apparatus network and to have roles in vesicular trafficking (16, 30, 44). However, we have observed no visible changes in Golgi apparatus appearance in cells ectopically expressing eEF1A2 or PI4KIIIβ (not shown). Taken together, these results indicate that PI4KIIIβ is sufficient to increase PI(4,5)P2 levels and activate the development of filopodia.
FIG. 6.
PI4KIIIβ is sufficient to induce production of filopodia. (A) Western blots showing PI4KIIIβ overexpression in Rat2 stable cell lines. (B) Rat2 cells stably overexpressing PI4KIIIβ have greater PI4KIIIβ lipid kinase activity, as seen by an increase in the PI4P product. The TLC is a representative sample of three independent in vitro kinase assays. (C) Left panels, PI4KIIIβ-overexpressing Rat2 stable cells show more and longer filopodia than vector controls. Right panels, quantification of filopodia (n = 134 and 118 for vector stable cells and PI4KIIIβ stable cells, respectively). Filled columns indicate cells with at least 10 filopodia greater than or equal to 3 μm in length. Open columns indicate cells with any number of filopodia less than 3 μm in length. Significance (P < 0.05, Student's t test) is indicated by an asterisk. (D) PI4KIIIβ-overexpressing Rat2 stable cells show more membrane-bound PI(4,5)P2 than control cells. The PI(4,5)P2 localizes to the plasma membrane as well as to parts of filopodia. Scale bars represent 10 μm.
DISCUSSION
eEF1A2 is a transforming gene, highly expressed in ∼50% of breast, ovarian, and lung tumors (2, 24, 24a, 25, 40). eEF1A2 expression also stimulates actin remodeling and cell invasion and migration (1). Here, we propose a pathway of actin remodeling in which eEF1A2 stimulates the extrusion of filopodia through PI4KIIIβ and PI4KIIIβ-mediated accumulation of PI(4,5)P2. This leads to filopodium creation through a Cdc42-dependent pathway. We also identify a region within the eEF1A2 protein that is necessary for its interaction with PI4KIIIβ.
We have previously found that eEF1A2 increases cellular PI4P abundance in a PI4KIIIβ-dependent manner (21). Here we report that eEF1A2 expression increases PI(4,5)P2 abundance as well. eEF1A2 mutants that are unable to interact with PI4KIIIβ activate neither the generation of PI(4,5)P2 nor the production of filopodia. Furthermore, PI4KIIIβ RNA interference (RNAi) ablation attenuates the eEF1A2-mediated increase in PI(4,5)P2 abundance, and PI4KIIIβ expression is sufficient to increase PI(4,5)P2 levels at both the cytosol and the cell membrane. The simplest interpretation of these findings is that the increased PI(4,5)P2 abundance upon eEF1A2 expression is a direct consequence of the larger PI4P precursor pool created by PI4KIIIβ activation. The Km of mammalian PI4P5 kinases, the enzymes responsible for converting PI4P to PI(4,5)P2, are in the 10 to 45 μM range (20). Thus, an increase in PI4P concentration will stimulate PI(4,5)P2 accumulation wherever PI4P concentrations are in the magnitude of 10 μM or less. In eukaryotic cells, phosphatidylinositol concentrations range from 0.5 to 2.5 μmol/g (31), with an overall concentration of ∼10 to 500 pM in cells with a volume range of 500 to 3,000 μm3 (10). PI4P comprises ∼10 to 20% of total phosphatidylinositol, suggesting that the overall cellular PI4P abundance is in the 1 to 100 pM range. However, plasma membrane concentrations of PI4P may be much higher. Stephens et al., for example, estimated that PI4P concentrations in the inner plasma membranes of unstimulated neutrophils approach 3 mM (38). Here, we find that eEF1A2 expression doubles both the total cellular PI4P and the PI(4,5)P2 abundance in cells. Furthermore, increased levels of PI(4,5)P2 are distributed along the plasma membrane, as well as in the cytoplasm. In vitro, purified eEF1A2 doubles the Vmax of recombinant PI4KIIIβ (21). Thus, PI4P concentration is likely to be limiting in the generation of PI(4,5)P2 in our cell lines. Broadly consistent with this idea, previous reports have suggested the existence of an intracellular pool of PI(4,5)P2 in mammalian cells that was under the control of a wortmannin-sensitive PI4 kinase (32), later identified as PI4KIIIβ (6).
