Abstract
The anaphase-promoting complex (APC) regulates the eukaryotic cell cycle by targeting specific proteins for proteasomal degradation. Its activity must be strictly controlled to ensure proper cell cycle progression. The co-activator proteins Cdc20 and Cdh1 are required for APC activity and are important regulatory targets. Recently, budding yeast Acm1 was identified as a Cdh1 binding partner and APCCdh1 inhibitor. Acm1 disappears in late mitosis when APCCdh1 becomes active and contains conserved degron-like sequences common to APC substrates, suggesting it could be both an inhibitor and substrate. Surprisingly, we found that Acm1 proteolysis is independent of APC. A major determinant of Acm1 stability is phosphorylation at consensus cyclin-dependent kinase sites. Acm1 is a substrate of Cdc28 cyclin-dependent kinase and Cdc14 phosphatase both in vivo and in vitro. Mutation of Cdc28 phosphorylation sites or conditional inactivation of Cdc28 destabilizes Acm1. In contrast, inactivation of Cdc14 prevents Acm1 dephosphorylation and proteolysis. Cdc28 stabilizes Acm1 in part by promoting binding of the 14-3-3 proteins Bmh1 and Bmh2. We conclude that the opposing actions of Cdc28 and Cdc14 are primary factors limiting Acm1 to the interval from G1/S to late mitosis and are capable of establishing APC-independent expression patterns similar to APC substrates.
Many proteins involved in regulating cell division are only present during defined windows of the cell cycle. Cell cycle-dependent expression generally results from a combination of transcriptional regulation and ubiquitin proteolysis. Proteolysis provides a rapid and irreversible mechanism to eliminate proteins at specific times to promote transitions from one cell cycle stage to the next. Proteolysis is achieved by the covalent modification of specific substrate proteins with polyubiquitin chains, catalyzed by the joint action of ubiquitin-conjugating enzymes and ubiquitin ligases, which leads to their recognition and destruction by the 26 S proteasome (1). Two large ubiquitin ligase complexes, the Skp1-cullin-F box protein (SCF)3 complex and the anaphase-promoting complex (APC), are responsible for the bulk of cell cycle-regulated ubiquitin proteolysis in eukaryotic cells. The SCF and APC are thought to function complementarily during cell division, and mounting evidence suggests that each regulates the activity of the other (2, 3).
APC initiates the proteolysis of B-type cyclins, securin, replication regulators, mitotic kinases, proteins involved in spindle assembly, the Skp2 F-box component of SCF, and numerous other proteins (4). The core APC is present throughout the cell cycle, but its activity is mostly limited to mitosis and G1 by a variety of regulatory mechanisms, emphasizing that fine control over APC activity is of critical importance to proper execution of the cell cycle. APC activity requires a WD40 repeat family co-activator protein, either Cdc20 or Cdh1, and availability of these co-activators determines the period of APC activity (5–7). Cdc20 expression is cell cycle-dependent. It appears during S phase and then is destroyed by APCCdh1 in late mitosis (5, 8–10). Cdh1 on the other hand is present throughout the cell cycle, but its ability to interact with APC is controlled by cyclin-dependent kinase (CDK) phosphorylation (11–13) and, at least in budding yeast, cytoplasmic sequestration (14). The ability of Cdc20 to activate APC is also controlled by phosphorylation, for example in response to checkpoint signals (15, 16). An additional mechanism of APCCdh1 regulation involves autoubiquitination (17, 18).
Finally, in recent years binding of inhibitors to the co-activator proteins has emerged as an important mode of APC control. Cdc20 is bound and inhibited by the mitotic checkpoint complex containing Mad2, Mad3/BubR1, and Bub3 in response to unattached kinetochores to prevent premature securin degradation and initiation of anaphase (19). Vertebrate Emi1 binds to both Cdc20 and Cdh1 as well as the APCCdh1 complex and prevents association of APC with substrates from late G1 until early M (20, 21). Fission yeast mes1 protein inhibits the APCCdc20-mediated degradation of mitotic cyclin following meiosis I by blocking substrate binding to Cdc20 (22), and Erp1/Emi2 is the cytostatic factor responsible for meiosis II arrest in vertebrate eggs by binding Cdc20 and inhibiting APCCdc20 (23, 24). In many cases, these inhibitors seem to act by preventing association of co-activators or APC-co-activator complexes with their substrates. In the case of Emi1 and Mad3/BubR1, evidence suggests that inhibition occurs via a pseudosubstrate mechanism (21, 25) in which the inhibitors structurally mimic APC substrates and competitively inhibit binding of true substrates.
We recently identified an inhibitor of APCCdh1 in budding yeast called Acm1 (26). Although the specific biological role of Acm1 is still not clear, when overexpressed it can suppress the lethal APCCdh1 activity that results from lack of inhibitory Cdh1 phosphorylation sites and inhibit the function of Cdh1 in promoting mitotic exit (26). These observations are consistent with Acm1 acting as an inhibitor of mitotic cyclin proteolysis in vivo, and indeed Acm1 inhibits APC-catalyzed ubiquitination of Clb2 in vitro (27). Acm1 is tightly regulated during the mitotic cell cycle, appearing in late G1 and disappearing abruptly in late mitosis, mirroring the window of high CDK activity and APCCdh1 inactivity. This pattern of expression is reminiscent of many APCCdh1 substrates, and Acm1 has conserved sequence elements common to APC substrates, including several destruction boxes (RXXL) and a KEN box (10, 28). These degron sequences are commonly required for substrate recognition and ubiquitination by APC (29). However, we report here that Acm1 proteolysis is actually independent of APC. We show instead that the budding yeast CDK, Cdc28, and the dual specificity phosphatase Cdc14 have opposing roles in regulating the phosphorylation and APC-independent proteolysis of Acm1. Our results reveal an alternative APC-independent mechanism for establishing cell cycle-dependent expression coinciding with the period of high CDK activity, and emphasize the existence of additional late mitotic proteolytic mechanisms.
EXPERIMENTAL PROCEDURES
Cloning, Strain Construction, and Mutagenesis—All plasmids used are listed in Table 1. Details of plasmid constructions are available upon request. In general, centromeric expression plasmids were constructed from a common plasmid series (30). ACM1-5A and ACM1-5E alleles encoding mutants with alanine (5A) or glutamate (5E) substitutions at the five consensus CDK sequences (residues 3, 31, 48, 102, and 161) and the cdh1-11A allele were generated with the QuikChange Multi site-directed mutagenesis kit (Stratagene). Individual point mutations in Acm1 were generated using the standard QuikChange kit. pHLP117, expressing 3HA-ACM1 from its natural promoter, was described previously (26). To construct pHLP183 expressing Cdc28 from its natural promoter, CDC28 with 350 bp of 5′- and 230 bp of 3′-flanking sequence was amplified by PCR from genomic DNA and cloned into the EcoRI and SalI sites of pBM258. pHLP183 complements the temperature sensitivity of a cdc28-4 strain. All constructions involving a PCR step and all site-specific mutations were confirmed by DNA sequencing.
TABLE 1.
