Abstract
The nuclear receptor CAR (constitutive active/androstane receptor) is a drug-sensing transcription factor, regulating the hepatic genes that encode various drug-metabolizing enzymes. We have now characterized the novel regulatory mechanism by which the signal molecule EGR1 (early growth response 1) determines CAR-mediated activation of the human CYP2B6 (cytochrome P450 2B6) gene. The CYP2B6 enzyme metabolizes commonly used therapeutics and also activates pro-drugs. The CAR directly binds to the distal enhancer element of the CYP2B6 promoter, which is essential in converging to its drug-sensing function onto promoter activity. However, this binding alone is not sufficient to activate the CYP2B6 promoter; the promoter requires EGR1 to enable CAR to activate the CYP2B6 promoter. Upon stimulation by protein kinase C, EGR1 directly binds to the proximal promoter and coordinates the nearby HNF4α (hepatocyte-enriched nuclear factor 4α) with CAR at the distal enhancer element to activate the promoter. Thus, synergy of drug activation and the stimulation of cellular signal are necessary for CAR to activate the CYP2B6 gene.
The liver is endowed with a defense mechanism against toxicity and carcinogenicity caused by xenobiotics, including therapeutic drugs. The underlying defense mechanism is xenobiotic-induced activation of various genes that encode xenobiotic-metabolizing enzymes and proteins, such as CYPs (cytochromes P450) and transporters, increasing hepatic capability of xenobiotic metabolism and excretion. The nuclear receptor CAR (constitutive active/androstane receptor) plays key roles in the regulation of these genes (1). Although CAR is primarily characterized as a xenobiotic-activated nuclear receptor, its ability to regulate the transcription critically depends on the status of endogenous stimuli. Recent studies have begun to decipher the molecular mechanisms by which these cellular signals regulate the CAR-mediated trans-activation. For example, insulin and growth factors are known to repress the CAR-mediated induction of the CYP gene. The insulin response forkhead transcription factor 1, acting as co-repressor, directly binds to CAR and represses CAR-mediated transcription (2). Hepatocyte growth factor inhibits the nuclear translocation of CAR to repress the induction of the CYP gene in mouse primary hepatocytes (3). Overexpression of the Rho-guanine nucleotide exchange factor ECT2 (epithelial cell transforming gene 2) represses drug-induced nuclear translocation of CAR in the mouse liver (4). However, the mechanism for how xenobiotics and endogenous cell signals cross-talk to activate the nuclear receptor is extremely complex and remains elusive.
A good example for this complexity is the phenomenon in which co-treatment with CAR ligand TCPOBOP and the protein phosphatase inhibitor okadaic acid (OA)2 synergistically activates the human CYP2B6 gene in Ym17 cells (HepG2 cells that stably express V5His-tagged mouse CAR) (5). Human CYP2B6 is a clinically important enzyme that metabolizes many commonly used therapeutics and activates anti-cancer pro-drugs, such as cyclophosphamid and ifosfamide (6, 7). Two distinct DNA sequences within the CYP2B6 promoter are required for CAR to synergistically activate the promoter activity: the distal phenobarbital-responsive enhancer module (PBREM) and the proximal OA response element now called OAREKI (5, 8). The PBREM is well characterized as being the CAR response enhancer to which CAR directly binds to activate PBREM (DR4)-bearing genes in response to xenobiotics (9, 10). OAREKI consists of a 52-bp DNA sequence and is activated by OA treatment; the activation mechanism of this process remains unknown. We have now dissected the 52-bp sequence into three distinct motifs (DR1, E-box, and CACCC) and identified HNF4α (hepatocyte-enriched nuclear factor 4α) and EGR1 (early growth response 1) as the transcription factors that regulate the DR1 and CACCC motifs, respectively. Moreover, EGR1 is characterized as being the OA-induced factor and is required to enable CAR to cross-talk with HNF4α in activating the CYP2B6 promoter. EGR1 is known to be induced by various endogenous stimuli, such as the phorbol 12-myristate 13-acetate (PMA)-activated protein kinase C (PKC) pathway (11, 12). Treatment with the PKC activator PMA has resulted in the induction of EGR1 in HepG2 cells, and the co-treatment with PMA and TCPOBOP has resulted in the synergic up-regulation of the CYP2B6 gene. Thus, the direct CAR binding to PBREM needs to be coordinated with the EGR1-mediated signal transduction for CAR to activate the CYP2B6 gene. Thus, the present characterization of EGR1 as being the essential factor required for CAR to activate transcription has significant pharmacological implications in drug therapy as well as introducing a new concept of CAR biology.
EXPERIMENTAL PROCEDURES
Reagents—TCPOBOP and Me2SO were purchased from Sigma; OA was from Calbiochem; CITCO and PMA were from Biomol (Plymouth Meeting, PA); poly(dI-dC)·(dI-dC) was from GE Healthcare Bio-Science Corp. The plasmids pGL3basic and pcDNA3.1-V5-His-TOPO were obtained from Promega (Madison, WI) and Invitrogen, respectively. Normal rabbit IgG and antibodies against HNF4α (H-171) (for supershift assay) and EGR1 ((588)X) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); antibody against HNF4α (for ChIP assay) was from Perseus Proteomics Inc. (Tokyo, Japan); V5 antibody was from Invitrogen.
Cloning and Plasmids—–1.8 kb and –1.6 kb promoters of the CYP2B6 gene, PBREM/–307 and PBREM/–213, were cloned into basic firefly luciferase reporter plasmid pGL3-basic as described previously (5). Deletion mutants were constructed in the contest of –1.8 kb promoter or PBREM/–307 using the QuikChange® site-directed mutagenesis kit (Cedar Creek, TX) and proper primers; ΔOAREKI, ΔDR1, and ΔCA delete the region of –268/–217, –227/–217, and –268/–244, respectively. The cDNAs encoding human HNF4α and EGR1 were amplified from HepG2 and Caco-2 RNAs, respectively, and were cloned into the pcDNA3.1-V5-His-TOPO plasmid (Invitrogen). All other plasmids were previously constructed in this laboratory.