The mechanism of PI4KIIIβ activation by eEF1A2 is unknown. Because purified eEF1A2 increases the lipid kinase activity of PI4KIIIβ in vitro, it is likely that eEF1A2 induces a conformational change in PI4KIIIβ that increases its activity. The small GTPase Ras activates PI3K via a change in conformation, so this method of activation is not unprecedented (33). We have mapped the residues necessary for eEF1A2-PI4KIIIβ interaction to amino acid residues 163 to 320. It is likely that amino acid residues therein are likely to directly bind PI4KIIIβ and induce an as-yet-uncharacterized structural change in the protein that stimulates kinase activity.
The eEF1A2 protein is diffusely cytoplasmic (21, 24), whereas PI4KIIIβ is localized largely in the Golgi apparatus (3, 16, 44, 45). Thus, it was somewhat of a surprise to us that PI4KIIIβ can stimulate plasma membrane PI(4,5)P2 accumulation and activate the generation of filopodia there. Inactivation of PI4KIIIβ in both yeast and mammalian cells impairs Golgi structure and function (11, 16, 43). However we have not observed gross Golgi apparatus abnormalities in eEF1A2- or PI4KIIIβ-overexpressing cells (not shown). Furthermore, the increased cytosolic PI(4,5)P2 accumulation we observed with the PI(4,5)P2 antibody did not appear to be like the distribution observed for the Golgi apparatus. Since eEF1A2 or PI4KIIIβ expression stimulates production of filopodia, we favor the idea that the PI(4,5)P2 generated upon eEF1A2 or PI4KIIIβ expression has primary importance at the cell membrane. eEF1A2-PI4KIIIβ interaction, mediated through amino acid residues 163 to 320 in eEF1A2, and activation could occur on organelle membranes in the cytosol. The resulting PI4P would then be shuttled rapidly to the plasma membrane to generate filopodia. It may even be possible that the PI4P is converted to PI(4,5)P2, coincident with its transport to the plasma membrane. This scenario may explain why we observed a general increase in overall membrane-bound PI(4,5)P2 levels and not distinct pools of the lipid on the membrane. PI4KIIIβ's ability to affect the plasma membrane may not require substantial localization of the protein there. For example, PI4KIIIα is localized primarily to the endoplasmic reticulum in mammalian cells (8, 45), but it controls plasma membrane PI4P levels (5) and is part of the P2X7 ion channel (22). Thus, the interaction between eEF1A2 and PI4KIIIβ could occur away from the plasma membrane, even though the PI(4,5)P2 generated would be functional there. Alternatively, while eEF1A2 and PI4KIIIβ proteins are found in the cytoplasm, the interaction and activation of the kinase may occur transiently, near the plasma membrane.
While we propose that eEF1A2-mediated activation of PI4KIIIβ leads to PI(4,5)P2 accumulation due to a larger PI4P substrate pool, other possibilities do exist. For example, PI4P could also be activating PTEN, the 5′-phosphatase responsible for converting PI(3,4,5)P3 to PI(4,5)P2, thereby increasing PI(4,5)P2 abundance. Alternatively, PI4P could be directly activating a PI4P5K. Monophosphoinositide can influence the activity of other proteins involved in bi-PI and tri-PI generation. For example, Pendaries et al. have recently reported that an increase in cellular PI5P levels upon Shigella flexneri infection leads to the activation of a class 1A PI3-kinase, resulting in an increased level of active Akt, a downstream target of PI3-kinase (34a). Thus, it is possible that the generated pool of PI4P could be indirectly regulating PI(4,5)P2 levels.
We have previously reported that eEF1A2 expression can cause activation of the Akt serine threonine kinase (1). The Akt/PKB is a regulator of cell proliferation, insulin responsiveness, and apoptosis (27, 34). Akt activation is dependent, in major part, on plasma membrane levels of PI(3,4,5)P3. The mechanism by which eEF1A2 activates Akt is currently unknown, but we speculate that eEF1A2 activates Akt indirectly, since we have been unable to coimmunoprecipitate or colocalize eEF1A2 with Akt (not shown). We hypothesize that eEF1A2 can increase membrane abundance of PI(3,4,5)P3 in the same manner that it does with PI(4,5)P2.