Yeast strains and plasmids used in this study
|
Strain
|
Relevant genotype
|
Source
|
||||
| W303 background | ||||||
| YKA150 | MATabar1::URA3 | 60 | ||||
| YKA247 | MATabar1::URA3 acm1::KanMX4 | 26 | ||||
| YKA254 | MATaacm1::KanMX4 | This study | ||||
| YKA415 | MATabar1::URA3 cdc23-1 | This study | ||||
| HCY114 | MATacdc14-1 | This study | ||||
| HCY115 | MATaleu2::GAL-HA-CDC14:LEU2 | This study | ||||
| HCY116 | MATaleu2::GAL-HA-CDC14C283S:LEU2 | This study | ||||
| DLY3033 | MATacdc15-2 | Daniel Lew, Duke University | ||||
| RJD2632 | MATacdc28-as1 | Raymond Deshaies, California Institute of Technology | ||||
|
5397
|
MATacdc28-4
|
Steven Reed, Scripps
|
||||
| BY4741 background | ||||||
| YKA170 | MATacdh1::KanMX4 CLB2-3HA:HIS3 | This study | ||||
| YKA226 | MATa3HA-ACM1 | 26 | ||||
| YKA242 | MATabar1::hisG acm1::URA3 CDH1-3FLAG:KanMX4 | This study | ||||
| YKA244 | MATacdh1::KanMX4 | This study | ||||
| YKA245 | MATabar1::URA3 cdh1::KanMX4 3HA-ACM1 | 26 | ||||
| YKA249 | MATabar1::hisG 3HA-BMH1 | 26 | ||||
| YKA253 | MATabar1::hisG acm1::KanMX4 | This study | ||||
| YKA295 | MATabar1::hisG acm1::KanMX4 3FLAG-BMH1 | This study | ||||
|
YKA407
|
MATabar1::hisG acm1::KanMX4
pdr5::URA3
|
This study
|
||||
| Plasmid name | Origin | Marker | Promoter | Expressed protein | ||
| pHLP101 | CEN/ARS | LEU2 | ADH | HA-Acm1 | ||
| pHLP102 | CEN/ARS | LEU2 | ADH | HA-Acm1-5A | ||
| pHLP103 | CEN/ARS | LEU2 | ADH | HA-Acm1-5E | ||
| pHLP107 | CEN/ARS | LEU2 | ADH | 3FLAG-Acm1 | ||
| pHLP109 | CEN/ARS | LEU2 | GAL1 | HA-Acm1 | ||
| pHLP110 | CEN/ARS | LEU2 | GAL1 | HA-Acm1-5A | ||
| pHLP111 | CEN/ARS | LEU2 | GAL1 | HA-Acm1-5E | ||
| pHLP112 | CEN/ARS | URA3 | GAL1 | 3FLAG-Acm1 | ||
| pHLP113 | CEN/ARS | URA3 | GAL1 | 3FLAG-Acm1-5A | ||
| pHLP117 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1 | ||
| pHLP130 | CEN/ARS | LEU2 | ADH | 3FLAG-Cdh1 | ||
| pHLP135 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-S37A | ||
| pHLP136 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-S48A | ||
| pHLP137 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-S102A | ||
| pHLP138 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-S202A | ||
| pHLP139 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-T161A | ||
| pHLP150 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-S3A | ||
| pHLP169 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-5A/T161 | ||
| pHLP183 | CEN/ARS | URA3 | CDC28 | Cdc28 | ||
| pHLP185 | CEN/ARS | LEU2 | GAL1 | HA-Acm1-T161A | ||
| pHLP209 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-5A | ||
| pHLP212 | CEN/ARS | LEU2 | GAL1 | 3HA-Acm1 | ||
| pHLP222 | CEN/ARS | LEU2 | ACM1 | 3HA-Acm1-5E | ||
| pHLP231 | CEN/ARS | TRP1 | GAL1 | 3FLAG-Cdh1 | ||
| pHLP232 | CEN/ARS | TRP1 | GAL1 | 3FLAG-Cdh1-11A | ||
| pESCLeu-mycCDC14 | 2 μm | LEU2 | GAL1 | myc-Cdc14 | ||
| pESCTrp-FIN1myc | 2 μm | TRP1 | GAL1 | Fin1-myc | ||
Yeast strains (Table 1) were engineered by PCR-mediated gene disruption or epitope tag insertion or linearized plasmid integration using standard procedures described elsewhere (31–33). Deletion strains were confirmed by PCR and tagged strains by PCR, DNA sequencing, and immunoblotting.
Protein Purification—Immunoaffinity purification of 3FLAG-Cdh1 and associated proteins has been described (26). 3FLAG-Acm1 was purified by the same procedure. Clb2–3HA/Cdc28 was immunoaffinity-purified from extracts of strain YKA170 using anti-HA antibody resin in 50 mm Tris-HCl (pH 8.0), 250 mm NaCl, 10% glycerol, 0.1% Triton X-100, 20 mm sodium fluoride, 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 μm pepstatin, and 100 μm leupeptin. After extensive washing with this buffer, the complex was eluted in kinase buffer (10 mm HEPES (pH 7.5), 10 mm MgCl2, 50 mm NaCl, 10% glycerol, 0.5 mm dithiothreitol, 0.05% Triton X-100) by competition with 100 μg/ml HA peptide (Sigma), adjusted to 40% glycerol, and stored at –20 °C.
Recombinant 3FLAG-Acm1 was expressed in Escherichia coli, and cells were lysed by sonication in buffer A (50 mm sodium phosphate (pH 7.5), 500 mm NaCl, 10% glycerol) containing 1% Triton X-100, 1 mm PMSF, and 1 μm pepstatin. Extracts were cleared by centrifugation for 30 min at 35,000 × g and incubated with anti-FLAG antibody resin for 2 h. Resin was washed extensively with buffer A containing 0.1% Triton X-100 and 3FLAG-Acm1 eluted twice by competition with 250 μg/ml 3xFLAG peptide (Sigma). Pooled elutions were flash-frozen in small aliquots and stored at –80 °C. Acm1 concentration was estimated by densitometry of Coomassie Blue-stained polyacrylamide gels using bovine serum albumin to generate a standard curve.
6His-Cdc14 was expressed in E. coli, and cells were lysed by sonication in 20 mm Tris-HCl (pH 7.9), 500 mm NaCl, and 5 mm imidazole. Extract was cleared by centrifugation and loaded onto a 5-ml Hi-Trap Ni2+ column (GE Healthcare). The column was washed with buffer containing 20 mm imidazole and processed with a 100-ml linear gradient from 20 to 300 mm imidazole. Pooled fractions containing 6His-Cdc14 were diluted 1:3 with 10 mm Tris-HCl (pH 7.5), 10 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol, loaded on a Mono Q column (GE Healthcare), and eluted with a linear gradient from 10 mm to 1 m NaCl. Peak 6His-Cdc14 fractions were pooled and stored at 4 °C. A Net1 fragment (residues 1–600) with a His6 tag on the C terminus was purified by Ni2+ affinity chromatography as described for 6His-Cdc14 and dialyzed into 20 mm Tris-HCl (pH 7.5), 137 mm NaCl, 2.6 mm KCl, 0.1% β-mercaptoethanol. Cdc14 and Net1 concentrations were determined by the Bradford method (34) using a bovine serum albumin standard curve.
Phosphorylation Site Mapping—SDS-polyacrylamide gel slices containing Acm1 (roughly 0.1–1 μg) were excised, destained, and treated overnight with either 20 μg/ml sequencing grade trypsin (Promega) in 50 mm ammonium bicarbonate, 20 μg/ml Lys-C (Roche Applied Science) in 25 mm Tris-HCl (pH 8.6), 1 mm EDTA, or 50 μg/ml Glu-C (Roche Applied Science) in 50 mm sodium phosphate (pH 7.8) at 37 °C. Peptides were extracted twice with acetonitrile, lyophilized, and resuspended in 5% acetonitrile, 0.1% trifluoroacetic acid immediately prior to LC/MS-MS analysis.
Peptides were separated on an Agilent 1100 capillary HPLC system using Zorbax C18 trap and 75-μm × 150-mm capillary columns. For LC/MALDI, peptides were eluted with a gradient of increasing acetonitrile in 0.1% trifluoroacetic acid at 800 nl/min, mixed 1:1 with 5 mg/ml α-cyano-4-hydroxycinnamic acid supplied by a syringe pump, and spotted in 0.8-μl fractions on a sample plate using an Agilent LC micro spotting system. Fractions were analyzed in positive MS and MS-MS modes on an Applied Biosystems 4700 mass spectrometer. For electrospray LC/MS-MS analysis, peptides were eluted with a gradient of increasing acetonitrile in 0.1% formic acid at 300 nl/min and injected into a Thermo Scientific LTQ ion trap using a nanoelectrospray source.
Peptides were initially identified by automated searching of a data base of Saccharomyces cerevisiae proteins with either Mascot (Matrix Science) for 4700 data or Sorcerer (Sage-N Research) for LTQ data. All spectra from putative phosphopeptides were then interpreted manually to confirm correct peptide identification and localization of phosphorylated residues.
Cell Growth and Immunoblotting—We used standard yeast growth conditions and media. For G1 arrests, α-factor peptide (Genscript Corp.) was used at 5 μg/ml for BAR1 or 50 μg/liter for bar1Δ. Hydroxyurea (HU; Sigma) was used at 10 mg/ml for S arrest, and nocodazole (Sigma) was used at 7.5 μg/ml for BY4741 strains, or 15 μg/ml for W303 strains, for early mitotic arrest. For late mitotic arrests, cdc15-2 or cdc14-1 cells were grown at 23 °C and then shifted to 37 °C. Arrests were confirmed by microscopic examination of cell morphology and in some cases by flow cytometry.
G1 block and release experiments (in BAR1 strains) were based on previously published procedures (35) and performed as described (26). Typically, α-factor was added back after 60 min to trap cells in the following G1. For block and release in cdc14-1 strains, cultures were arrested with α-factor at 23 °C and released at 37 °C in pre-warmed medium. Flow cytometry controls were prepared and analyzed as described previously (26).
The following antibodies were used. Monoclonal anti-HA 12CA5 and anti-Myc 9E10 were from Roche Applied Science. Monoclonal anti-FLAG M2, rabbit anti-glucose-6-phosphate dehydrogenase (G6PD), and EZview anti-FLAG M2 and anti-HA-7-agarose affinity resins were from Sigma. Goat anti-Cdc28, rabbit anti-Clb2, and horseradish peroxidase-conjugated donkey anti-goat were from Santa Cruz Biotechnology. Horseradish peroxidase-conjugated anti-rabbit and anti-mouse were from GE Healthcare. All immunoblots were developed using ECL Plus reagents (GE Healthcare).
