Abstract
Shortly after the synthesis of the two cells required for sporulation in Bacillus subtilis, the membranes of the larger mother cell begin to migrate around and engulf the smaller forespore cell. At the completion of this process the leading edges of the migrating membrane meet and fuse, releasing the forespore into the mother cell cytoplasm. We developed a fluorescent membrane stain-based assay for this membrane fusion event, and we isolated mutants defective in the final stages of engulfment or membrane fusion. All had defects in spoIIIE, which is required for translocation of the forespore chromosome across the polar septum. We isolated one spoIIIE mutant severely defective in chromosome translocation, but not in membrane fusion; this mutation disrupts the ATP/GTP-binding site of SpoIIIE, suggesting that ATP binding and hydrolysis are required for DNA translocation but not for the late engulfment function of SpoIIIE. We also correlated relocalization of SpoIIIE-green fluorescent protein from the sporulation septum to the forespore pole with the completion of membrane fusion and engulfment. We suggest that SpoIIIE is required for the final steps of engulfment and that it may regulate or catalyze membrane fusion events.
An early step in the Bacillus subtilis sporulation pathway, asymmetric cell division, creates the small forespore and larger mother cell. Soon after this polar division, mother cell- and forespore-specific σ factors direct the transcription of genes required for the next stage of sporulation, engulfment, during which the mother cell membranes move up and around the forespore, finally meeting and fusing, thereby releasing the forespore into the mother cell cytoplasm (see Fig. 1A). After engulfment, the spore completes development within the mother cell and is released after mother cell lysis (reviewed in ref. 1). Five proteins have been implicated in engulfment, four of which, SpoIID, SpoIIM, SpoIIP, and SpoIIB, are involved in degradation of septal peptidoglycan at the onset of engulfment [see Fig. 1A, stages IIi to IIii (2–5)]. Mutants lacking SpoIID, SpoIIM, or SpoIIP fail to initiate membrane migration, and the forespore breaks through the septum, bulging into the mother cell. Similarly, spoIIB mutants initially show bulging of the forespore into the mother cell; however, they are able to degrade the septal peptidoglycan in an uncoordinated manner and complete engulfment, producing viable spores (A. R. Perez, personal communication). The fifth protein, SpoIIQ, appears to function during the late stages of engulfment (6), although more recent work suggests that SpoIIQ is only conditionally required for engulfment (Y. L. Sun, personal communication).
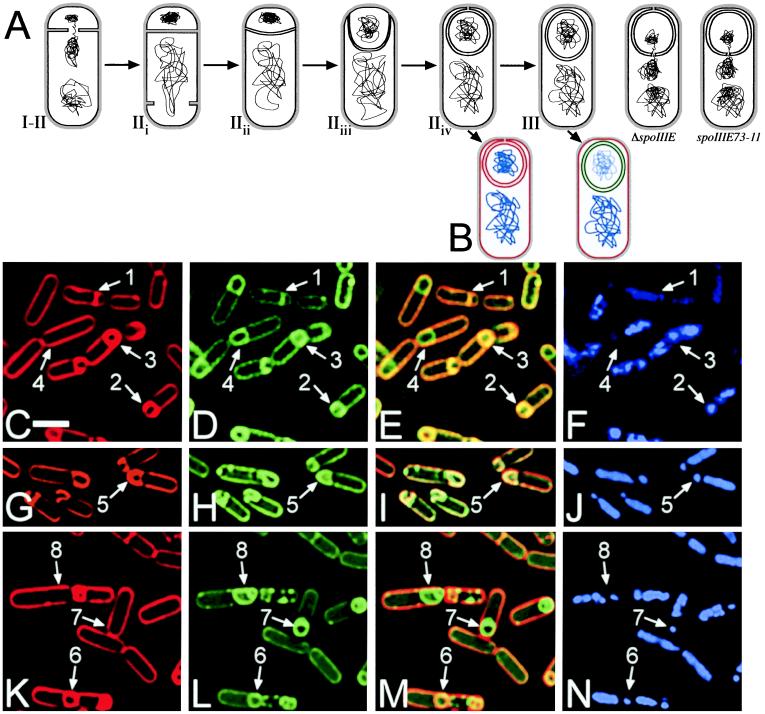
Figure 1.