Cell Cultures—HepG2-derived Ym17 and Yh18 cells were produced previously (5). These and HepG2 cells were cultured in minimal essential medium supplement with 10% (v/v) fetal bovine serum, antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin) and 2 mm glutamine. COS-1 cells were cultured in Dulbecco's modified Eagle's medium high glucose supplemented with the same amount of fetal bovine serum and antibiotics and 1 mm sodium pyruvate. To prepare nuclear extracts, cells were harvested from four confluent 145-cm2 dishes for each treatment using the procedure described previously (13).
Cell-based Transfection Assays—Cells were plated on a 24-well plate at a density of 3.5 × 105 cells/well. About 24 h postplating, cells were transfected with a given plasmid or co-transfected with a combination of multiple plasmids using FuGENE6 (Roche Applied Science) for 24 h prior to drug treatments. The doses of TCPOBOP, OA, and CITCO were 250, 10, and 250 nm, respectively. After an additional 48 h of incubation, cells were harvested for luciferase assays using the Dual-Luciferase reporter assay system (Promega). For PMA experiments, cells were treated with Me2SO or TCPOBOP for 24 h prior to adding PMA (150 nm). At various time points after adding PMA, cells were harvested for subsequent luciferase assays.
Gel Shift Assays—Double-stranded oligonucleotides containing the DR1 (–229/–217) and CACCC (–253/–242) sequence (5′-GATCTGGACTTTGATAT-3′ and 5′-GATCCACCCACACATT-3′, respectively) were used as probes. The underlined sequences indicate additional sequences that were filled in with DNA polymerase Klenow fragment (New England Biolabs, Beverly, MA) in the presence of [α-32P]dATP. 2–5 μg of nuclear proteins were used for each reaction. For the competition assay, a 50-fold excess of unlabeled probes was added into reaction mixture. The sequence of mutant probes for DR1 and CACCC were 5′-GATCTGGACTTAGATAT-3′ and 5′-GATCCAGGCACACATTC-3′, respectively. For supershift assays, nuclear extracts were incubated with 1 μg of a given antibody for 1 h on ice prior to adding probes.
Real Time PCR—Preparation of total RNA and the subsequent synthesis of first strand cDNA were performed using TRIzol® reagent (Invitrogen) and high capacity cDNA archive kit (Applied Biosystems, Foster City, CA), respectively, and were subjected to real time PCR analysis using the 7900HT fast real time PCR system (Applied Biosystems). For CYP2B6 mRNA, probe (Hs00167937_g1 from Applied Biosystems) and TaqMan Universal PCR Master Mix were used to perform PCR amplification. Forward and reverse primers used for EGR1 mRNA were 5′-CCGCAGAGTCTTTTCCTGACA-3′ and 5′-CCACAAGGTGTTGCCACTGT-3′, respectively, and PCR amplification was performed with SYBR Green Master Mix (Applied Biosystems). The levels of CYP2B6 and EGR1 mRNAs were normalized by β-actin mRNA that was measured using a predeveloped Taqman assay for human β-actin (Applied Biosystems).
Western Blot—Two hundred μg of nuclear proteins prepared from drug-treated Ym17 cells were applied on 20 μlofSP Sepharose™ XL Reagents (GE Healthcare Bio-Science Corp.) equilibrated with 20 mm Tris-HCl (pH 7.5) containing 0.1 m NaCl, 10% glycerol, 0.2 mm EDTA, 1 mm NaMoO4, and 0.5% Nonidet P-40. After washing the resin with equilibrating buffer three times, bound proteins were eluted into 30 μl by increasing NaCl to 0.5 m, separated on a 10% polyacrylamide gel, and transferred onto Immobilon-P membrane (Millipore, Bedford MA). These membranes were incubated overnight at 4 °C with anti-EGR1 antibody (1:5000 dilutions) in 5% (w/v) nonfat milk in Tris-buffered saline, 0.2% (v/v) Tween 20. Subsequently, they were incubated with horseradish peroxidase-conjugated anti-rabbit IgG (1:5000 dilution; Santa Cruz Biotechnology). Protein bands were visualized by ECL Plus Western blotting detection reagents (GE Healthcare).
siRNA Knockdown—siRNAs were obtained from Dharmacon Research (Lafayette, CO). Ym17 cells plated on a 6-well plate at a density of 1.4 × 106 cells/well were transfected with 200 pmol of ON-TARGET Plus SMART pool EGR1 (catalog number L-006526-00) or ON-TARGET Plus siCONTROL™ nontargeting pool (catalog number D-001810-10) using Lipofectamine™ 2000 (Invitrogen). After 24 h, the cells were subjected to treatment with drugs for an additional 48 h. Total RNAs and nuclear extracts were for real time PCR and gel shift assays to verify siRNA knockdown of both EGR1 mRNA and protein. The same total RNAs were also used in real time PCR for the CYP2B6 mRNA.
ChIP Assays—ChIP assay was performed as described in our previous report (8) using a ChIP assay kit (Upstate Biotechnologies, Charlottesville, VA). Briefly, Ym17 cells were crosslinked in serum-free minimal essential medium containing 1% of formaldehyde, washed by cold PBS, and were homogenized in wash buffer containing 0.3% Nonidet P-40. Cross-linked DNA-protein complexes were sonicated to sheared chromatin in SDS lysis buffer and were centrifuged to lysates. After it was precleared by shaking with protein A, the lysate was incubated with 5 μg of either antibody or normal IgG and was added by protein A to precipitate immunocomplexes. From decross-linked and protease-digested immunocomplexes, DNA was purified by the QIAquick PCR purification kit (Qiagen, Valencia, CA) and was amplified by PCR using LA Taq polymerase (Takara, Otsu Shiga, Japan). The amplicon (–338 to –99 region) was resolved on a 1.5% agarose gel.