Protein Stability Assays—For all protein stability measurements, expression from the GAL1 promoter was induced with 2% galactose in YP-Raf (20g/liter peptone, 10 g/liter yeast extract, 2% raffinose), typically for 2 h. Transcription and translation were terminated by addition of glucose (2%) and cycloheximide (0.5–1.0 mg/ml), respectively. Samples were withdrawn at the indicated time points and processed for immunoblot analysis as described previously (26). Other details are described in the figure legends. For experiments in the cdc28-as1 strain, cells were first arrested with HU and then treated with 5 μm 1-NM-PP1 ((36) kindly provided by Dr. Kavita Shah, Purdue University) for 1 h prior to galactose induction. Using centromeric plasmids, expression from the GAL1 promoter results in only slighter higher Acm1 levels than expression from the natural ACM1 promoter (data not shown). Thus, our experimental conditions closely mimic natural Acm1 levels.
Co-immunopurification (co-IP)—Co-IP experiments were performed as described (26) using co-IP buffer (50 mm sodium phosphate (pH 7.5), 100 mm NaCl, 10% glycerol, 0.1% Triton X-100, 5 mm EDTA). PMSF (0.5 mm) and pepstatin (1 μm) were added during cell lysis.
Phosphatase and Kinase Assays—Kinase assays (25 μl) contained 4 μl of immunoaffinity-purified Clb2–3HA/Cdc28 (or control purification), ∼200 nm recombinant 3FLAG-Acm1, and 1 mm ATP in kinase buffer and were performed at 30 °C for 30 min. Phosphatase assays contained ∼100 nm immunoaffinity-purified 3FLAG-Acm1 from yeast and the indicated concentration of recombinant 6His-Cdc14 in 50 mm imidazole (pH 6.9), 1 mm EDTA, 1 mm dithiothreitol and were performed at 30 °C for the indicated time. All reactions were stopped by addition of SDS loading dye, boiled, and processed by SDS-PAGE on 12% polyacrylamide gels (37:1 cross-linking ratio) using standard Tris-glycine-SDS running buffer. Phosphorylated and dephosphorylated Acm1 species were detected by immunoblotting and distinguished based on differences in electrophoretic mobility.
Peptide Competition Assay—Crude Acm1 peptides (154ISLPSFITPPRNSK167) with and without a phosphate on Thr-161 were synthesized by Genscript Corp. and were further purified by reverse phase HPLC, lyophilized, and stored in phosphate-buffered saline. Concentrations were determined by amino acid analysis. 3HA-Bmh1 from yeast extracts was immobilized on anti-HA beads and washed extensively with co-IP buffer. Beads were then incubated with different concentrations of the synthetic peptides for 30 min at 30 °C in the same buffer and pelleted by centrifugation, and the dissociation of 3FLAG-Acm1 from 3HA-Bmh1 was monitored by anti-FLAG immunoblotting of the supernatants.
RESULTS
Acm1 Is a Phosphoprotein and a Substrate of Cdc28—In our initial identification of Acm1 as a Cdh1 interaction partner, a prominent peptide was detected containing phosphorylation at Thr-161. To determine whether Acm1 is phosphorylated in vivo at additional sites, we conducted an extensive tandem MS analysis of purified Acm1. We used several site-specific proteases and both electrospray and MALDI instruments to obtain 100% sequence coverage. Acm1 is phosphorylated on at least nine different residues (Table 2). Three of these, including Thr-161, are part of consensus recognition sequences for CDK ((S/T)PX(K/R)). The same phosphorylation sites were detected in Acm1 from cells arrested in S or late M phases. Two sites, Thr-161 and Ser-202, appeared to be extensively phosphorylated based on comparison of the ion signals for unmodified and phosphorylated peptides during LC/MS analyses (Fig. 1A). The remaining sites appeared to be phosphorylated with relatively low stoichiometry, and phosphorylation was not detected at two additional consensus CDK sequences, including one (Ser-3) that is highly conserved in ACM1 orthologs. Ser-3-containing peptides were either not detected or exhibited very low intensity in all of the MS analyses, leaving open the possibility that it too is phosphorylated.
TABLE 2.
Acm1 phosphopeptides and phosphorylation sites identified by MS
|
Peptide
sequencea
|
Phosphorylation site(s)
|
Detected by
|
|
|---|---|---|---|
| MALDI | ESI | ||
| 37SQIDTDYALR46 | Ser-37 | Yes | Yes |
| 35RRSQIDTDYALR46 | Ser-37 | Yes | Yes |
| 35RRSQIDTDYALRRSPIK51 | Ser-37,b Ser-48c | Yes | No |
| 100NLSPAKICPYE110 | Ser-102c | Yes | Yes |
| 124DLSVDEFK131 | Ser-126 | Yes | Yes |
| 120IALKDLSVDEFK131 | Ser-126 | No | Yes |
| 117GGRIALKDLSVDE129 | Ser-126 | Yes | Yes |
| 154ISLPSFITPPR164 | Ser-155, Thr-161c,d | Yes | Yes |
| 153KISLPSFITPPR164 | Ser-155, Thr-161c,d | Yes | No |
| 154ISLPSFITPPRNSK167 | Thr-161c | Yes | Yes |
| 153KISLPSFITPPRNSK167 | Thr-161,c,d Ser-166 | Yes | Yes |
| 168ISIFFTSK175 | Thr-173 and/or Ser-174 | No | Yes |
| 201LSFHVYEDE209 | Ser-202 | No | Yes |
| 200KLSFHVYEDE209 | Ser-202 | Yes | Yes |
| 195KKVVRKLSFHVYE207 | Ser-202 | Yes | Yes |
| 197VVRKLSFHVYEDE209 | Ser-202 | Yes | Yes |
Confirmed phosphorylation sites are underlined.
The peptide containing Ser-37 and Ser-48 phosphorylation was not observed in doubly phosphorylated form. Rather the monophosphorylated peptide was a mixture of species containing phosphorylation at either Ser-37 or Ser-48.
This denotes phosphorylation at a consensus CDK site.
Ser-155 and Ser-166 were never observed on monophosphorylated peptides (i.e. in the absence of Thr-161 phosphorylation).
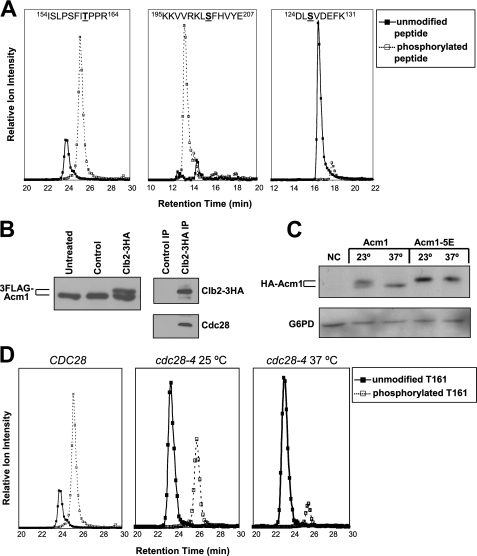
FIGURE 1.
Acm1 is a phosphoprotein and an in vivo substrate of Cdc28. A, extracted ion chromatograms for the doubly charged unmodified and phosphorylated precursor ions of the indicated peptides obtained from electrospray LC/MS analyses. Phosphorylated residues are underlined. Data are from 3FLAG-Acm1 (from pHLP107) purified from yeast arrested in S phase with HU. The 1st two panels illustrate peptides with high phosphate occupancy, suggesting extensive modification at Thr-161 and Ser-202. The last panel illustrates a peptide with low phosphate occupancy at Ser-126. Similar results were obtained from extracted ion chromatograms generated by MALDI LC/MS analysis. B, recombinant 3FLAG-Acm1 was treated with purified Clb2–3HA·Cdc28 complex or a control purification from yeast lacking the HA3 tag. Reaction products were processed by SDS-PAGE and detected by anti-FLAG immunoblotting. Panels on the right are control immunoblots for Clb2–3HA and Cdc28 in the IP samples. C, cdc28-4 strain containing pHLP109 or pHLP111 expressing HA-Acm1 or HA-Acm1–5E, respectively, from PGAL1 was arrested in S phase with HU at 23 °C. Galactose induction was performed at either 23 or 37 °C for 2 h, and proteins were detected by anti-HA immunoblotting. NC, negative control with empty vector. G6PD is a loading control. D, extracted ion chromatograms for unmodified and phosphorylated forms of 3FLAG-Acm1 peptide 154ISLPSFITPPR164 from HU-arrested CDC28 or cdc28-4 cells at permissive (25 °C) and nonpermissive (37 °C) temperatures. The CDC28 chromatogram is repeated from A to allow direct comparison to cdc28-4 chromatograms.