(A) Stages of engulfment, showing completion of chromosome translocation and polar septation (I–IIi), early engulfment (IIii), membrane movement across the cell pole (IIiii), and a stage of engulfment newly defined here, mother cell membrane fusion (IIiv). The two cells at the right illustrate the proposed defects in ΔspoIIIE and spoIIIE73-11. (B) Schematic of the membrane fusion assay. Red lines represent membranes stained with FM 4-64 and MTG, green lines indicate membranes stained with MTG only, and blue lines show DAPI staining of chromosomes. (C–N) Sporangia of PY79 (C–F), KP141 (G–J), and KP541 (K–N) stained with FM 4-64 (C, G, and K), MTG (D, H, and L), and DAPI (F, J, and N) 3 hr after the initiation of sporulation (t3); E, I, and M are an overlay of FM 4-64 and MTG staining. Arrows 1 and 2 show cells at stage IIi and IIii–iii respectively. Arrows 3, 5, and 6 point to cells at stage IIiv which have completed membrane migration but have not yet completed membrane fusion as shown by the FM 4-64 (C, G, and K) and MTG (D, H, and L) staining. Finally, arrows 4, 7, and 8 indicate sporangia that have completed membrane movement and fusion; only the mother cell membranes stain with FM 4-64 (C and K), but MTG stains both the forespore and mother cell membranes (D and L). The reduced forespore chromosome content of spoIIIE mutants can be seen in N and J (arrows 5–8). F shows that after the completion of membrane fusion the forespore chromosome stains less intensely with DAPI in wild-type sporangia (compare arrows 3 and 4). (White bar in C is 2 μm.)
We performed genetic and visual screening to isolate additional engulfment mutants. The visual screening relied on the fluorescent stain FM 4-64, which allows visualization of membrane movement throughout engulfment (7), and which provides an assay for membrane fusion at the completion of engulfment (described below). Our screen yielded a variety of engulfment mutants, but we were particularly interested in those that had very late engulfment or membrane fusion defects. These mutants could give us new insight into late stages of membrane migration and also into how engulfment is coupled to the activation of postengulfment transcription factors (1). Finally, in contrast to eukaryotic systems, little is known about the mechanism of membrane fusion in bacteria, a process that might be elucidated by such late engulfment mutants. We isolated several mutants defective in the very last stages of membrane migration or in membrane fusion, all of which mapped to the well-characterized spoIIIE gene.
SpoIIIE is one of a family of conserved proteins including Escherichia coli FtsK, a bifunctional protein essential for both cell division and chromosome segregation (8–11). SpoIIIE is required to translocate the forespore chromosome across the polar septum during sporulation (12) and, like FtsK, can free chromosomes trapped in vegetative septa (13). We present evidence of a second, distinct, function for SpoIIIE in the final steps of engulfment or membrane fusion.
Methods
Strains, Growth Conditions, and Membrane Fusion Assay.
Sporulation was induced by resuspension (14) unless otherwise noted. In the fusion assay, 0.5 ml of sporulating cells were pelleted and resuspended in 0.11 ml of the original culture medium; 2 μl of this cell suspension was placed on a slide and mixed with 1 μl of a stain mix containing 5 μg/ml FM 4-64, 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), and 30 μg/ml MitoTracker Green FM (MTG) (all from Molecular Probes) diluted in sporulation salts. The cells were immobilized and images were collected with an optical sectioning, deconvolution microscope using standard rhodamine, DAPI, and fluorescein filter sets, as described (7). Strains used for this work are PY79 (15) derivatives: KP92 (spoIIIE36) (12), KP141 (ΔspoIIIE∷spc) (16), KP158 (ΔspoIIIE∷neo), KP209 (thrC∷sspE(2G)lacZΩtet; a gift of P. Stragier, Institute Pasteur), KP211 (ΔspoIIIE∷spc, thrC∷ sspE(2G)lacZΩtet), KP213 (spoIIIE36, thrC∷sspE(2G)lacZΩtet), KP502 (cotC∷spc), KP503 (spoIIIG+Ωspc), KP527 (Δ(spo0J-soj)∷spc), KP537 (spoIIIE73-11-gfpΩspc), KP538 (spoIIIE36-gfpΩspc), KP539 (spoIIIE73-11, thrC∷ sspE(2G)lacZΩtet), KP540 (ΔspoIIIE∷neo, Δ(spo0-soj)∷spc), KP541 (spoIIIE73-11), KP542 (spoIIIE-gfpΩspc), KP554 (sspB-lacZΩcat, cotE-gfpΩkan) (17, 18), KP555 (thr∷cotD-lacZΩmls, cotE-gfpΩkan; the lacZ fusion was a gift of C. D. Webb and R. Losick), KP556 (thr∷cotD-lacZΩmls, cotE-gfpΩkan, amyE∷cat), KP557 (thr∷cotD-lacZΩmls, cotE-gfpΩkan, amyE∷cat, spoIIIE∷spc). E. coli strain KJ622 (TG1, pcnB uvc24-1) was used for cloning.
Mutagenesis and Screening.