The ability of PI4KIIIβ to increase production of filopodia also suggests that transcriptional or posttranscriptional control of PI4KIIIβ abundance could be an important facet of both actin remodeling and PI signaling control. To the best of our knowledge, however, no extracellular stimulus is known to increase the abundance of PI4KIIIβ message or protein. It is also unclear whether the ability of eEF1A2 to activate PI4KIIIβ and PI signaling contributes to its ability to transform cells in vitro or to enhance their tumorigenicity. Moreover, eEF1A2 might have effects on other aspects of cell physiology that depend on PI(4,5)P2. For example, hydrolysis of PI(4,5)P2 by phospholipase C yields diacylglycerol and inositol trisphosphate (IP3). IP3 is known to release calcium from its storage pools in the endoplasmic reticulum (15). Similarly, PI(4,5)P2 is known to mediate endocytosis/exocytosis and vesicular transport (42). Thus, eEF1A2 could have multiple effects on plasma membrane physiology.
There may be other pathways of eEF1A2-dependent actin remodeling; for example, purified eEF1A proteins bundle actin in vitro, and in Dictyostelium sp. they become localized in filopodia extended in response to cyclic AMP stimulation (7a, 7b). This suggests that direct actin bundling by eEF1A2 could also contribute to filopodium production. The localization of eEF1A2 to filopodium bases and within filopodia suggests that both actin bundling and phospholipid regulation may contribute to actin remodeling.
In summary, we have shown that eEF1A2 regulates the formation of filopodia through the binding and activation of PI4KIIIβ. This activation is required to generate a pool of cytosolic and membrane-bound PI(4,5)P2. The membrane PI(4,5)P2 then induces formation of filopodia through a Cdc42-dependent mechanism(s). Moreover, we also show that PI4KIIIβ itself can regulate formation of filopodia, a novel function for this enzyme. This work provides additional evidence for the link between protein translation, phosphatidylinositol signaling, actin remodeling, and oncogenesis.
Acknowledgments
We thank Heidi McBride, Dixie Pinke, Sanaa Noubir, and Zemin Yao for helpful discussions and critical review of the manuscript. We thank Heidi McBride and Astrid Schauss for use of and assistance with confocal microscopy. We thank T. Balla for the generous gift of many plasmids.
S.J. is supported by a studentship from the CIHR Institute of Gender and Health and the Ontario Women's Health Council. This work was supported by grants from the National Cancer Institute of Canada and by funds from the Canadian Cancer Society and the Canadian Institutes of Health Research.
Footnotes
Published ahead of print on 12 May 2008.
REFERENCES
- 1.Amiri, A., F. Noei, S. Jeganathan, G. Kulkarni, D. E. Pinke, and J. M. Lee. 2007. eEF1A2 activates Akt and stimulates Akt-dependent actin remodeling, invasion and migration. Oncogene 263027-3040. [DOI] [PubMed] [Google Scholar]
- 2.Anand, N., S. Murthy, G. Amann, M. Wernick, L. A. Porter, I. H. Cukier, C. Collins, J. W. Gray, J. Diebold, D. J. Demetrick, and J. M. Lee. 2002. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 31301-305. [DOI] [PubMed] [Google Scholar]
- 3.Balla, A., G. Vereb, H. Gulkan, T. Gehrmann, P. Gergely, L. M. Heilmeyer, Jr., and M. Antal. 2000. Immunohistochemical localisation of two phosphatidylinositol 4-kinase isoforms, PI4K230 and PI4K92, in the central nervous system of rats. Exp. Brain Res. 134279-288. [DOI] [PubMed] [Google Scholar]
- 4.Balla, T. 2005. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 1182093-2104. [DOI] [PubMed] [Google Scholar]
- 5.Balla, T. 1998. Phosphatidylinositol 4-kinases. Biochim. Biophys. Acta 143669-85. [DOI] [PubMed] [Google Scholar]
- 6.Balla, T., G. J. Downing, H. Jaffe, S. Kim, A. Zolyomi, and K. J. Catt. 1997. Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J. Biol. Chem. 27218358-18366. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, D. M., J. Peters, and C. M. Abbott. 1998. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1alpha, encoded by the Eef1a2 gene. Proc. Natl. Acad. Sci. USA 954463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Condeelis, J. 1995. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem. Sci. 20169-170. [DOI] [PubMed] [Google Scholar]