Acm1 was previously identified in a proteomic screen as an in vitro substrate of the yeast mitotic CDK, Clb2-Cdc28 (37). We found that treatment of recombinant 3FLAG-Acm1 with immunoaffinity-purified Clb2-Cdc28 resulted in a slow mobility form of Acm1 during SDS-PAGE (Fig. 1B) that was sensitive to phosphatase (not shown and see below), confirming in vitro phosphorylation by Cdc28. To determine whether Cdc28 phosphorylates Acm1 in vivo, we monitored HA-Acm1 by immunoblotting in yeast containing the temperature-sensitive cdc28-4 allele (Fig. 1C). At the permissive temperature (23 °C), HA-Acm1 migrated as a pair of bands similar to those detected in the in vitro kinase assay. The slower mobility form was absent when HA-Acm1 was expressed at the nonpermissive temperature (37 °C). Acm1 mutants containing glutamic acid or alanine substitutions at the five potential CDK sites (Acm1–5E and Acm1–5A) migrated as single bands that were not affected by temperature (Fig. 1C and not shown). We also purified 3FLAG-Acm1 expressed from PGAL1 in a cdc28-4 strain arrested in S phase and monitored the prominent Thr-161 and Ser-202 phosphorylation sites by MS. Even at 25 °C phospho-Thr-161 was sharply reduced compared with a strain expressing wild-type Cdc28 (Fig. 1D). This is consistent with the documented catalytic deficiency of the Cdc28–4 mutant even at permissive temperature (38). At 37 °C phospho-Thr-161 was reduced even further in the cdc28-4 strain. Phospho-Ser-202, which is not part of a consensus CDK recognition sequence, was not significantly altered in the cdc28-4 strain (not shown). Collectively, these results lead us to conclude that Acm1 is a direct in vivo substrate of Cdc28.
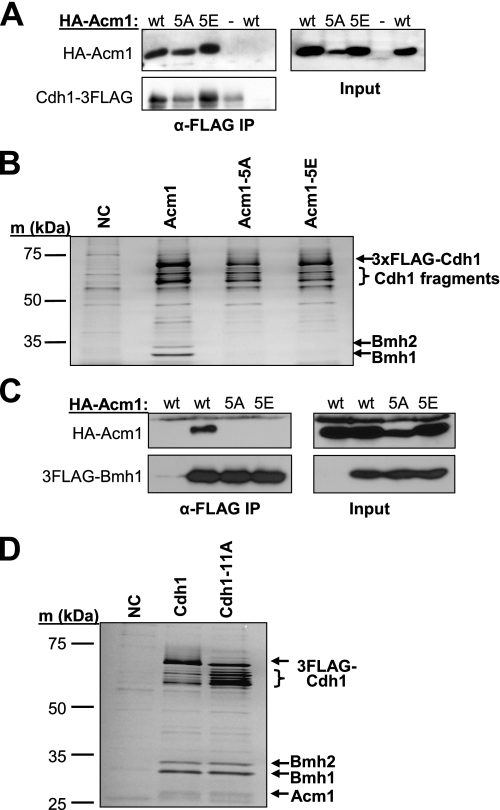
Cdc28 Phosphorylation Sites Are Required for Acm1 Stability and Interaction with Bmh1 and Bmh2—To determine the function of CDK phosphorylation sites on Acm1, we further studied the Acm1–5A and Acm1–5E CDK site mutants. Our previous work demonstrated that Acm1 is required for association of Cdh1 with the 14-3-3 proteins Bmh1 and Bmh2, suggesting that it interacts directly with each (26). Both Acm1–5A and Acm1–5E associated with Cdh1 similar to wild-type Acm1 in a co-IP assay (Fig. 2A). In contrast, neither Acm1–5A nor Acm1–5E associated with Bmh1 or Bmh2 (Fig. 2, B and C). This result suggested that Acm1 phosphorylation at one or more CDK consensus sequences was required to create a binding site for the 14-3-3 proteins and that the glutamate side chain is not an effective phosphate mimic for 14-3-3 binding. Cdh1 is also heavily phosphorylated by Cdc28 at multiple sites (11, 13, 39). However, the ability of the Cdh1-11A mutant containing alanine substitutions at all 11 possible Cdc28 phosphorylation sites to associate with Acm1, Bmh1, and Bmh2 was indistinguishable from that of wild-type Cdh1 (Fig. 2D). Thus, Cdc28 phosphorylation of Cdh1 is not required for assembly of this inhibitory complex.
FIGURE 2.
Role of Acm1 and Cdh1 CDK sites in protein-protein interactions. A, indicated HA-Acm1 proteins were expressed from PADH on CEN plasmids in acm1Δ CDH1–3FLAG cells (YKA242) and Cdh1–3FLAG purified on anti-FLAG beads. The presence of co-purifying HA-Acm1 was monitored by anti-HA immunoblotting. wt, wild-type Acm1; 5A, Acm1–5A; 5E, Acm1–5E. Input represents whole cell extracts used for the co-IPs. B, same as A, except 3FLAG-Cdh1 was expressed from pHLP130 in acm1Δ cells (YKA247), and proteins eluted from anti-FLAG beads were visualized by silver staining. Bmh1 and Bmh2 were identified by MS. HA-Acm1 proteins co-migrated with a nonspecific contaminant in this experiment and are not shown. The presence of each HA-Acm1 protein was confirmed by immunoblotting as in A (not shown). NC, negative control from cells lacking 3FLAG-Cdh1. C, endogenous 3FLAG-Bmh1 was immunopurified from yeast extracts (YKA295), and co-purifying HA-Acm1 and mutants expressed as in A were detected by anti-HA immunoblotting. D, 3FLAG-Cdh1 and the 3FLAG-Cdh1-11A mutant expressed from pHLP231 and pHLP232, respectively, were purified using anti-FLAG beads from whole cell extracts of log phase cultures. Eluted proteins were separated by SDS-PAGE and stained with silver. The identity of labeled bands was confirmed by MS. NC, negative control from cells lacking the FLAG3 tag.
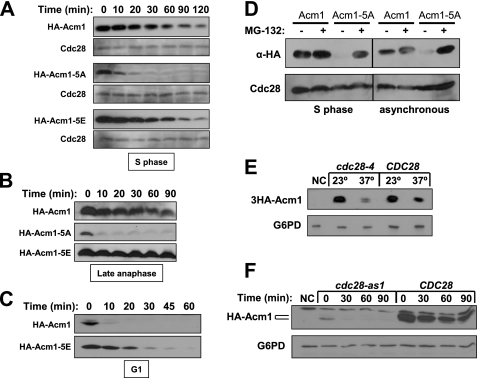
The level of Acm1–5A detected in extracts (see Fig. 2, A and C) was consistently lower than Acm1 and Acm1–5E, suggesting that it might be less stable. We performed promoter shutoff assays to determine whether Acm1–5A has a shorter half-life than Acm1. In S phase (Fig. 3A), early mitosis (not shown), and late mitosis (Fig. 3B), wild-type Acm1 and Acm1–5E were stable, whereas the Acm1–5A mutant was degraded very rapidly. As expected from its expression profile, wild-type Acm1 was highly unstable in α-factor-arrested G1 cells (when Cdc28 activity is absent), but Acm1–5E was significantly more stable (Fig. 3C), demonstrating that mimicking constitutive Cdc28 phosphorylation protects Acm1 from G1 proteolysis. Because the stability experiments require induced expression from the GAL1 promoter, we also compared expression of Acm1 and Acm1–5A expressed from the natural ACM1 promoter on a single-copy plasmid. In a synchronized cell cycle, we never detected 3HA-Acm1–5A (not shown). Fig. 3D shows that in S phase or asynchronous cells 3HA-Acm1–5A was essentially undetectable unless the proteasome was inhibited with MG-132. Stable wild-type 3HA-Acm1 on the other hand was unaffected by MG-132 treatment. This result argues that our stability assays do not reflect artifacts of unnatural Acm1 expression from the GAL1 promoter.
FIGURE 3.
Cdc28 phosphorylation protects Acm1 from proteolysis. A, expression of HA-Acm1, HA-Acm1–5A, and HA-Acm1–5E from PGAL1 on CEN plasmids in YKA150 was induced with galactose after HU arrest and then shut off by addition of glucose and cycloheximide. Protein levels in whole cell extracts were monitored at the indicated times after shutoff by anti-HA immunoblotting. Cdc28 is a loading control. B, identical to A, except cdc15-2 cells (DLY3033) were used and arrested in late anaphase at 37 °C. C, identical to A, except cells were arrested in G1 with α-factor. D, 3HA-Acm1 or 3HA-Acm1–5A were expressed from the ACM1 promoter on pHLP117 or pHLP209, respectively, in pdr5Δ cells (YKA407) growing asynchronously, or arrested in S with HU. Cultures were treated with MG-132 or left untreated and extracts analyzed by anti-HA immunoblotting. Cdc28 is a loading control. E, cdc28-4 cells expressing 3HA-Acm1 from the ACM1 promoter on pHLP117 at 23 °C were treated with HU and then incubated at the permissive (23 °C) or nonpermissive (37 °C) temperature for 2 h. 3HA-Acm1 level was compared by anti-HA immunoblotting. The CDC28 cells contain pHLP183, which complements the cdc28-4 defect. F, cdc28-as1 cells expressing HA-Acm1 from PGAL1 (pHLP109) were arrested in S with HU, treated with 1-NM-PP1 to specifically inhibit Cdc28-as1, and induced with galactose. HA-Acm1 level at the indicated time points after shutoff was monitored by immunoblotting. CDC28 cells also harbor pHLP183. In both E and F G6PD is a loading control, and NC is a negative control lacking tagged Acm1.