KP554 and KP555 cells were grown in LB to an OD600 = 0.5 then mutagenized at 30°C for 30 min with nitrosoguanidine (80 μg/ml) as described in ref. 19, except that cells were washed with citrate buffer and allowed to recover in LB at 37°C for 1 hr, yielding ≈50% killing. The cells were plated at a density of 300–400 colonies per plate on DS medium (Difco) containing 40 μg/ml magenta phosphate and 60 μg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) and incubated for 1–2 days at 37°C, and colonies that produced alkaline phosphatase (which were pink) but failed to activate σG or σK (which were not blue) were purified on LB. The plate phenotypes were confirmed, and each mutant was sporulated by nutrient exhaustion in overnight DS liquid cultures at 30°C and 37°C, stained, and microscopically screened as described in Results and Discussion. Mutations of interest were congressed (see below) into an unmutagenized background before further characterization (20).
Mapping of spoIIIE Mutations.
Phage PBS1 transducing lysates were grown on KP502 and KP503 as described (20). These strains carry spectinomycin resistance markers at 168° and 135° on the B. subtilis chromosome, respectively, which flank spoIIIE (142°). Membrane-fusion-defective mutants were transduced with these lysates, selecting for spectinomycin resistance. All yielded Spo+ transductants when transduced with these phage, indicating that the mutations were near spoIIIE.
We confirmed that the mutations were in the spoIIIE gene by exploiting the phenomenon of congression, whereby B. subtilis cells, during transformation, will incorporate unselected DNA into their chromosome, at low frequency, in addition to an unlinked selected marker. We transformed fusion-defective mutants with either KP556 (amyE∷cat) or KP557 (amyE∷cat, ΔspoIIIE∷spc) chromosomal DNA, selected for chloramphenicol resistance, and screened for Spo+ transformants. With KP556 donor DNA, 5–10% of transformants were Spo+, indicating that the wild-type gene had been introduced by congression and was able to rescue the sporulation defect. However, with KP557 (ΔspoIIIE∷spc) donor DNA no Spo+ transformants occurred, indicating that the donor DNA was unable to rescue the sporulation defect, suggesting that all of the mutants had mutations in spoIIIE. Similar rescue experiments with other spo mutants (i.e., spoIID) showed an identical frequency of rescue with both donor DNA preparations. Finally, the entire spoIIIE73-11 gene was sequenced by using automated fluorescent sequencing, from a PCR template produced with the primers GCC TGT AAA CGG CTG CTT TC and GCA TCG GCT CGA GAG AAG AGA GCT CAT CAT ATT TCT C, which amplify the spoIIIE gene, revealing a single G-to-A change at nucleotide 1399, numbered from the first base of the spoIIIE initiation codon.
Construction of SpoIIIE-Green Fluorescent Protein (GFP) Fusion Protein.
We created our SpoIIIE-GFP fusion protein by using the vector pPL52 (gift of P. Levin, Massachusetts Institute of Technology), which contains a spectinomycin-resistance gene for selection in B. subtilis, and unique EcoRI and XhoI restriction sites in front of the gfp gene (mutant S58T). We amplified 500 bp at the 3′ end of spoIIIE by using the PCR primers CCT GTG GAG AAT TCC GTT CAG CGT ATC TTC ACA GA and GCA TCG GCT CGA GAG AAG AGA GCT CAT CAT ATT TCT C (boldface indicates the two restriction sites). This fragment, when ligated into pPL52, replaced the stop codon of spoIIIE with the in-frame gfp gene, encoding a SpoIIIE-GFP fusion protein. This construct was integrated onto the B. subtilis chromosome by a single recombination replacing spoIIIE with spoIIIE-gfp. We confirmed that spoIIIE-gfp produced a functional gene product by performing a fusion assay during sporulation and by assaying for heat-resistant spores (not shown). GFP fusions to the mutants spoIIIE36 and spoIIIE73-11 were made by transforming mutant strains with wild-type spoIIIE-gfp chromosomal DNA, selecting for spectinomycin resistance, screening for the rare Spo− colonies, and confirming that these had the same membrane fusion phenotype as the original strains.
Visualization of SpoIIIE-GFP with FM 4-64.
FM 4-64 fluoresces faintly green in addition to its strong red fluorescence, so we could not add FM 4-64 to cell suspensions when visualizing SpoIIIE-GFP. To perform the FM 4-64-based membrane fusion assay at the same time we followed SpoIIIE localization, we first collected images of SpoIIIE-GFP and chromosomes, then performed the membrane fusion assay on the slide. We stained cells with DAPI at 0.3 μg/ml and immobilized them with a poly(l-lysine)-treated coverslip placed slightly off center so that approximately 2–3 mm of the coverslip projected from the edge of the slide. When this slide was then mounted on an inverted microscope the coverslip served as a ledge to which we could add 5 μl of a stain mix containing 5 μg/ml FM 4-64 and 30 μg/ml MTG. This mixture required 5–15 min to diffuse under the coverslip and stain the cells.
Preparation and Staining for Immunofluorescence.