- 7b.Dharmawardhane, S., M. Demma, F. Yang, and J. Condeelis. 1991. Compartmentalization and actin binding properties of ABP-50: the elongation factor-1 alpha of Dictyostelium. Cell Motil. Cytoskeleton 20279-288. [DOI] [PubMed] [Google Scholar]
- 8.Ekblad, L., and B. Jergil. 2001. Localization of phosphatidylinositol 4-kinase isoenzymes in rat liver plasma membrane domains. Biochim. Biophys. Acta 1531209-221. [DOI] [PubMed] [Google Scholar]
- 9.Faix, J., and K. Rottner. 2006. The making of filopodia. Curr. Opin. Cell Biol. 1818-25. [DOI] [PubMed] [Google Scholar]
- 10.Frame, K. K., and W.-S. Hu. 1990. Cell volume measurement as an estimation of mammalian cell biomass. Biotechnol. Bioeng. 36191-197. [DOI] [PubMed] [Google Scholar]
- 11.Godi, A., A. Di Campli, A. Konstantakopoulos, G. Di Tullio, D. R. Alessi, G. S. Kular, T. Daniele, P. Marra, J. M. Lucocq, and M. A. De Matteis. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6393-404. [DOI] [PubMed] [Google Scholar]
- 12.Grignani, F., T. Kinsella, A. Mencarelli, M. Valtieri, D. Riganelli, F. Grignani, L. Lanfrancone, C. Peschle, G. P. Nolan, and P. G. Pelicci. 1998. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 5814-19. [PubMed] [Google Scholar]
- 13.Gupton, S. L., and F. B. Gertler. 2007. Filopodia: the fingers that do the walking. Sci. STKE 2007re5. [DOI] [PubMed] [Google Scholar]
- 14.Hansson, M. D., K. Rzeznicka, M. Rosenback, M. Hansson, and N. Sirijovski. 2008. PCR-mediated deletion of plasmid DNA. Anal. Biochem. 375373-375. [DOI] [PubMed] [Google Scholar]
- 15.Haucke, V. 2005. Phosphoinositide regulation of clathrin-mediated endocytosis. Biochem. Soc. Trans. 331285-1289. [DOI] [PubMed] [Google Scholar]
- 16.Hausser, A., P. Storz, S. Martens, G. Link, A. Toker, and K. Pfizenmaier. 2005. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat. Cell Biol. 7880-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heilmeyer, L. M., Jr., G. Vereb, Jr., G. Vereb, A. Kakuk, and I. Szivak. 2003. Mammalian phosphatidylinositol 4-kinases. IUBMB Life 5559-65. [DOI] [PubMed] [Google Scholar]
- 18.Hershey, J. W. 1991. Translational control in mammalian cells. Annu. Rev. Biochem. 60717-755. [DOI] [PubMed] [Google Scholar]
- 19.Higgs, H. N., and T. D. Pollard. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70649-676. [DOI] [PubMed] [Google Scholar]
- 20.Ishihara, H., Y. Shibasaki, N. Kizuki, T. Wada, Y. Yazaki, T. Asano, and Y. Oka. 1998. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem. 2738741-8748. [DOI] [PubMed] [Google Scholar]
- 21.Jeganathan, S., and J. M. Lee. 2007. Binding of elongation factor eEF1A2 to phosphatidylinositol 4-kinase beta stimulates lipid kinase activity and phosphatidylinositol 4-phosphate generation. J. Biol. Chem. 282372-380. [DOI] [PubMed] [Google Scholar]
- 22.Kim, M., L. H. Jiang, H. L. Wilson, R. A. North, and A. Surprenant. 2001. Proteomic and functional evidence for a P2X7 receptor signalling complex. EMBO J. 206347-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozma, R., S. Ahmed, A. Best, and L. Lim. 1995. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol. Cell. Biol. 151942-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulkarni, G., D. A. Turbin, A. Amiri, S. Jeganathan, M. A. Andrade-Navarro, T. D. Wu, D. G. Huntsman, and J. M. Lee. 2007. Expression of protein elongation factor eEF1A2 predicts favorable outcome in breast cancer. Breast Cancer Res. Treat. 10231-41. [DOI] [PubMed] [Google Scholar]
- 24a.Lee, J. M. 2003. The role of protein elongation factor eEF1A2 in ovarian cancer. Reprod. Biol. Endocrinol. 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24b.Lee, S., L. A. Wolfraim, and E. Wang. 1993. Differential expression of S1 and elongation factor-1 alpha during rat development. J. Biol. Chem. 26824453-24459. [PubMed] [Google Scholar]
- 25.Li, R., H. Wang, B. N. Bekele, Z. Yin, N. P. Caraway, R. L. Katz, S. A. Stass, and F. Jiang. 2006. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene 252628-2635. [DOI] [PubMed] [Google Scholar]