To provide independent evidence that Cdc28 phosphorylation stabilizes Acm1, we initially monitored 3HA-Acm1 expressed from its natural promoter in HU-arrested CDC28 and cdc28-4 cells before and after shift to 37 °C (Fig. 3E). The level of 3HA-Acm1 was dramatically reduced in cdc28-4 cells following the shift to 37 °C, supporting a role for Cdc28 in stabilizing Acm1. However, temperature itself seemed to affect Acm1 stability because 3HA-Acm1 was also reduced at 37 °C in CDC28 cells (although not as acutely as in cdc28-4 cells). To avoid temperature effects, we performed HA-Acm1 stability assays in HU-arrested cdc28-as1 cells in which Cdc28 activity can be chemically inhibited by the ATP analog 1-NM-PP1 (36). The initial HA-Acm1 level achieved in cdc28-as1 cells treated with 1-NM-PP1 was relatively low, was detected exclusively as the fast mobility form, and disappeared rapidly after expression was terminated (Fig. 3F). In contrast, the initial HA-Acm1 level was much higher in CDC28 control cells, existed primarily as a slower mobility form, and was mostly stable throughout the time course. Collectively these results reveal that phosphorylation of Acm1 by Cdc28 stabilizes it during S phase and mitosis when CDK activity is high and raises the possibility that dephosphorylation of Acm1 might contribute to its rapid proteolysis in late mitosis.
Acm1 Is a Substrate of Cdc14 Phosphatase—Cdc14 phosphatase is activated in late anaphase in budding yeast to reverse the action of mitotic CDK and help establish the period of low CDK activity necessary for mitotic exit (40). One of its critical substrates is Cdh1, which becomes activated upon dephosphorylation to promote the APC-dependent destruction of Clb2 and other proteins (11). We speculated that Acm1 might be a direct target of Cdc14 because Acm1 is tightly associated with Cdh1 during mitosis and is phosphorylated at several CDK consensus sequences, the preferred substrate of Cdc14 phosphatases (41, 42).
To test if Acm1 is recognized by Cdc14 as a substrate in vivo, we used a Cdc14 active site point mutant, Cdc14-C283S, as a substrate trap. Studies with protein-tyrosine phosphatases have shown that certain active site mutants that lack catalytic activity are nevertheless able to form stable complexes with their cognate phospho-substrates (43, 44). These mutant-substrate complexes can be isolated by biochemical means. HA-Cdc14 or HA-Cdc14-C283S was co-expressed with 3FLAG-Acm1 or 3FLAG-Acm1–5A and interactions assessed by co-IP. HA-Cdc14-C283S, but not HA-Cdc14, was detected strongly in 3FLAG-Acm1 IP samples (Fig. 4A). Neither protein was detected in 3FLAG-Acm1–5A IP samples, demonstrating that the interaction requires Cdc28 phosphorylation sites. The lack of interaction between Cdc14-C283S and Acm1–5A also excludes any indirect interaction mediated by Cdh1 because Acm1–5A interacts normally with Cdh1 under our co-IP conditions (Fig. 2A). The same results were observed when the reverse anti-HA IPs were performed (not shown). We conclude that Cdc14 can specifically recognize the Cdc28-phosphorylated form of Acm1 as a substrate in vivo.
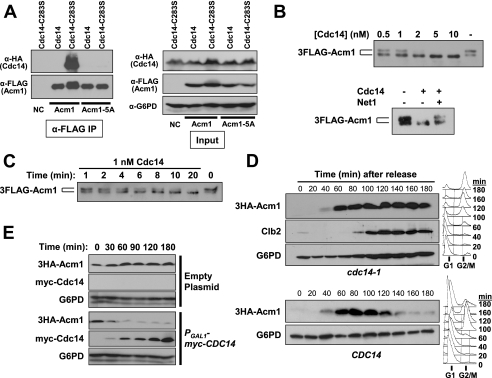
FIGURE 4.
Cdc14 dephosphorylates Acm1 and is required for Acm1 proteolysis. A, interaction of 3FLAG-Acm1 (expressed from pHLP112) or the 3FLAG-Acm1–5A mutant (from pHLP113) with either HA-tagged Cdc14 (strain HCY115) or Cdc14-C283S (strain HCY116) was monitored by anti-FLAG co-IP. Expression of all tagged proteins was induced with galactose in asynchronous mid-log cultures. Co-purification of Cdc14 proteins was detected by anti-HA immunoblotting. Input represents initial extracts. B, immunoaffinity purified 3FLAG-Acm1 expressed from pHLP107 in YKA247 was treated with the indicated concentrations of recombinant 6His-Cdc14 for 5 min at 30 °C. In the lower panel, 100 nm of a recombinant N-terminal fragment of the Cdc14-specific inhibitor Net1 was added to an Acm1 phosphatase reaction containing 10 nm 6His-Cdc14 to rule out contaminating E. coli phosphatase activity. C, dephosphorylation of the same 3FLAG-Acm1 substrate by 1 nm recombinant 6His-Cdc14 was measured over the indicated time. D, CDC14 and cdc14-1 cells (strains W303 and HCY114, respectively) containing pHLP117 expressing 3HA-Acm1 from the ACM1 promoter were arrested with α-factor at 23 °C and released into fresh medium at 37 °C. 3HA-Acm1 was monitored over time by anti-HA immunoblotting. Cell cycle stage was monitored by flow cytometry (right panels) and Clb2 immunoblotting. E, YKA245 (cdh1Δ 3HA-ACM1) containing a PGAL1-myc-CDC14 2 μm expression plasmid or an empty control plasmid was arrested with nocodazole and myc-Cdc14 expression induced with galactose. The level of endogenous 3HA-Acm1 was then monitored over time by immunoblotting. G6PD is a loading control.
We also tested if Cdc14 is able to dephosphorylate Acm1 in vitro. 3FLAG-Acm1 purified from an asynchronous yeast culture was used as a substrate in phosphatase assays with recombinant 6His-Cdc14. Dephosphorylation of Acm1 was detected as loss of the Cdc28-dependent slow mobility SDS-PAGE band defined in Fig. 1. The slow mobility form of Acm1 was completely eliminated by 6His-Cdc14 concentrations as low as 2 nm in a 5-min reaction (Fig. 4B). Cdc14 specificity was confirmed by sensitivity of the reaction to the Cdc14 inhibitor Net1. In a time course, 1 nm 6His-Cdc14 completely dephosphorylated 3FLAG-Acm1 in 20 min (Fig. 4C). This concentration of Cdc14 is similar to that reported previously for dephosphorylation of the Cdc14 targets Swi5 and Sic1 (40, 45) and even lower than that reported for dephosphorylation of Cdh1 itself (11). Thus, Acm1 is a potent in vitro substrate of Cdc14.
Cdc14 Triggers Acm1 Proteolysis—We also speculated that dephosphorylation by Cdc14 might trigger the rapid proteolysis of Acm1 that occurs in late mitosis because our data suggested that lack of CDK phosphorylation renders Acm1 highly unstable. To determine whether Cdc14 activity contributes to Acm1 proteolysis in vivo, we first analyzed the fate of 3HA-Acm1 in the absence of Cdc14 function. A cdc14-1 strain arrests in late anaphase at 37 °C (46). We arrested cdc14-1 and CDC14 cells in G1 at 23 °C and released them into fresh media at 37 °C so they would progress synchronously through the cell cycle (Fig. 4D). The cdc14-1 cells arrested in late mitosis with stable Clb2, as expected. 3HA-Acm1 also remained stable in these cells, and furthermore, it exhibited the same phosphorylation pattern observed in S phase cells (not shown). 3HA-Acm1 was destroyed as cells exited mitosis in the CDC14 strain. These results demonstrate that Cdc14 function is essential for Acm1 dephosphorylation and proteolysis in late mitosis.
Next, we overexpressed Cdc14 in nocodazole-arrested cdh1Δ cells to see if it would destabilize endogenous 3HA-Acm1 at a point where it is normally stable (Fig. 4E). Induction of myc-Cdc14 overexpression with galactose resulted in a rapid and substantial decrease in 3HA-Acm1. cdh1Δ cells were used to minimize indirect effects of Cdc14 triggering mitotic exit events by reducing CDK activity (11, 40). However, this result also provides evidence that APCCdh1 is not responsible for Acm1 proteolysis (see below).