Cells were essentially treated as described (16) except that we used 0.5 mg/ml lysozyme for permeabilization, polyclonal rabbit anti-GFP primary antibodies (CLONTECH, 1:10,000 dilution), and Oregon green-conjugated anti-rabbit secondary antibodies (Molecular Probes, 2.5 μg/ml). Samples were equilibrated in Slow Fade (Molecular Probes) containing 0.2 μg/ml DAPI and 5 μg/ml FM 4-64. We found that FM 4-64 rapidly bleached by ultraviolet light in fixed cells. Therefore, before collecting images of Oregon green fluorescence we bleached the FM 4-64, using the DAPI filter set to ensure that there was no FM 4-64 fluorescence remaining, which might otherwise be visualized together with Oregon green.
β-Galactosidase Assay.
β-Galactosidase assays were performed as described (16, 19).
Results and Discussion
Assay for Membrane Fusion During B. subtilis Sporulation.
The fusion assay relies on the fact that FM 4-64 intercalates into the outer surface of biological membranes but is unable to cross the lipid bilayer, therefore only membrane surfaces directly exposed to FM 4-64 will stain. During engulfment, both the mother cell and forespore membranes are accessible to FM 4-64; however, once engulfment and membrane fusion are complete, the two membranes surrounding the forespore are isolated from the external environment. Thus, after membrane fusion at the completion of engulfment, only the mother cell cytoplasmic membrane stains with FM 4-64, while the forespore membranes remain unstained (Fig. 1B right cell, red). However, if the engulfing membranes fail to fuse at the cell pole, the forespore membranes will stain with FM 4-64 (Fig. 1B, left cell). We used a second, membrane-permeant, stain (MTG) to visualize fully engulfed and fused sporangia (Fig. 1B right cell, green). As shown in Fig. 1 C–E, the fusion assay differentiates cells at the stage of polar septation (arrow 1) from those in the process of engulfment (arrow 2) and allows us to separate those cells that have not yet completed membrane fusion (arrow 3) from those that have (arrow 4). Although the fusion assay relies on fluorescence microscopy, it directly probes the continuity of the mother cell and forespore membranes, providing a physical assay for the completion of engulfment.
Using this fusion assay, we followed membrane movement and fusion throughout sporulation in wild-type B. subtilis, taking samples every 15 min, starting 90 min after the onset of sporulation (= t1.5). Consistent with published results (7, 21), we found that sporangia that had completed membrane migration but not fusion began to appear about 90 min after the onset of sporulation. These sporangia display uniformly bright FM 4-64 staining around the forespore (Fig. 1C, arrow 3), in contrast to engulfing sporangia, which show a gap in the FM 4-64 fluorescence (Fig. 1C, arrow 2). We occasionally observed asymmetric engulfment (≈15% of sporangia), where the membranes meet on one side of the forespore (not shown). Fully fused sporangia (Fig. 1C, arrow 4), appear about 15 min after the completion of membrane migration (see supplemental Fig. 5A for quantitation at www.pnas.org), a delay that may reflect either the limited resolution of the light microscope (which makes it difficult to distinguish between sporangia that have completed membrane migration and those that have not) or an actual delay in membrane fusion after the completion of membrane movement.
Visual Screen for Engulfment Mutants.
The known engulfment mutants have defects in activation of two post-engulfment sigma factors, σG and σK (1), and it seemed likely that new engulfment mutants would have a similar defect. Therefore, we screened for new engulfment mutants with two enzymatic reporters, one expressed at the onset of sporulation, alkaline phosphatase, and another expressed after the completion of engulfment, either sspB-lacZ (under the control of σG), or cotD-lacZ (under the control of σK). We screened ≈20,000 mutagenized colonies and isolated 660 mutants defective in σG or σK activation, some of which were defective in engulfment or membrane fusion. To identify such mutants, we performed a visual screening in the fluorescence microscope, using the membrane fusion assay described above. We employed a σE-controlled cotE-gfp fusion to ensure that strains with an apparent engulfment defect were not simply delayed in the onset of sporulation. While CotE-GFP is synthesized shortly after polar septation, it becomes brightly fluorescent only after the completion of engulfment in wild-type sporangia. Engulfment-defective mutants show brightly fluorescent GFP in sporangia that have failed to complete engulfment. In addition to several classes of mutants defective in various stages of engulfment (to be described elsewhere), we identified six mutants with defects in the final steps of engulfment. All six mutations mapped to spoIIIE (Methods).