RESULTS
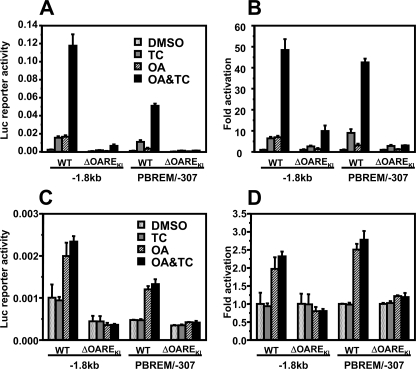
Defining OAREKI—Our previous characterization of the OA response activity and subsequent sequence motif analysis delineated to OA response element (now called OAREKI) to the –268/–217 region of the CYP2B6 promoter (Fig. 1). The 5′- and 3′-regions of OAREKI constitute the CA-rich and DR1 motifs, respectively, which are flanked by the E-box. For further functional characterization of OAREKI, two primary constructs of the CYP2B6 promoter were used: –1.8 kb CYP2B6-Luc containing both PBREM and OAREKI and PBREM/–307 CYP2B6-Luc in which the –1682/–308 region was internally deleted in the context of –1.8 kb CYP2B6-Luc (Fig. 1). Similar to the –1.8 kb promoter, PBREM/–307 was synergistically activated by co-treatment with TCPOBOP and OA in Ym17 cells (Fig. 2, A and B). Although PBREM/–307 exhibited lower basal promoter activity compared with the –1.8 kb promoter (Fig. 2A), PBREM/–307 was synergized at the same rate (43–48-fold) as the –1.8 kb promoter (Fig. 2B). Deletion of OAREKI abrogated OA response activity and synergy in both the –1.8 kb and PBREM/–307 promoters (Fig. 2, A and B). In addition, these deletion promoters diminished their ability to be activated by TCPOBOP, although they retained some degree of the activation. To examine whether CAR is required for OA to activate the CYP2B6 promoter, similar experiments were performed using HepG2 cells (Fig. 2, C and D). Since HepG2 cells lack CAR, no activation by TCPOBOP was observed. Both –1.8 kb and PBREM/–307 promoters were activated by OA 2-fold in HepG2 cells, although the rates of OA activation were lower compared with those observed in Ym17 cells. These results indicated that the synergistic activation of the CYP2B6 promoter can be regulated by the two elements PBREM and OAREKI and that OAREKI is the element responsible for regulating the activation by OA. In addition, OAREKI can be activated by OA in the absence of CAR, whereas the low activation rates by OA in HepG2 cells imply that some form of cross-talk between CAR and OAREKI is involved in the regulation of OA activation.
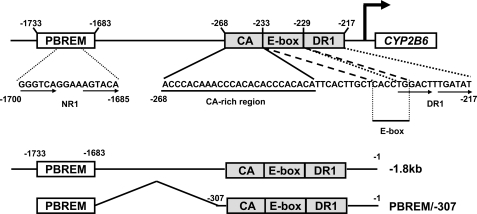
FIGURE 1.
Map of the CYP2B6 promoter. The CAR binding site NR1 (a DR4 motif) resides within the distal PBREM (–1733/–1683), whereas the proximal OAREKI (–268/–217) can be divided into three consensus sequences: CA-rich (–268/–245), E-box (–233/–228), and DR1 (–229/–217). Two principal constructs of the CYP2B6 promoter used for the present work were –1.8 kb and PBREM/–307 promoter; the former contains the whole 1.8 kb sequence, and the latter deletes a region (–1682/–308) between PBREM and OAREKI. From these two principal promoters, various deletion mutants were constructed, as described under “Experimental Procedures”: –1.6 kb, –1.8 kb ΔOAREKI, PBREM/–307 ΔCA, PBREM/–307 ΔDR1, PBREM/–307 ΔOAREKI, and PBREM/–213.
FIGURE 2.
OAREKI required for CAR to activate the CYP2B6 promoter. The –1.8 kb promoter, its deletion mutant (–1.8 kb ΔOAREKI), PBREM/–307, or its deletion mutant (PBREM/–307ΔOAREKI) was co-transfected with phRL-SV40 into Ym17 (A and B) or HepG2 (C and D) cells. The cells were treated with Me2SO (DMSO), 250 nm TCPOBOP (TC), 10 nm OA, or TCPOBOP plus OA (OA&TC) and were harvested for measuring luciferase activity. Reporter activity was calculated by dividing firefly luciferase with Renilla luciferase activities (A and C). -Fold activation was calculated by taking control activity (Me2SO) as 1 (B and D). WT, wild type.
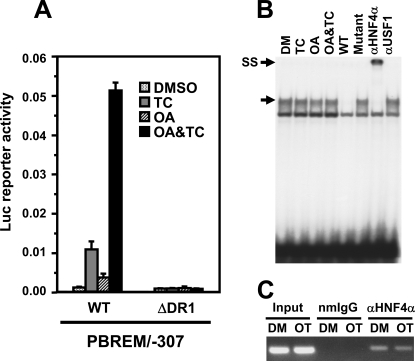
Defining DR1 Function—The –229/–217 (TGGACTTTGATAT) in the 3′-region of OAREKI appeared to be the DR1 motif and resembled various HNF4α binding sites (14, 15). Removal of the –227/–217 from PBREM/–307 resulted in the complete elimination of the promoter activity, no response to TCPOBOP and OA and no synergy (Fig. 3A). This DR1 motif formed a specific protein-DNA complex with nuclear protein prepared from Ym17 cells (Fig. 3B). Moreover, supershift by HNF4α antibody of this complex formed between the –229/–217 probe and nuclear protein confirmed that HNF4α was the protein that bound to the DR1 (Fig. 3B). ChIP assays confirmed that HNF4α occupied the OAREKI of the CYP2B6 promoter before co-treatment with TCPOBOP and OA and remained occupied after the co-treatment (Fig. 3C). Two insights became evident; HNF4α was required for both TCPOBOP and OA to activate PBREM/–307, and OA treatment did not increase HNF4α binding to the DR1 probe. These results indicated that HNF4α is essential for the activation and synergy of the CYP2B6 promoter but is not the OA response transcription factor. Although HNF4α is considered as a constitutively activated nuclear receptor, the present results showed that this receptor requires an additional regulation to activate the promoter activity.