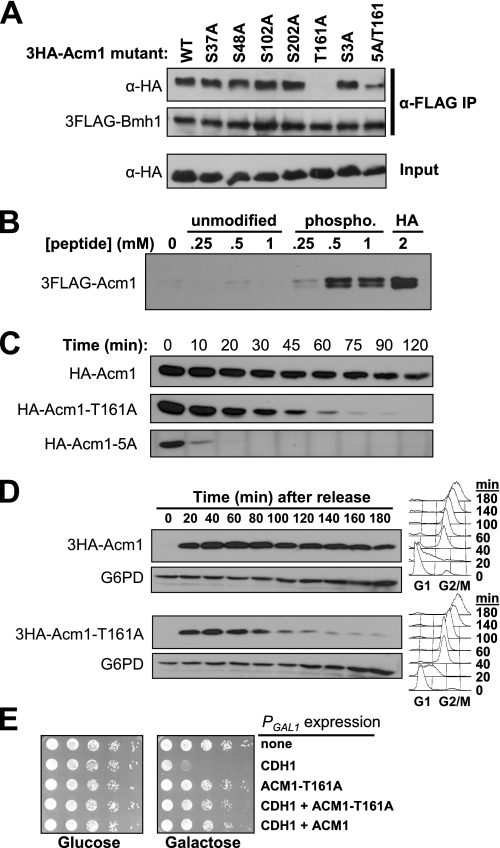
The 14-3-3 Protein Bmh1 Binds Directly to Phosphorylated Thr-161 on Acm1 and Contributes to Acm1 Stability—The observations that CDK phosphorylation sites on Acm1 promote its stability and are required for Bmh1/Bmh2 binding raised the possibility that Bmh1 and Bmh2 might help protect Acm1 from proteolysis. We screened a panel of six individual 3HA-Acm1 phosphorylation site mutants (the three CDK sites and two most prominent non-CDK sites identified by MS and the conserved CDK sequence at Ser-3) for defects in binding to 3FLAG-Bmh1. Most of the single mutations had no effect on association of 3HA-Acm1 with 3FLAG-Bmh1 (Fig. 5A). T161A was the notable exception. Binding of 3HA-Acm1-T161A to 3FLAG-Bmh1 was undetectable. 3FLAG-Bmh1 interacted with Acm1 when Thr-161 was the only one of the six sites present (Acm1–5A/T161), albeit with reduced efficiency. This suggested that phospho-Thr-161 alone was sufficient for Bmh1 binding and that one or more additional phosphorylation sites may contribute to optimal binding as well. It is noteworthy that the sequence surrounding Thr-161 (FIpTPP) is both highly conserved in ACM1 orthologs (26) and closely resembles the mode II 14-3-3 consensus binding sequence (RX(Y/F)XpSXP) revealed in a peptide library screen (47).
FIGURE 5.
Bmh1 binding to phosphorylated Thr-161 contributes to Acm1 stability. A, co-IP of wild-type 3HA-Acm1 and the indicated mutants (expressed from the ACM1 promoter on CEN plasmids) with endogenous 3FLAG-Bmh1 (YKA295) isolated on anti-FLAG beads from mid-log yeast extracts was detected by anti-HA immunoblotting. B, endogenous 3HA-Bmh1 (YKA249) with bound 3FLAG-Acm1 (expressed from pHLP107) was isolated from yeast extracts and challenged with the indicated concentration of unmodified and phosphorylated synthetic peptide as described under “Experimental Procedures.” Dissociation of 3FLAG-Acm1 was monitored by anti-FLAG immunoblotting. The HA lane is for comparison and represents protein eluted from the resin by competition with the antigenic HA peptide. C, stability of HA-Acm1, HA-Acm1-T161A, and HA-Acm1–5A in nocodazole-arrested cells was monitored at the indicated time points after PGAL shutoff. D, cdc14-1 cells (HCY114) expressing 3HA-Acm1 or 3HA-Acm1-T161A from the ACM1 promoter on CEN plasmids were arrested at 23 °C in G1 and released into fresh medium at 37 °C. 3HA-Acm1 was monitored over time by immunoblotting. Flow cytometry (right panels) was used to monitor cell cycle progression and confirm mitotic arrest. G6PD is a loading control. E, 10-fold serial dilutions of YKA247 cultures expressing the indicated combinations of 3FLAG-Cdh1 (pHLP231), HA-Acm1 (pHLP109), and HA-Acm1-T161A (pHLP185) from PGAL1 were spotted on rich media plates containing either glucose or galactose and grown at 30 °C for several days.
Phospho-Thr-161 could be bound directly by Bmh1 and Bmh2 or it could act allosterically, perhaps by inducing a conformational change in Acm1 that exposes an independent binding site. To test for direct binding, we performed competition assays with synthetic Acm1 peptides containing Thr-161 or phospho-Thr-161 (Fig. 5B). 3HA-Bmh1 with bound 3FLAG-Acm1 was isolated from yeast whole cell extracts on anti-HA affinity beads. The immobilized complex was challenged with the phosphorylated and unmodified synthetic peptides and dissociation of 3FLAG-Acm1 monitored by immunoblotting. The phosphopeptide, but not the unmodified peptide, efficiently dissociated 3FLAG-Acm1 from immobilized 3HA-Bmh1. The required peptide concentration seemed high but was similar or even lower than that used for dissociation of 14-3-3 interactions on affinity beads in other studies (48–50). It likely reflects a requirement for additional sequence elements and secondary phosphorylation sites to promote high affinity binding of the dimeric 14-3-3 proteins, as has been proposed (47, 51). We conclude that Bmh1 (and likely Bmh2) directly binds phosphorylated Thr-161 on Acm1. To our knowledge this is the first reported case of CDK directly regulating 14-3-3 binding to a target protein.
To determine whether Bmh1/Bmh2 binding contributes to Acm1 stability, we initially followed Acm1-T161A levels during a synchronized cell cycle. The cell cycle expression profiles of Acm1-T161A and Acm1 were very similar (data not shown). To more directly assess differences in stability, we performed promoter shutoff assays (Fig. 5C). The half-life of Acm1-T161A was clearly much shorter than wild-type Acm1, although not nearly as short as the Acm1–5A mutant. Thus, Thr-161 contributes to Acm1 stability, most likely by promoting the phosphorylation-dependent binding of Bmh1 and Bmh2. Because our results suggest Cdc14 dephosphorylation of Acm1 triggers its proteolysis in late mitosis, we examined Acm1-T161A stability in cdc14-1 cells. At 37 °C, wild-type 3HA-Acm1 remains stable as synchronous G1-released cells arrest in late anaphase (Fig. 5D). In contrast, the 3HA-Acm1-T161A mutant undergoes nearly complete proteolysis, bypassing the requirement for Cdc14. These results suggest that Bmh1 and Bmh2 binding may be necessary to maintain the stability of Acm1 until Cdc14 is activated in late mitosis. Normal binding of Bmh1 and Bmh2 to Acm1 does not appear to be required for APC inhibition because the Acm1-T161A mutant was as effective as wild-type Acm1 at suppressing the lethality of overexpressed Cdh1 (Fig. 5E), an in vivo assay for APCCdh1 inhibition by Acm1 that we described previously (26). We conclude that the primary function of Bmh1/Bmh2 binding to Acm1 is to protect Acm1 from inopportune proteolysis.
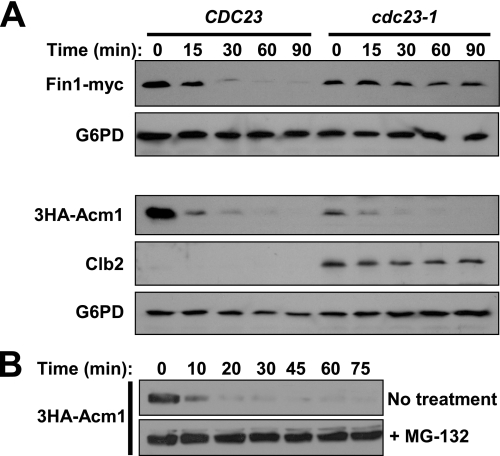
Acm1 Proteolysis Is Independent of the APC—The cell cycle expression profile of Acm1 is reminiscent of many APC substrates. That Acm1 associates with the APC co-activator Cdh1, contains conserved APC degron-like sequences, and appears to compete with some substrates for binding to Cdh1 (26) further suggests that Acm1 proteolysis could be mediated by APC. To test this directly, we measured Acm1 stability in G1-arrested cdc23-1 cells by PGAL promoter shutoff. APC is inactive at 37 °C in cdc23-1 cells. We used the recently described APC substrate Fin1 as a positive control (52). In wild-type CDC23 cells both 3HA-Acm1 and Fin1-myc were rapidly degraded after addition of glucose and cycloheximide at 37 °C, as expected (Fig. 6A). In cdc23-1 cells Fin1-myc was stabilized as expected for an APC substrate, yet 3HA-Acm1 was still destroyed. In fact, 3HA-Acm1 was reproducibly detected at a lower level in G1-arrested cdc23-1 cells than CDC23 cells at the zero time point. We probed for Clb2 in the 3HA-Acm1 samples to ensure APC activity was deficient. Clb2 is stabilized and persists in a G1 arrest even at the permissive temperature in cdc23-1 cells (53). In contrast to APC, inhibition of the proteasome with MG-132 resulted in stable 3HA-Acm1 in G1-arrested cells after shutoff (Fig. 6B). We conclude that Acm1 proteolysis is APC-independent but proteasome-dependent.
FIGURE 6.
Acm1 proteolysis is APC-independent. A, Fin1-myc or 3HA-Acm1 (pHLP212) expression from PGAL1 was induced with galactose in CDC23 and cdc23-1 cells (YKA150 and YKA415, respectively) after arrest in G1 with α-factor. Expression was terminated with glucose and cycloheximide after shifting the temperature to 37 °C, and the tagged proteins or Clb2 were monitored at the indicated time points by immunoblotting. G6PD is a loading control. The level of Clb2 detected in cdc23-1 samples was comparable with that from asynchronous CDC23 cells (not shown). B, PGAL shutoff assay similar to A was performed in a pdr5Δ strain (YKA407) containing pHLP212. Cells were arrested in G1 and either treated with 50 μm MG-132 or left untreated. HA-Acm1 level was monitored at the indicated time points after shutoff by anti-HA immunoblotting.