These results led us to test a ΔspoIIIE mutant to see whether it exhibited a late engulfment defect. By t3, the forespore membranes of most ΔspoIIIE mutant sporangia were stained by both FM 4-64 and MTG (Fig. 1 G–I, arrow 5), with only 0.7% of ΔspoIIIE mutant sporangia having completed membrane fusion (versus 50% in wild type). By t4 only 0.3% were fused (versus 74% in wild type). We also noted that ΔspoIIIE strains had a consistent delay in the completion of membrane movement around the forespore, as by t3, only 22% of ΔspoIIIE mutant sporangia showed complete membrane migration (Fig. 1G; versus 61% in wild type), as further discussed below (see supplemental Fig. 5B at www.pnas.org for quantitative data). The chromosome translocation defect characteristic of spoIIIE mutants is evident by comparing the DAPI staining of wild-type sporangia (Fig. 1F, arrows 2 and 3) to that of ΔspoIIIE sporangia (Fig. 1J, arrow 5); a similar phenotype is seen in all of the late-engulfment mutants we isolated.
In addition to a chromosome translocation defect, ΔspoIIIE strains show aberrant compartmentalization of both σF and σE (12, 16), which could be responsible for the late-engulfment defect of the ΔspoIIIE strain. To test whether this was the case, we performed a membrane fusion assay on the well characterized spoIIIE36 mutant, which shows normal compartmentalization of σF and σE activities but still displays defective chromosome translocation (16, 22). This mutant also has a late engulfment defect, with 5% of sporangia having fused by t3, and 15% fused by t4, but no apparent delay in the rate of membrane migration, with 62% of sporangia showing a fully engulfed but unfused phenotype by t3 (supplemental Fig. 5B at www.pnas.org). Therefore, the defect in σF and σE compartmentalization in ΔspoIIIE mutants may delay membrane movement, but it is not responsible for the late-engulfment defect.
The Roles of SpoIIIE in DNA Translocation and Late Engulfment Are Separable.
The failure of spoIIIE mutants to complete membrane fusion at the end of engulfment may be a secondary consequence of the lack of a complete chromosome in the forespore. The final stages of engulfment or membrane fusion may require forespore expressed genes located on the origin-distal part of the chromosome, which fail to enter the forespore in a spoIIIE mutant, and which therefore cannot be expressed. We addressed this possibility three ways.
First, we identified a spoIIIE mutant from our library, spoIIIE73-11, on the basis of the characteristic spoIIIE chromosome translocation defect (Fig. 1N arrows 6–8), that was capable of completing engulfment and membrane fusion (Fig. 1 K–M, arrows 7 and 8). By t3, 21% of spoIIIE73-11 sporangia have completed engulfment and membrane fusion. This frequency rises to 46% 1 hr later (vs. 74% in wild type). Thus spoIIIE73-11 mutant sporangia complete membrane fusion better than spoIIIE36 and ΔspoIIIE, although at a decreased rate relative to wild type.
The increased ability of spoIIIE73-11 mutants to complete membrane fusion in comparison to other spoIIIE mutants may be a result of a subtle difference in the chromosome translocation defect. If a gene involved in membrane fusion lies just outside the origin-proximal 30% of the chromosome normally present in the forespore in spoIIIE mutants, then a slight increase in the amount of chromosomal DNA translocated into the forespore could have a significant impact on the efficiency of membrane fusion. To test this possibility we evaluated expression of the forespore-specific σF-transcribed sspE(2G)-lacZ fusion integrated at the thrC locus, which is just outside the origin-proximal 30% of the chromosome, but which, infrequently, enters the forespore in spoIIIE mutants (23). Strains bearing this fusion along with either spoIIIE36 or spoIIIE73-11 showed a low, but detectable, level of β-galactosidase activity (10.9 and 11.4 units, respectively, vs. 720 units in wild type at t3.5, and 3.5 units in a strain without a lacZ fusion). In contrast, a ΔspoIIIE mutant, in which σF becomes active in the mother cell, showed a high level of β-galactosidase activity with this fusion (530 units at t3.5). We also confirmed that the lacZ fusion was being expressed specifically in the forespore in both spoIIIE36 and spoIIIE73-11 mutants by using immunofluorescence microscopy (not shown). Thus, it appears that the increased ability of spoIIIE73-11 mutants to complete membrane fusion is not due to a difference in the DNA translocation defect. This result suggests that the chromosome translocation and membrane fusion functions of SpoIIIE are (at least somewhat) separable.
To further explore the possibility that the chromosome translocation defect is directly responsible for the late engulfment defect of spoIIIE mutants, we employed a Δ(spo0J-soj) mutant. Previous work has shown that the Δ(spo0J-soj) mutant allows origin-distal regions of the chromosome to enter the forespore in spoIIIE mutant sporangia (24). This is likely because of the increased chromosome copy number of Δ(spo0J-soj) mutants at the onset of polar septation, which thereby traps random chromosomal regions in the forespore, along with the origin-proximal portion (25). If the late-engulfment defect of ΔspoIIIE mutants is due to the absence of origin-distal genes from the forespore, then Δ(spo0J-soj) ΔspoIIIE sporangia should complete membrane fusion at an increased frequency, similar to that seen for the expression of origin-distal σF promoters in this background, approximately 5% (24). We screened more than 800 Δ(spo0J-soj) ΔspoIIIE sporangia for completion of engulfment and membrane fusion at t3, and we saw no change in the frequency of membrane fusion. We also confirmed that the Δ(spo0J-soj) mutation itself had no affect on engulfment or membrane fusion.