FIGURE 3.
HNF4α as a constitutive factor regulating OAREKI. A, transient transfection assays to show HNF4α activity. Ym17 cells were co-transfected by PBREM/-307 (WT) or its DR1 deletion mutant (ΔDR1) and phRL-SV40 into Ym17 cells and were subsequently treated with Me2SO (DMSO), 250 nm TCPOBOP (TC), 10 nm OA, or TCPOBOP plus OA (OA&TC). Reporter activity was calculated by dividing firefly luciferase activity with Renilla luciferase activity. B, gel shift assays to show binding of HNF4α to DR1 motif within OAREKI. Double-stranded 32P-end-labeled DR1 probe (–229/–217) was incubated with nuclear extracts (2 μg) prepared from Ym17 cells treated with Me2SO (DM), 250 nm TCPOBOP, 10 nm OA, or OA plus TCPOBOP for 36 h. For competition assays, a 50-fold excess of unlabeled DR1 probe (–229/–217, wild type) or its mutated probe (Mutant) was added into the reaction mixture. Anti-HNF4α antibody (αHNF4α) or unrelated antibody (αUSF1) was used for supershift assays. The arrows indicate specific shifted band and supershifted band (SS). C, ChIP assays to show HNF4α interacts with CYP2B6 promoter regardless of drug treatment. Cross-linked chromatin-protein complexes were immunoprecipitated with anti-HNF4α (αHNF4α) or normal mouse IgG (nmIgG), and PCR amplicon (–338 to –99) was resolved on a 1.5%, agarose gel.
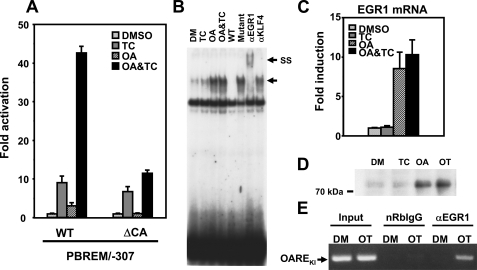
Identifying EGR1 as the OA Response Factor—When the CA-rich region (–268/–244) was deleted, PBREM/–307 retained the ability to be activated by TCPOBOP, but OA was no longer able to activate this deletion promoter (Fig. 4A). Following gel shift assays using various fragments representing the CA-rich region and the nuclear extracts from Ym17 cells, the –253/–242 (CACCCACACATT) was found to specifically bind with a nuclear protein (Fig. 4B). This protein binding to the –253/–242 probe appeared only after OA treatment and remained after TCPOBOP and OA co-treatment. Based on the nucleotide sequence CACCCACACA, various transcription factors, such as SP1, AP1, Kruppel-like factors, and EGR1, were selected (16, 17) and tested for their binding to the –253/–242 probe; only the EGR1 antibody supershifted the specific band (Fig. 4B). No supershift occurred with anti-KLF4 (Kruppel-like factor 4) antibody (Fig. 4B) and also with KLF9, KLF3, SP1, and AP1 (data not shown). EGR1 mRNA and protein levels were induced after OA treatment (Fig. 4, C and D), and concomitantly, the binding of ERG1 to the endogenous CYP2B6 promoter was increased in ChIP assays (Fig. 4E). Thus, EGR1 was clearly shown to be the OA response transcription factor. Unlike HNF4α, EGR1, however, plays no role in promoter activation by TCPOBOP.
FIGURE 4.
EGR1 as the OA response factor required for OAREKI activation. A, transient transfection assays to show requirement of the CA-rich region for OAREKI activity. Ym17 cells were co-transfected by PBREM/–307 (WT) or its CA-rich deletion mutant (ΔCA) with phRL-SV40 and were subsequently treated with Me2SO (DMSO), 250 nm TCPOBOP (TC), 10 nm OA, or TCPOBOP plus OA (OA&TC). Reporter activity was calculated by dividing firefly luciferase activity with Renilla luciferase activity. -Fold activation was calculated by taking the control activity (Me2SO; DMSO) as 1. B, Gel shift assays to show OA-responsive binding of EGR1 to the CA-rich region. Double-stranded 32P-end-labeled CA-rich probe (–253/–242) was incubated with nuclear extracts (5 μgof protein) prepared from Ym17 cells treated with Me2SO (DM), TCPOBOP (TC), OA, or OA plus TCPOBOP (OA&TC) for 36 h. For the competition assay, a 50-fold excess of unlabeled CA-rich probe (–253/–242, wild type) or its mutated probe (Mutant) was added into the reaction mixture. Anti-EGR1 antibody (αEGR1) or anti-KLF4 antibody (αKLF4) as negative control was used for supershift assays. The arrows indicate specific shifted band and supershifted band (SS). C, RT-PCR to show induction of EGR1 mRNA by OA treatment. Ym17 cells were treated with Me2SO, 250 nm TCPOBOP, 10 nm OA, or TCPOBOP plus OA (OT). Total cellular RNAs were prepared from these Ym17 cells and were subjected to quantitative real time RT-PCR of EGR1 mRNA. The levels of EGR1 mRNA were normalized by β-actin mRNA levels, and -fold induction was calculated to the levels in Me2SO-treated cells. D, Western blot to show increase of EGR1 protein in nuclear extracts of Ym17 cells treated with 250 nm TCPOBOP, 10 nm OA, or TCPOBOP plus OA (OT). E, ChIP assays to show that EGR1 increased its binding to the CYP2B6 promoter. Ym17 cells were treated with Me2SO (DM) or TCPOBOP plus OA for 36 h. As described under “Experimental Procedures,” cross-linked chromatin-protein complexes were immunoprecipitated with anti-EGR1 antibody (αEGR1) or normal rabbit IgG (nRbIgG). The DNA region, including OAREKI, was amplified by PCR (amplicon –338 to –99) and was resolved on a 1.5% agarose gel.