DISCUSSION
Acm1 Stability Is Dependent on Its Phosphorylation Status—In this study we have demonstrated a role for CDK phosphorylation in stabilizing the budding yeast APC inhibitor Acm1 during S and M phases of the cell cycle. We have also shown that Acm1 is a direct in vivo substrate of Cdc14 phosphatase and that dephosphorylation by Cdc14 is the primary trigger for rapid Acm1 proteolysis in late mitosis. This is consistent with the proposal that Cdc14 reverses the bulk of CDK phosphorylation as cells exit mitosis and resets the cell cycle for another round of division (40). Cdc14 may act indirectly to promote Acm1 proteolysis as well. Cdc14 dephosphorylates Swi5, Sic1, and Cdh1 leading to inhibition of Cdc28 and complete cyclin destruction required for mitotic exit. This elimination of CDK activity, a crucial factor for stabilization of Acm1, probably contributes to its destruction.
CDK phosphorylation was previously shown to stabilize the replication licensing factor Cdc6 by protecting its degrons from recognition by APCCdh1 (54). Mitotic phosphorylation was also suggested to stabilize the Xenopus Aurora A kinase by blocking recognition by APCCdh1 (55). We initially thought Acm1 stability might be regulated in a similar manner, but our results demonstrate that Acm1 is not an APC substrate (see below). Instead, they reveal a novel mechanism for generating a cell cycle expression profile that mirrors the window of high CDK activity from late G1 until late M. This mechanism includes a combination of cell cycle-dependent transcription of ACM1 (56), the relative balance of CDK and Cdc14 activity, and a proteolytic mechanism other than APC that is active in late mitosis and G1.
Acm1 Is Not a Substrate of APC—The timing of Acm1 destruction in late mitosis, the presence of conserved degron sequences that are commonly required for APC substrate ubiquitination and destruction, and its association with the co-activator Cdh1 all suggested that Acm1 might be an APC substrate in addition to an inhibitor. However, our results invariably suggest that Acm1 proteolysis is independent of APC. Acm1 proteolysis was unaffected in an APC-deficient strain (Fig. 6A), and it was still degraded in response to Cdc14 overexpression in nocodazole-arrested cells lacking Cdh1 (Fig. 4E). We also failed to see any difference in the proteolysis of Acm1 in CDH1 cdc15-2 and cdh1Δ cdc15-2 cells after synchronous release from a late anaphase block (not shown).
Furthermore, during a separate mechanistic study on Acm1, we found that mutations in its conserved destruction box and KEN box sequences disrupted Cdh1 binding and APC inhibition but had no effect on Acm1 proteolysis in vivo.4 Why does Acm1 contain these degron-like sequences then? One possibility is that Acm1 inhibits APC via a pseudosubstrate mechanism similar to the vertebrate Emi1 and budding yeast Mad3 proteins (21, 25). Both Emi1 and Mad3 contain conserved degron sequences that are essential for interaction with APC co-activator proteins, APC inhibition, and biological function. We are currently testing this possibility for Acm1. Despite Acm1 proteolysis being independent of APC, it was absolutely dependent on activity of the 26 S proteasome, suggesting that another ubiquitin ligase is responsible for its late mitotic degradation.
Implications for Mechanism of Acm1 Proteolysis—Our data suggest that multiple proteolytic mechanisms may be active toward Acm1 during the cell cycle. The constitutive instability of the Acm1–5A mutant lacking CDK phosphorylation sites and the instability of Acm1 in the absence of Cdc28 function suggest that Acm1 is susceptible to a proteolytic activity that is not cell cycle-dependent. On the other hand, the Acm1–5E mutant mimicking constitutive phosphorylation nevertheless exhibits a much shorter half-life in G1 compared with S and M (Fig. 3), and is still fully degraded as synchronized cells exit mitosis (data not shown). Thus, the proteolytic machinery that acts on Acm1 in late mitosis appears to have a cell cycle-regulated component that may be less sensitive to Acm1 phosphorylation status.
Why is the Acm1–5E mutant not constitutively stable if Cdc14-catalyzed dephosphorylation is required for Acm1 proteolysis? The answer may lie in the function of the 14-3-3 proteins Bmh1 and Bmh2. The Acm1–5E mutant does not interact with Bmh1 and Bmh2, and a single point mutation in a phosphorylation site required for Bmh1 binding (Acm1-T161A) substantially reduced the stability of Acm1 but only had a noticeable effect on the profile of naturally expressed Acm1 when cells were arrested in late anaphase with inactive Cdc14 (Fig. 5). These observations suggest that 14-3-3 binding may either protect the phosphorylation status of Acm1 and/or prevent access of the ubiquitin ligase to Acm1 specifically in late mitosis prior to the activation of Cdc14. This could be important, for example, to protect Acm1 from other phosphatases as CDK activity begins to drop after APC initiates the destruction of B-type cyclins at anaphase onset or to protect Acm1 from a ubiquitinating activity that is activated prior to Cdc14. This model suggests that the mere presence of phosphate groups at CDK sites is sufficient to protect Acm1 during most of its expression window but that an additional protective measure is necessary either very briefly in late mitosis or when activation of Cdc14 is delayed. The relative stability of Acm1-T161A compared with Acm1–5A, both of which fail to bind Bmh1 but differ in the number of CDK sites present, supports this idea.
Regulation of 14-3-3 Proteins by CDK—14-3-3 proteins interact with literally dozens of phosphorylated target proteins and modulate their functions in a variety of ways. Many different kinases have been shown or suggested to create phosphorylated binding sites for 14-3-3 proteins on specific substrates (57). However, identification of Thr-161 on Acm1 as a 14-3-3-binding site is apparently the first reported case of CDK directly promoting 14-3-3 binding to a substrate. 14-3-3 proteins have well defined roles in regulation of the eukaryotic cell cycle and checkpoints, including regulation of CDK activity itself (58, 59). It is currently unclear to what extent CDK activity controls other 14-3-3 functions in yeast, but preliminary analyses5 suggesting that CDK-mediated phosphorylation of Fin1 creates Bmh1/Bmh2-binding sites indicate that this mode of regulation may not be restricted to Acm1. It is not known whether similar phenomena exist in higher eukaryotes.
If Bmh1/Bmh2 binding to Acm1 protects its phosphorylation status, then how does Cdc14 specifically gain access to Acm1 to dephosphorylate it? Two plausible possibilities are as follows: 1) that an independent event causes dissociation of Bmh1 and Bmh2 from phosphorylated Acm1 so that Cdc14 (and maybe other phosphatases) can gain access, and 2) that Cdc14 phosphatase specifically recognizest he Bmh1/Bmh2-Acm1 complex as a substrate and can displace Bmh1 and Bmh2 as it dephosphorylates Acm1. Our data do not currently favor one possibility over the other.
Implications for Acm1 Function—Acm1 clearly acts as an inhibitor of APCCdh1 activity (26, 27). The biological significance of this inhibition is still uncertain, and it is not known whether Acm1 proteolysis is necessary for full activation of APCCdh1 in late mitosis. The current model for activation of APCCdh1 exclusively invokes dephosphorylation of Cdh1 by Cdc14, which allows it to associate with the core APC and complete the destruction of mitotic cyclins (11–13). It is now evident that budding yeast Cdh1 is tightly bound to Acm1 prior to Cdc14 activation and that Cdc14 also acts on Acm1 to promote its destruction, thereby relieving any inhibitory effects. What happens if Acm1 is not destroyed and its inhibitory effect is not relieved? Overexpression of Acm1 is toxic in sic1Δ cells (26) suggesting that it can block APC activation in late mitosis at an unnatural expression level. However, the inability to isolate a constitutively stable Acm1 mutant has prevented us from rigorously testing if destruction of the natural level of Acm1 is important for full activation of APCCdh1. Identification of the ubiquitin ligase that targets Acm1 should help address this problem.
Acknowledgments
We thank Kavita Shah for generously donating the 1-NM-PP1 and Ying Wang for the Fin1-Myc expression plasmid. We are grateful to David Morgan, Angelika Amon, Steven Reed, and Raymond Deshaies for providing yeast strains. Efrain Sanchez, Anindya Chatterjee, and Juan Martinez made important contributions to cloning and creation of site-directed mutants.
This work was supported in part by an American Heart Association scientist development grant (to M. C. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SCF, Skp1-cullin-F box protein complex; MALDI, matrix-assisted laser desorption ionization; LC, liquid chromatography; MS, mass spectrometry; MS-MS, tandem MS; IP, immunopurification; CDK, cyclin-dependent kinase; APC, anaphase-promoting complex/cyclosome; G6PD, glucose-6-phosphate dehydrogenase; 1-NM-PP1, 4-amino-1-(tert-butyl)-3-(1′-naphthylmethyl)pyrazolo[3,4-d] pyrimidine; PMSF, phenylmethylsulfonyl fluoride; HPLC, high pressure liquid chromatography; HU, hydroxyurea; HA, hemagglutinin.