Taking the following three results together, a spoIIIE mutant capable of completing the membrane fusion event at the end of engulfment at high efficiency but unable to translocate its chromosome into the forespore, the apparently identical chromosome translocation defect in spoIIIE36 and spoIIIE73-11 mutants, and the observation that there is no change in the late-engulfment phenotype in ΔspoIIIE mutants bearing the Δ(spo0J-soj) mutation, suggests that SpoIIIE has two functions during sporulation: DNA translocation during polar septation and some role in the completion of engulfment or membrane fusion. These results also indicate that the third stage of engulfment, movement of the mother cell membranes across the forespore pole, is composed of two separate steps, membrane movement across the pole (Fig. 1A IIiii) and membrane fusion (Fig. 1A IIiv), which requires SpoIIIE.
Relocalization of SpoIIIE Protein to the Cell Pole Correlates with Membrane Fusion.
Previously, immunofluorescence microscopy was used to demonstrate that SpoIIIE initially localizes between the forespore and mother cell chromosomes, presumably in the middle of the polar septum, and then relocalizes to the pole of the sporangium (26), where it could play a role in the final steps of engulfment. To correlate SpoIIIE localization with the completion of engulfment, we constructed a C-terminal SpoIIIE-GFP protein. We found that SpoIIIE-GFP initially localized to the middle of the sporulation septum (Fig. 2, arrow 1), then moved as a discrete focus around the forespore usually, but not always, associated with the leading edge of the engulfing membranes (Fig. 2, arrow 2). When membrane migration was complete, the focus localized to the cell pole (Fig. 2, arrow 3). After the completion of membrane fusion, the SpoIIIE complex remained intact for a short time (Fig. 2, arrow 4) before delocalizing (Fig. 2, arrow 5).
Figure 2.
SpoIIIE-GFP localization (B and C, green) in KP542 sporangia stained with FM 4-64 (A and B, red) and DAPI (C and D, blue) at t3. SpoIIIE initially localizes to the polar septum (arrow 1), then moves with the engulfing membranes (arrow 2) to the cell pole just before membrane fusion (arrow 3), where it remains after fusion is complete (arrow 4) until the protein delocalizes (arrow 5). (White bar in A is 2 μm.) A larger field can be seen in supplemental Fig. 6 at www.pnas.org.
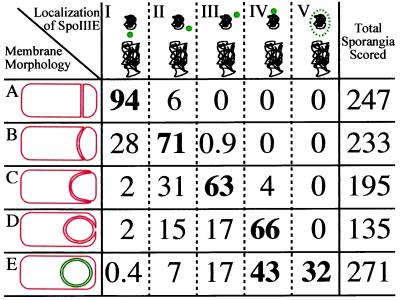
Scoring of the SpoIIIE-GFP localization pattern at different stages of engulfment and after membrane fusion (Fig. 3; a field is shown in supplemental Fig. 6, at www.pnas.org) revealed that 66% of sporangia that had completed membrane movement but not fusion (Fig. 3, class D) had a polar focus of SpoIIIE-GFP, whereas 17% showed a focus near but not at the cell pole, which may reflect sporangia that underwent asymmetric engulfment (observed in about 15% of all sporangia). Delocalized SpoIIIE-GFP was seen in 32% of sporangia that had completed membrane fusion (Fig. 3, class E), but it was never observed in sporangia in which membrane fusion was incomplete. Thus, SpoIIIE relocalizes to the cell pole before the completion of engulfment and delocalizes only after membrane fusion is complete.
Figure 3.
Correlation of stage of engulfment with SpoIIIE-GFP localization. Membrane morphology: class A, polar septation (Fig. 1C, arrow 1); class B, <50% forespore engulfment, membranes have migrated <50% around the forespore chromosome; class C, >50% forespore engulfment (Fig. 2A, arrow 2); class D, apparently engulfed but not fused (Fig. 2A, arrow 3); and class E, engulfed and fused (Fig. 2A, arrow 4). Localization of SpoIIIE-GFP (green; chromosomes in black): I, at polar septum; II, <50% around the forespore; III, >50% around the forespore; IV, localized to the cell pole; and V, delocalized. The values in the table represent the percentage of sporangia at a given stage of engulfment showing the indicated pattern of SpoIIIE-GFP localization. Those in boldface reflect the predominant SpoIIIE-GFP localization pattern(s) for each stage of engulfment.