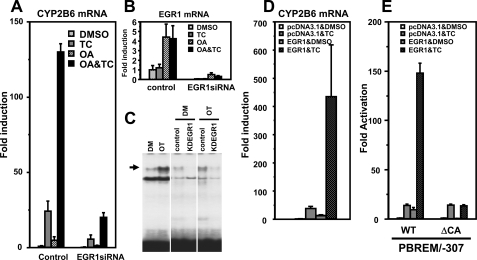
Defining EGR1 Function—Various experiments were employed to confirm that EGR1 is, in fact, the OA response transcription factor responsible for the synergistic activation of the endogenous CYP2B6 gene. First, siRNA was used to knock down EGR1 in Ym17 cells, proving the effective attenuation of both drug induction and synergy of the CYP2B6 gene (Fig. 5A). siRNA treatment dramatically reduced the EGR1 mRNA levels as well as the levels of EGR1 protein as observed with gel shift assays (Fig. 5, B and C). Second, OA treatment was replaced by epitotic expression of EGR1 to mimic the synergistic activation of the CYP2B6 gene. In the presence of pcDNA3.1, TCPOBOP induced the CYP2B6 gene ∼40-fold, whereas transfection of pcDNA3.1-EGR1 exhibited the ∼13-fold induction of the gene. Moreover, pcDNA3.1-EGR1 was transfected, the induction of the CYP2B6 gene was synergized up to 450-fold after TCPOBOP treatment (Fig. 5D). This synergistic induction of the CYP2B6 gene was directly reflected upon the synergistic activation of the CYP2B6 promoter by EGR1 and TCPOBOP (Fig. 5E). Co-transfection of pcDNA3.1-EGR1 synergistically up-regulated CAR-mediated activation of the CYP2B6 promoter in HepG2 cells (supplemental Fig. 1). These EGR1-mediated inductions and synergistic activations were reminiscent of that by OA and TCPOBOP (Figs. 4A and 5A). Thus, EGR1 is the OA response transcription factor required for the synergistic activation of the CYP2B6 gene following co-treatment with TCPOBOP.
FIGURE 5.
EGR1 required for CAR to activate the CYP2B6 promoter. A–C, siRNA knockdown to show that EGR1 is required for expression of the CYP2B6 gene. Ym17 cells were transfected with EGR1 siRNA or control scramble siRNA and were treated with Me2SO (DMSO), 250 nm TCPOBOP (TC), 10 nm OA, or TCPOBOP plus OA (OA&TC). Total cellular RNAs were prepared from these cells and were subjected to quantitative real time RT-PCR for the CYP2B6 (A) and EGR1 (B) mRNAs. C, gel shift assays to show decrease of EGR1 protein by siRNA. Nuclear extracts were prepared from Ym17 cells transfected with control siRNA (control) and EGR1 siRNA (KDEGR1) and were subjected to gel shift assays using CA-rich probe (–253/–242). The arrows indicate a specific shifted band. D, RT-PCR to demonstrate EGR1 synergizing TCPOBOP activation of the CYP2B6 gene. Ym17 cells were transfected with pcDNA3.1-EGR1 plasmid or empty plasmid (pcDNA3.1) and were treated with Me2SO (DMSO) or TCPOBOP. Total cellular RNAs were prepared from these cells and were subjected to quantitative real time RT-PCR of CYP2B6 mRNA. The levels of CYP2B6 mRNA were normalized by β-actin mRNA levels. -Fold induction was calculated relative to the levels in pcDNA3.1-transfected and Me2SO-treated cells. E, transient transfections to show that EGR1 is required for the CA-rich region to activate the CYP2B6 promoter. Ym17 cells were co-transfected by PBREM/–307 (WT) or its CA-rich deletion mutant (ΔCA) with phRL-SV40, pcDNA3.1 or pcDNA3.1-EGR1. Luciferase reporter activity was measured, and -fold activation was calculated by taking the control activity (pcDNA3.1 with Me2SO) as 1.
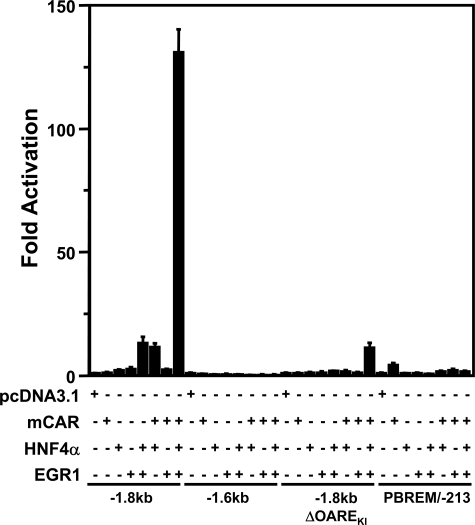
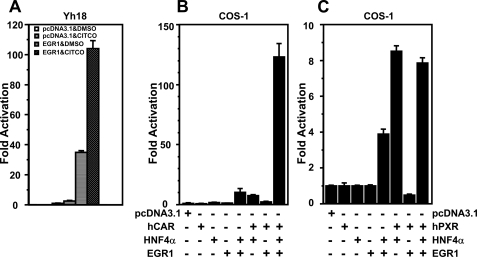
Using CAR, HNF4α, and EGR1, we attempted to reconstitute the synergistic activation of the CYP2B6 promoter. For this purpose, untagged mouse CAR (mCAR) was used as a substitute for TCPOBOP treatment, since, unlike the V5 His-tagged mCAR that is stably expressed in Ym17 cells, untagged mCAR possesses a high constitutive activity and does not need TCPOBOP to activate the CYP2B6 promoter. Instead of HepG2 cells, COS-1 cells, which do not express endogenous HNF4α, were employed as the host cells; the levels of endogenous EGR1 in COS-1 cells are as low as those observed in HepG2 cells. Under these conditions, reconstitution experiments were performed by expressing only CAR, HNF4α, or EGR1 and by co-expressing them in various combinations (Fig. 6). mCAR, HNF4α, or EGR1 exhibited practically no activation activity of the –1.8 kb promoter. HNF4α plus EGR1 and mCAR plus HNF4α activated the –1.8 kb promoter ∼10-fold, whereas mCAR plus EGR1 exhibited no activation activity. Subsequently, the co-expression of mCAR, HNF4α, and EGR1 all together synergized the promoter activity over 130-fold (Fig. 6). This synergistic activation depended on the presence of both PBREM and OAREKI, as indicated by the fact that neither the –1.6 kb (which lacks PBREM), the –1.8 kb (ΔOAREKI), nor the PBREM/–213 (in which PBREM is directly placed the front of the CYP2B6 –213 bp promoter) could respond to these transcription factors. In addition to mCAR, human CAR (hCAR) was also capable of synergizing the –1.8 kb promoter; epitotic expression of EGR1 synergistically activated the –1.8 kb promoter after co-treatment with the hCAR ligand CITCO (18) in Yh18 cells that stably express hCAR (Fig. 7A), and the co-expression of hCAR with HNF4α and EGR1 also resulted in synergistic promoter activation in COS-1 cells (Fig. 7B). In addition, this activity of CAR appeared to be specific to the nuclear receptor, since human PXR failed to synergize the promoter activity in the presence of HNF4α and EGR1 (Fig. 7C).