- 26.Lidke, D. S., K. A. Lidke, B. Rieger, T. M. Jovin, and D. J. Arndt-Jovin. 2005. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J. Cell Biol. 170619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LoPiccolo, J., C. A. Granville, J. J. Gills, and P. A. Dennis. 2007. Targeting Akt in cancer therapy. Anticancer Drugs 18861-874. [DOI] [PubMed] [Google Scholar]
- 28.Lund, A., S. M. Knudsen, H. Vissing, B. Clark, and N. Tommerup. 1996. Assignment of human elongation factor 1alpha genes: EEF1A maps to chromosome 6q14 and EEF1A2 to 20q13.3. Genomics 36359-361. [DOI] [PubMed] [Google Scholar]
- 29.McLaughlin, S., and D. Murray. 2005. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438605-611. [DOI] [PubMed] [Google Scholar]
- 30.Meyers, R., and L. C. Cantley. 1997. Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J. Biol. Chem. 2724384-4390. [DOI] [PubMed] [Google Scholar]
- 31.Michell, R. H. 1975. Inositol phospholipids and cell surface receptor function. Biochim. Biophys. Acta 41581-147. [DOI] [PubMed] [Google Scholar]
- 32.Nakanishi, S., K. J. Catt, and T. Balla. 1995. A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc. Natl. Acad. Sci. USA 925317-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pacold, M. E., S. Suire, O. Perisic, S. Lara-Gonzalez, C. T. Davis, E. H. Walker, P. T. Hawkins, L. Stephens, J. F. Eccleston, and R. L. Williams. 2000. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103931-943. [DOI] [PubMed] [Google Scholar]
- 34.Parcellier, A., L. A. Tintignac, E. Zhuravleva, and B. A. Hemmings. 2007. PKB and the mitochondria: AKTing on apoptosis. Cell Signal. 2021-30. [DOI] [PubMed] [Google Scholar]
- 34a.Pendaries, C., H. Tronchere, L. Arbibe, J. Mounier, O. Gozani, L. Cantley, M. J. Fry, F. Gaits-Iacovoni, P. J. Sansonetti, and B. Payrastre. 2006. PtdIns(5)P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. Embo J. 251024-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potter, M., A. Bernstein, and J. M. Lee. 1998. The wst gene regulates multiple forms of thymocyte apoptosis. Cell Immunol. 188111-117. [DOI] [PubMed] [Google Scholar]
- 36.Shultz, L. D., H. O. Sweet, M. T. Davisson, and D. R. Coman. 1982. “Wasted”, a new mutant of the mouse with abnormalities characteristic to ataxia telangiectasia. Nature 297402-404. [DOI] [PubMed] [Google Scholar]
- 37.Steketee, M. B., and K. W. Tosney. 2002. Three functionally distinct adhesions in filopodia: shaft adhesions control lamellar extension. J. Neurosci. 228071-8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephens, L. R., T. R. Jackson, and P. T. Hawkins. 1993. Agonist-stimulated synthesis of phosphatidylinositol(3,4,5)-trisphosphate: a new intracellular signalling system? Biochim. Biophys. Acta 117927-75. [DOI] [PubMed] [Google Scholar]
- 39.Subauste, M. C., M. Von Herrath, V. Benard, C. E. Chamberlain, T. H. Chuang, K. Chu, G. M. Bokoch, and K. M. Hahn. 2000. Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J. Biol. Chem. 2759725-9733. [DOI] [PubMed] [Google Scholar]
- 40.Tomlinson, V. A., H. J. Newbery, N. R. Wray, J. Jackson, A. Larionov, W. R. Miller, J. M. Dixon, and C. M. Abbott. 2005. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varnai, P., X. Lin, S. B. Lee, G. Tuymetova, T. Bondeva, A. Spat, S. G. Rhee, G. Hajnoczky, and T. Balla. 2002. Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J. Biol. Chem. 27727412-27422. [DOI] [PubMed] [Google Scholar]
- 42.Varnai, P., B. Thyagarajan, T. Rohacs, and T. Balla. 2006. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol. 175377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walch-Solimena, C., and P. Novick. 1999. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1523-525. [DOI] [PubMed] [Google Scholar]
- 44.Weixel, K. M., A. Blumental-Perry, S. C. Watkins, M. Aridor, and O. A. Weisz. 2005. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J. Biol. Chem. 28010501-10508. [DOI] [PubMed] [Google Scholar]
- 45.Wong, K., R. Meyers dd, and L. C. Cantley. 1997. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 27213236-13241. [DOI] [PubMed] [Google Scholar]