We predicted that SpoIIIE73-11 would show a localization pattern similar to wild-type SpoIIIE because it is capable of completing membrane fusion, whereas SpoIIIE36, which is more strongly defective in membrane fusion, has been shown by immunofluorescence microscopy to remain at the polar septum (26). We therefore constructed C-terminal GFP fusions to both proteins and followed their localization throughout engulfment. Both SpoIIIE36-GFP and SpoIIIE73-11-GFP initially localized to the polar septum, and failed to relocalize to the forespore pole during engulfment (not shown). Indeed, we frequently saw sporangia that had completed membrane fusion with a single focus of SpoIIIE73-11-GFP between the forespore and mother cell chromosomes.
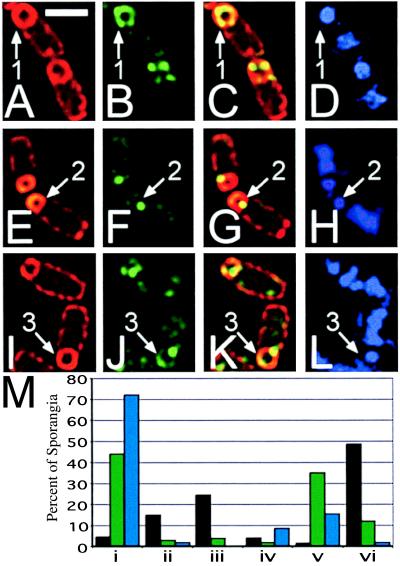
To explore the possibility that GFP fluorescence alone was unable to detect all of the SpoIIIE-GFP, either because a portion of the protein was not fluorescent or because it was below the detection limits of our deconvolution microscope, we localized both SpoIIIE73-11-GFP and SpoIIIE36-GFP by immunofluorescence microscopy using primary antibodies directed against GFP. Samples were prepared from cultures at t3, at which time membrane fusion should be ongoing in spoIIIE73-11 mutant sporangia. We scored localization of SpoIIIE-GFP in sporangia that had completed engulfment by the criterion of FM 4-64 staining; these included both fused and unfused sporangia, which cannot be distinguished in immunofluorescence microscopy. Of the wild-type sporangia that had completed engulfment, 50% showed delocalized SpoIIIE-GFP immunostaining (Fig. 4 A–D, arrow 1), whereas 25% showed a single polar focus of SpoIIIE-GFP (scored in Fig. 4M iii, black bar). In contrast, of the engulfed spoIIIE36 sporangia, 72% showed a single focus of SpoIIIE-GFP immunostaining at the polar septum (Fig. 4 E–H, arrow 2; scored in M i, blue bar), although 15% showed an additional focus or smear of fluorescence at the cell pole that was not observed by GFP fluorescence alone (Fig. 4M v, blue bars). In engulfed spoIIIE73-11 sporangia, 44% had a single focus of SpoIIIE-GFP immunostaining at the polar septum (Fig. 4M i, green bar), whereas 35% showed an additional focus or smear of fluorescence at the cell pole (Fig. 4 I–L, arrow 3; M iii and v, green bars). Both mutants showed a low frequency of sporangia with delocalized SpoIIIE-GFP immunostaining around the forespore (12% for spoIIIE73-11, 2% for spoIIIE36; Fig. 4M vi). For both mutants the percentage of sporangia with some form of polar localization pattern (either a discrete polar focus or delocalized fluorescence) exceeds the frequency of fully fused sporangia (47% polar localization vs. 33% fusion in fully engulfed sporangia for spoIIIE73-11, 17% vs. 8% for spoIIIE36), which is consistent with localization of SpoIIIE to the cell pole preceding membrane fusion. Therefore, it appears that both mutant proteins can localize to the cell pole, although SpoIIIE73-11 does so more efficiently.
Figure 4.
Cells of KP542 (wild type; A–D), KP538 (spoIIIE36; E–H), and KP537 (spoIIIE73-11; I–L) immunostained with anti-GFP antibodies (B, C, F, G, J, and K, green) FM 4-64 (A, B, E, F, I, and J, red) and DAPI (D, H, and L) at t3. The predominant staining pattern for each strain is shown; fields are shown in supplemental Fig. 7 at www.pnas.org. Arrow 1 shows wild-type SpoIIIE-GFP completely surrounding the forespore of an engulfed sporangium. Arrow 2 shows the predominant septal immunostaining pattern of SpoIIIE36-GFP. Arrow 3 indicates the localization of SpoIIIE73-11-GFP to a septal focus and in a crescent at the forespore pole. (White bar in A is 2 μm.) (M) Scoring of SpoIIIE-GFP localization in fully engulfed sporangia, SpoIIIE-GFP (black bars), SpoIIIE73-11-GFP (green bars), SpoIIIE36-GFP (blue bars). i, sporangia with a single focus of fluorescence in the middle of the polar septum; ii, sporangia with a single focus of fluorescence on the side of the forespore; iii, sporangia with a single focus of fluorescence at the cell pole; iv, sporangia with two foci of fluorescence, one at the polar septum and one on the side of the forespore; v, sporangia with two foci of fluorescence, one at the septum and one at the cell pole; vi, sporangia with delocalized fluorescence or with more than two foci. At least 100 fully engulfed sporangia were scored for each strain; values represent percentages of scored sporangia. More detailed scoring is shown in supplemental Fig. 7 at www.pnas.org.