FIGURE 6.
Reconstitution of the CYP2B6 promoter activity by CAR, HNF4α, and EGR1. Four constructs (–1.8 kb, –1.6 kb, –1.8 kb ΔOAREKI, and PBREM/–213 were used to the CYP2B6 promoters. Each of these promoter constructs and phRL-SV40 were co-transfected with various combinations of pcDNA3.1, pCR3-mCAR, pcDNA3.1-HNF4α, and pcDNA3.1-EGR1 in COS-1 cells. The total amount of plasmid transfected into cells was adjusted by adding supplemental pcDNA3.1. -Fold activation of the luciferase reporter gene was calculated by taking the control activity (pcDNA3.1) as 1.
FIGURE 7.
EGR1 co-regulates CAR but not PXR. A, Yh18 (HepG2 cells stably expressing human CAR) were co-transfected with –1.8 kb promoter, phRL-SV40, and pcDNA3.1 or pcDNA3.1-EGR1 and were treated with Me2SO (DMSO) or CITCO (250 nm) for 48 h and assayed for luciferase activity. -Fold luciferase reporter activation was calculated by taking the control activity (pcDNA3.1 with Me2SO) as 1. B, COS-1 cells were co-transfected with –1.8 kb promoter, phRL-SV40, and various combinations of pcDNA3.1, pCR3-hCAR, pcDNA3.1-HNF4α, and pcDNA3.1-EGR1. The total amounts of plasmids transfected into cells were adjusted by adding supplementary pcDNA3.1. -Fold activation was calculated by taking the control activity (pcDNA3.1) as 1. C, instead of pCR3-hCAR, pCR3-hPXR was used for the experiments.
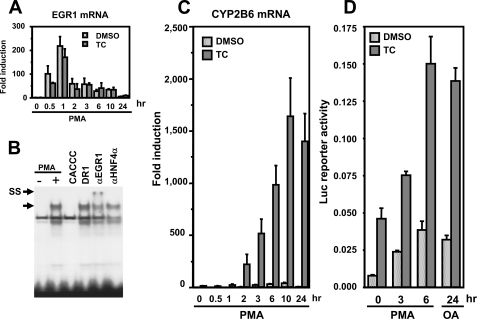
Characterizing PKC as an Induction Signal for EGR1—It has been shown that the EGR1 gene can be regulated through various cell signals, such as the PKC pathway, growth factor-ERK pathway, and stress from injury (11, 19–22). Instead a relatively nonspecific protein phosphatase inhibitor OA, the specific PKC activator PMA was employed to examine whether the PKC signal pathway could synergize CAR-mediated activation of the CYP2B6 promoter. PMA treatment increased the EGR1 mRNA level in a time-dependent manner, reaching its peak at 1 h after treatment in Ym17 cells (Fig. 8A). Gel shift and supershift assays showed that PMA induces the EGR1 protein and the binding to the –253/–242 probe (Fig. 8B). Although CYP2B6 mRNA was barely increased at any time points following treatment with PMA alone, a dramatic increase of mRNA was observed after co-treatment with PMA and TCPOBOP, starting at 2 h and reaching a peak at 10 h after co-treatment (Fig. 8C). At 24 h, TCPOBOP alone, PMA alone, and PMA plus TCPOBOP induced CYP2B6 mRNA 16-, 7-, and 1500-fold, respectively (Fig. 8C), revealing the strong ability of PMA to synergize drug induction of the CYP2B6 gene. Indeed, the –1.8 kb promoter was also synergistically activated by co-treatment with PMA and TCPOBOP; the ability of PMA to synergize the promoter activity was as strong as that of OA (Fig. 8D). These results indicated that PKC signal can synergize the CAR-activated transcription of the CYP2B6 gene.
FIGURE 8.
PKC regulates EGR1 to activate the CYP2B6 promoter. A, RT-PCR to show that EGR1 mRNA increased after PMA treatment. Ym17 cells were treated with Me2SO (DMSO), 250 nm TCPOBOP, Me2SO plus 150 nm PMA, or TCPOBOP plus PMA. The total duration of treatments was 24 h, within which PMA was added to cell cultures hours before the end of the 24-h treatment. Total cellular RNAs were prepared from the cells and subjected to quantitative real time RT-PCR of EGR1 mRNA. The levels of EGR1 mRNA were normalized by β-actin mRNA levels, and -fold induction was calculated relative to the levels in Me2SO-treated cells. B, gel shift assays to show increase of EGR1 protein after PMA treatment. Double-stranded 32P-end-labeled CA-rich probe (–253/–242) was incubated with nuclear extracts (5 μg of protein) prepared from Ym17 cells treated with Me2SO or PMA (150 nm) for 1 h. Competition and supershift assays were performed as described in the legend to Fig. 4B. The arrows indicate specific shifted band and supershifted band (SS). C, RT-PCR to show synergistic increase of CYP2B6 mRNA after co-treatment with PMA and TCPOBOP. Drug treatment of Ym17 cells and RT-PCR were performed as described in the legend of A. D, transient transfection to show PMA activation of the CYP2B6 promoter. The –1.8 kb promoter was co-transfected with phRL-SV40 into Ym17 cells. The cells were treated with Me2SO (DMSO), TCPOBOP (TC), and PMA as described in A. The separate Ym17 cells were treated with OA or OA plus TCPOBOP for 24 h. Luciferase reporter activity was calculated by dividing firefly luciferase activity by Renilla luciferase activity.
DISCUSSION
Until now, CAR has been understood to regulate gene transcription by directly binding to its enhancer elements or by indirectly interacting with other transcription factors. In this mechanism of gene transcription, transduction of a drug-induced signal to the CAR protein either directly or indirectly should be sufficient. Here we have demonstrated that although the signal activating CAR is essential, by itself it is not sufficient for regulating the CYP2B6 gene. The drug-activated CAR requires an additional endogenous cell signal to regulate the CYP2B6 gene; the endogenous signal we have now found for the regulation is an early growth signal via the PKC-EGR1 pathway. EGR1 directly binds to the proximal region of the CYP2B6 promoter and coordinates cross-talk of HNF4α with the drug-activated CAR that directly binds to the distal enhancer element. Contrary to the fact that both CAR and HNF4α are characterized as the constitutively activated nuclear receptors, their activities appear to be confined within tight regulations.
Our present study has unequivocally shown that PBREM/–307 is the minimal promoter that can be synergistically activated in the presence of CAR, HNF4α, and EGR1, delineating the xenobiotic activation activity of the CYP2B6 gene to two distinct elements PBREM and OAREKI and these three transcription factors. Although all three transcription factors are essential for the CYP2B6 gene to be activated, each factor appears to play a different role in this activation. HNF4α is the constitutive factor to govern the basal transcription activity of the CYP2B6 promoter, since the CYP2B6 promoter was slightly activated by co-expression of HNF4α and EGR1 or CAR but not by that of CAR and EGR1 in COS-1 cells. On the other hand, CAR and EGR1 are the regulated factors. CAR and its enhancer PBREM act as the xenobiotic sensors and are required for xenobiotic activation of the CYP2B6 gene. EGR1 is not a xenobiotic-sensing factor and is regulated by endogenous signals, such as PKC. Apparently, EGR1 may enable the xenobiotic sensor CAR to allow communicating with HNF4α, completing the process of xenobiotic activation of the CYP2B6 promoter. A number of xenobiotics, classically represented by phenobarbital, do not bind to CAR for activation. This regulatory mechanism that requires additional factors, such as EGR1 and HNF4α, for CAR activation may endow hepatocytes with tightness as well as flexibility toward xenobiotic responsiveness.
In the CYP2B6 promoter, CAR directly binds to the –1.7 kb upstream PBREM, whereas the binding site of HNF4α resides at the –229/–217 bp within OAREKI. Our previous ChIP study demonstrated that CAR indirectly interacts with OAREKI following co-treatment with OA and TCPOBOP, suggesting that the interaction of CAR with OAREKI is the most critical step in CAR-mediated promoter regulation (5, 8). Since there is no CAR binding site within OAREKI and gel shift assays confirmed it (5, 8), the interaction of CAR with OAREKI is of an indirect nature, through a DNA-binding protein. This protein may be HNF4α, since GST pull-down assays showed CAR binding to HNF4α, and DR1-based DNA affinity chromatography co-purified CAR with HNF4α.3 Although further work is necessary to prove this point, it will not be surprising to find that HNF4α is, in fact, the protein that mediates the indirect interaction of CAR with OAREKI. CAR-mediated induction by drugs such as phenobarbital (PB) is a liver-specific phenomenon, and HNF4α, the hepatocyte-enriched transcription factor, is a most critical transcription factor that generally determines liver development and function. The roles of HNF4α have repeatedly been shown also in the area of xenobiotic metabolism (23–25); expression of CAR and PB induction of CAR target genes was not observed in the liver of HNF4α KO mice (26, 27), and CAR can activate promoters of the human CYP3A4 and CYP2C8 genes only in the presence of HNF4α (28–30). Because both PBREM and OAREKI are essential for CAR to activate the CYP2B6 promoter and CAR interacts with both, it is necessary for the receptor to co-regulate them. Apparently, the binding of EGR1 to the CACCC site facilitates the interaction of CAR with OAREKI (probably via HNF4α) to complete the process of promoter activation; solving the exact molecular mechanism of this process will be the key subject of future investigations.
In conclusion, PKC activation signals HepG2 cells to increase EGR1, allowing CAR to confer its ability to sense xenobiotics and induce transcription of the CYP2B6 gene. In fact, PKC is previously suggested to be the signal that regulates the EGR1-mediated activation of the PDGFA gene in human head and neck cancer cells (11). Due to the extreme complexity of these signaling mechanisms that differ among the various cell systems, however, it is uncertain whether PKC can dictate the CAR-mediated transcription in human liver. An intriguing question raised is whether xenobiotics initiate EGR1 signal simultaneously when they activate CAR or the initiation of EGR1 is totally independent from xenobiotic action. Conversely, it is also an interesting question whether an enzyme activity of CYP2B6 is involved in an early growth response of hepatocytes. Our present discovery of the PKC-EGR1 as being the key signal necessary for CAR to activate the CYP2B6 gene has opened a new concept in CAR biology and its pharmacological and toxicological implications in understanding human susceptibility to xenobiotic exposure and efficacy of drug therapy.
Supplementary Material
This work was supported by the Intramural Research Program of the National Institutes of Health and NIEHS. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: OA, okadaic acid; TCPOBOP, 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene; PBREM, phenobarbital-responsive enhancer module; OAREKI, OA response element; DR, direct repeat; PMA, phorbol 12-myristate 13-acetate; PKC, protein kinase C; Me2SO, dimethyl sulfoxide (DMSO); CITCO, 6-(4-chloropheny)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; mCAR, mouse CAR; hCAR, human CAR; RT, reverse transcription; PB, phenobarbital.