Sequence Analysis of SpoIIIE73-11.
We sequenced the entire coding region of spoIIIE73-11 and found a single base change, which resulted in the replacement of the first conserved glycine residue of the Walker A ATP/GTP-binding site by serine (G467S). This residue is highly conserved among homologues of SpoIIIE, and similar mutations in other Walker ATP/GTPases abolish nucleotide binding (27). spoIIIE73-11 is severely defective in chromosome translocation, but not in the completion of engulfment and membrane fusion, suggesting that nucleotide binding and hydrolysis are essential for chromosome translocation, but not for the completion of membrane migration or fusion.
Our results suggest that SpoIIIE is involved in two separate processes during sporulation: translocation of the chromosome into the forespore during polar septation and the final step of engulfment. There are several possibilities for this unanticipated role of SpoIIIE in the completion of engulfment. First, SpoIIIE may be required for the final stages of membrane migration around the forespore prior to membrane fusion. Second, SpoIIIE could play a direct role in membrane fusion, either catalyzing or regulating this process. Finally, SpoIIIE could localize other proteins required for these processes to the engulfing membrane. If SpoIIIE does catalyze membrane fusion at the completion of engulfment, then it is tempting to speculate that it serves a similar function at the polar septum, promoting septal membrane fusion after the completion of chromosome translocation, possibly serving to coordinate these two processes.
Supplementary Material
Acknowledgments
We thank Joe Pogliano for performing the nitrosoguanidine mutagenesis, participating in the mutant screen, and for extensive discussions. We would also thank Chris Webb and Patrick Stragier for the gift of lacZ fusions and Petra Levin for the vector used in construction of our spoIIIE-gfp fusion protein. This work was supported by National Institutes of Health Grant GM-57045 and by the Arnold and Mabel Beckman Foundation and the Searle Scholars Program/The Chicago Community Trust. M.D.S. is a National Science Foundation predoctoral fellow.
Abbreviations
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- MTG
MitoTracker Green FM
References
- 1.Stragier P, Losick R. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 2.Coote J G. J Gen Microbiol. 1972;71:1–15. doi: 10.1099/00221287-71-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Smith K, Bayer M E, Youngman P. J Bacteriol. 1993;175:3607–3617. doi: 10.1128/jb.175.11.3607-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frandsen N, Stragier P. J Bacteriol. 1995;177:716–722. doi: 10.1128/jb.177.3.716-722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margolis P S, Driks A, Losick R. J Bacteriol. 1993;175:528–540. doi: 10.1128/jb.175.2.528-540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Londoño-Vallejo J-A, Fréhel C, Stragier P. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- 7.Pogliano J, Osborne N, Sharp M D, Abanes-de Mello A, Perez A, Sun Y-L, Pogliano K. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diez A A, Farewell A, Nannmark U, Nyström T. J Bacteriol. 1997;179:5878–5883. doi: 10.1128/jb.179.18.5878-5883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Lutkenhaus J. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Weihe E, Margolin W. J Bacteriol. 1998;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draper G C, McLennan N, Begg K, Masters M, Donachie W D. J Bacteriol. 1998;180:4621–4627. doi: 10.1128/jb.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L J, Errington J. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 13.Sharpe M E, Errington J. Proc Natl Acad Sci USA. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterlini J M, Mandelstam J. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Youngman P, Perkins J B, Losick R. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- 16.Pogliano K, Hofmeister A E M, Losick R. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolis P, Driks A, Losick R. Science. 1991;254:562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- 18.Webb C D, Decatur A, Teleman A, Losick R. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 20.Hoch J A. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- 21.Partridge S R, Errington J. Mol Microbiol. 1993;8:945–955. doi: 10.1111/j.1365-2958.1993.tb01639.x. [DOI] [PubMed] [Google Scholar]
- 22.Wu L J, Lewis P J, Allmansberger R, Hauser P M, Errington J. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 23.Wu L J, Errington J. Mol Microbiol. 1998;27:777–786. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]
- 24.Sharpe M E, Errington J. Mol Microbiol. 1996;21:501–509. doi: 10.1111/j.1365-2958.1996.tb02559.x. [DOI] [PubMed] [Google Scholar]
- 25.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C-H, Grossman A D, Wright A, Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 26.Wu L J, Errington J. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deyrup A T, Krishnan S, Cockburn B N, Schwartz N B. J Biol Chem. 1998;273:9450–9456. doi: 10.1074/jbc.273.16.9450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.