K. Inoue and M. Negishi, unpublished data.
References
- 1.Sueyoshi, T., and Negishi, M. (2001) Annu. Rev. Pharmacol. Toxicol. 41 123–143 [DOI] [PubMed] [Google Scholar]
- 2.Kodama, S., Koike, C., Negishi, M., and Yamamoto, Y. (2004) Mol. Cell. Biol. 24 7931–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koike, C., Moore, R., and Negishi, M. (2007) Mol. Pharmacol. 71 1217–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosseinpour, F., Timsit, Y., Koike, C., Matsui, K., Yamamoto, Y., Moore, R., and Negishi, M. (2007) FEBS Lett. 581 4937–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swales, K., Kakizaki, S., Yamamoto, Y., Inoue, K., Kobayashi, K., and Negishi, M. (2005) J. Biol. Chem. 280 3458–3466 [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Antona, C., and Ingelman-Sundberg, M. (2006) Oncogene 25 1679–1691 [DOI] [PubMed] [Google Scholar]
- 7.Schwartz, P. S., Chen, C. S., and Waxman, D. J. (2003) Cancer Gene Ther. 10 571–582 [DOI] [PubMed] [Google Scholar]
- 8.Inoue, K., Borchers, C. H., and Negishi, M. (2006) Biochem. J. 398 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sueyoshi, T., Kawamoto, T., Zelko, I., Honkakoski, P., and Negishi, M. (1999) J. Biol. Chem. 274 6043–6046 [DOI] [PubMed] [Google Scholar]
- 10.Honkakoski, P., Zelko, I., Sueyoshi, T., and Negishi, M. (1998) Mol. Cell. Biol. 18 5652–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Worden, B., Yang, X. P., Lee, T. L., Bagain, L., Yeh, N. T., Cohen, J. G., Van Waes, C., and Chen, Z. (2005) Cancer Res. 65 7071–7080 [DOI] [PubMed] [Google Scholar]
- 12.Wong, W. K., Ou, X. M., Chen, K., and Shih, J. C. (2002) J. Biol. Chem. 277 22222–22230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, K., Sueyoshi, T., Inoue, K., Moore, R., and Negishi, M. (2003) Mol. Pharmacol. 64 1069–1075 [DOI] [PubMed] [Google Scholar]
- 14.Hung, H. L., and High, K. A. (1996) J. Biol. Chem. 271 2323–2331 [DOI] [PubMed] [Google Scholar]
- 15.Tarumi, T., Kravtsov, D. V., Zhao, M., Williams, S. M., and Gailani, D. (2002) J. Biol. Chem. 277 18510–18516 [DOI] [PubMed] [Google Scholar]
- 16.Meester-Smoor, M. A., Molijn, A. C., Zhao, Y., Groen, N. A., Groffen, C. A., Boogaard, M., van Dalsum-Verbiest, D., Grosveld, G. C., and Zwarthoff, E. C. (2007) J. Mol. Endocrinol. 38 113–125 [DOI] [PubMed] [Google Scholar]
- 17.Black, A. R., Black, J. D., and Azizkhan-Clifford, J. (2001) J. Cell. Physiol. 188 143–160 [DOI] [PubMed] [Google Scholar]
- 18.Maglich, J. M., Parks, D. J., Moore, L. B., Collins, J. L., Goodwin, B., Billin, A. N., Stoltz, C. A., Kliewer, S. A., Lambert, M. H., Willson, T. M., and Moore, J. T. (2003) J. Biol. Chem. 278 17277–17283 [DOI] [PubMed] [Google Scholar]
- 19.Silva, P. N., Soares, J. A., Brasil, B. S., Nogueira, S. V., Andrade, A. A., de Magalhaes, J. C., Bonjardim, M. B., Ferreira, P. C., Kroon, E. G., Bruna-Romero, O., and Bonjardim, C. A. (2006) Biochem. J. 398 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khachigian, L. M., Lindner, V., Williams, A. J., and Collins, T. (1996) Science 271 1427–1431 [DOI] [PubMed] [Google Scholar]
- 21.Cao, X., Mahendran, R., Guy, G. R., and Tan, Y. H. (1992) J. Biol. Chem. 267 12991–12997 [PubMed] [Google Scholar]
- 22.Cao, X., Mahendran, R., Guy, G. R., and Tan, Y. H. (1993) J. Biol. Chem. 268 16949–16957 [PubMed] [Google Scholar]
- 23.Kamiya, A., Inoue, Y., and Gonzalez, F. J. (2003) Hepatology 37 1375–1384 [DOI] [PubMed] [Google Scholar]
- 24.Saborowski, M., Kullak-Ublick, G. A., and Eloranta, J. J. (2006) J. Pharmacol. Exp. Ther. 317 778–785 [DOI] [PubMed] [Google Scholar]
- 25.Barbier, O., Girard, H., Inoue, Y., Duez, H., Villeneuve, L., Kamiya, A., Fruchart, J. C., Guillemette, C., Gonzalez, F. J., and Staels, B. (2005) Mol. Pharmacol. 67 241–249 [DOI] [PubMed] [Google Scholar]
- 26.Hayhurst, G. P., Lee, Y. H., Lambert, G., Ward, J. M., and Gonzalez, F. J. (2001) Mol. Cell. Biol. 21 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding, X., Lichti, K., Kim, I., Gonzalez, F. J., and Staudinger, J. L. (2006) J. Biol. Chem. 281 26540–26551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamiyama, Y., Matsubara, T., Yoshinari, K., Nagata, K., Kamimura, H., and Yamazoe, Y. (2007) Drug Metab. Pharmacokinet. 22 287–298 [DOI] [PubMed] [Google Scholar]
- 29.Chen, Y., Kissling, G., Negishi, M., and Goldstein, J. A. (2005) J. Pharmacol. Exp. Ther. 314 1125–1133 [DOI] [PubMed] [Google Scholar]
- 30.Tegude, H., Schnabel, A., Zanger, U. M., Klein, K., Eichelbaum, M., and Burk, O. (2007) Drug Metab. Dispos. 35 946–954 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.