D.-E. Jeong, E. Choi, and M. C. Hall, manuscript in preparation.
H. Charbonneau and Y. Wang, unpublished results.
References
- 1.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Vodermaier, H. C. (2004) Curr. Biol. 14 R787–R796 [DOI] [PubMed] [Google Scholar]
- 3.Yamasaki, L., and Pagano, M. (2004) Curr. Opin. Cell Biol. 16 623–628 [DOI] [PubMed] [Google Scholar]
- 4.Peters, J. M. (2006) Nat. Rev. Mol. Cell Biol. 7 644–656 [DOI] [PubMed] [Google Scholar]
- 5.Kramer, E. R., Gieffers, C., Holzl, G., Hengstschlager, M., and Peters, J. M. (1998) Curr. Biol. 8 1207–1210 [DOI] [PubMed] [Google Scholar]
- 6.Visintin, R., Prinz, S., and Amon, A. (1997) Science 278 460–463 [DOI] [PubMed] [Google Scholar]
- 7.Fang, G., Yu, H., and Kirschner, M. W. (1998) Mol. Cell 2 163–171 [DOI] [PubMed] [Google Scholar]
- 8.Shirayama, M., Zachariae, W., Ciosk, R., and Nasmyth, K. (1998) EMBO J. 17 1336–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prinz, S., Hwang, E. S., Visintin, R., and Amon, A. (1998) Curr. Biol. 8 750–760 [DOI] [PubMed] [Google Scholar]
- 10.Pfleger, C. M., and Kirschner, M. W. (2000) Genes Dev. 14 655–665 [PMC free article] [PubMed] [Google Scholar]
- 11.Jaspersen, S. L., Charles, J. F., and Morgan, D. O. (1999) Curr. Biol. 9 227–236 [DOI] [PubMed] [Google Scholar]
- 12.Kramer, E. R., Scheuringer, N., Podtelejnikov, A. V., Mann, M., and Peters, J. M. (2000) Mol. Biol. Cell 11 1555–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zachariae, W., Schwab, M., Nasmyth, K., and Seufert, W. (1998) Science 282 1721–1724 [DOI] [PubMed] [Google Scholar]
- 14.Jaquenoud, M., van Drogen, F., and Peter, M. (2002) EMBO J. 21 6515–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang, Z., Shu, H., Oncel, D., Chen, S., and Yu, H. (2004) Mol. Cell 16 387–397 [DOI] [PubMed] [Google Scholar]
- 16.Searle, J. S., Schollaert, K. L., Wilkins, B. J., and Sanchez, Y. (2004) Nat. Cell Biol. 6 138–145 [DOI] [PubMed] [Google Scholar]
- 17.Listovsky, T., Oren, Y. S., Yudkovsky, Y., Mahbubani, H. M., Weiss, A. M., Lebendiker, M., and Brandeis, M. (2004) EMBO J. 23 1619–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rape, M., and Kirschner, M. W. (2004) Nature 432 588–595 [DOI] [PubMed] [Google Scholar]
- 19.Yu, H. (2002) Curr. Opin. Cell Biol. 14 706–714 [DOI] [PubMed] [Google Scholar]
- 20.Reimann, J. D., Gardner, B. E., Margottin-Goguet, F., and Jackson, P. K. (2001) Genes Dev. 15 3278–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. J., Summers, M. K., Hansen, D. V., Nachury, M. V., Lehman, N. L., Loktev, A., and Jackson, P. K. (2006) Genes Dev. 20 2410–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izawa, D., Goto, M., Yamashita, A., Yamano, H., and Yamamoto, M. (2005) Nature 434 529–533 [DOI] [PubMed] [Google Scholar]
- 23.Schmidt, A., Duncan, P. I., Rauh, N. R., Sauer, G., Fry, A. M., Nigg, E. A., and Mayer, T. U. (2005) Genes Dev. 19 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoji, S., Yoshida, N., Amanai, M., Ohgishi, M., Fukui, T., Fujimoto, S., Nakano, Y., Kajikawa, E., and Perry, A. C. (2006) EMBO J. 25 834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burton, J. L., and Solomon, M. J. (2007) Genes Dev. 21 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez, J. S., Jeong, D. E., Choi, E., Billings, B. M., and Hall, M. C. (2006) Mol. Cell. Biol. 26 9162–9176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dial, J. M., Petrotchenko, E. V., and Borchers, C. H. (2007) J. Biol. Chem. 282 5237–5248 [DOI] [PubMed] [Google Scholar]
- 28.King, R. W., Glotzer, M., and Kirschner, M. W. (1996) Mol. Biol. Cell 7 1343–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper, J. W., Burton, J. L., and Solomon, M. J. (2002) Genes Dev. 16 2179–2206 [DOI] [PubMed] [Google Scholar]
- 30.Mumberg, D., Müller, R., and Funk, M. (1995) Gene (Amst.) 156 119–122 [DOI] [PubMed] [Google Scholar]
- 31.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., and Boeke, J. D. (1998) Yeast 14 115–132 [DOI] [PubMed] [Google Scholar]
- 32.Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999) Yeast 15 963–972 [DOI] [PubMed] [Google Scholar]
- 33.Schneider, B. L., Seufert, W., Steiner, B., Yang, Q. H., and Futcher, A. B. (1995) Yeast 11 1265–1274 [DOI] [PubMed] [Google Scholar]
- 34.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 35.Amon, A. (2002) in Guide to Yeast Genetics and Molecular and Cell Biology (Guthrie, C., and Fink, G. R., eds) pp. 458–460, Academic Press, San Diego
- 36.Bishop, A. C., Ubersax, J. A., Petsch, D. T., Matheos, D. P., Gray, N. S., Blethrow, J., Shimizu, E., Tsien, J. Z., Schultz, P. G., Rose, M. D., Wood, J. L., Morgan, D. O., and Shokat, K. M. (2000) Nature 407 395–401 [DOI] [PubMed] [Google Scholar]
- 37.Ubersax, J. A., Woodbury, E. L., Quang, P. N., Paraz, M., Blethrow, J. D., Shah, K., Shokat, K. M., and Morgan, D. O. (2003) Nature 425 859–864 [DOI] [PubMed] [Google Scholar]
- 38.Surana, U., Robitsch, H., Price, C., Schuster, T., Fitch, I., Futcher, A. B., and Nasmyth, K. (1991) Cell 65 145–161 [DOI] [PubMed] [Google Scholar]
- 39.Hall, M. C., Warren, E. N., and Borchers, C. H. (2004) Cell Cycle 3 1278–1284 [DOI] [PubMed] [Google Scholar]
- 40.Visintin, R., Craig, K., Hwang, E. S., Prinz, S., Tyers, M., and Amon, A. (1998) Mol. Cell 2 709–718 [DOI] [PubMed] [Google Scholar]
- 41.Gray, C. H., Good, V. M., Tonks, N. K., and Barford, D. (2003) EMBO J. 22 3524–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser, B. K., Zimmerman, Z. A., Charbonneau, H., and Jackson, P. K. (2002) Mol. Biol. Cell 13 2289–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flint, A. J. (2003) in Handbook of Cell Signaling (Bradshaw, R. A., and Dennis, E. A., eds) pp. 671–676, Academic Press, San Diego
- 44.Flint, A. J., Tiganis, T., Barford, D., and Tonks, N. K. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traverso, E. E., Baskerville, C., Liu, Y., Shou, W., James, P., Deshaies, R. J., and Charbonneau, H. (2001) J. Biol. Chem. 276 21924–21931 [DOI] [PubMed] [Google Scholar]
- 46.Hartwell, L. H., Mortimer, R. K., Culotti, J., and Culotti, M. (1973) Genetics 74 267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaffe, M. B., Rittinger, K., Volinia, S., Caron, P. R., Aitken, A., Leffers, H., Gamblin, S. J., Smerdon, S. J., and Cantley, L. C. (1997) Cell 91 961–971 [DOI] [PubMed] [Google Scholar]
- 48.Kakiuchi, K., Yamauchi, Y., Taoka, M., Iwago, M., Fujita, T., Ito, T., Song, S. Y., Sakai, A., Isobe, T., and Ichimura, T. (2007) Biochemistry 46 7781–7792 [DOI] [PubMed] [Google Scholar]
- 49.Meek, S. E., Lane, W. S., and Piwnica-Worms, H. (2004) J. Biol. Chem. 279 32046–32054 [DOI] [PubMed] [Google Scholar]
- 50.Pozuelo Rubio, M., Geraghty, K. M., Wong, B. H., Wood, N. T., Campbell, D. G., Morrice, N., and Mackintosh, C. (2004) Biochem. J. 379 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yaffe, M. B. (2002) FEBS Lett. 513 53–57 [DOI] [PubMed] [Google Scholar]
- 52.Woodbury, E. L., and Morgan, D. O. (2007) Nat. Cell Biol. 9 106–112 [DOI] [PubMed] [Google Scholar]
- 53.Irniger, S., Piatti, S., Michaelis, C., and Nasmyth, K. (1995) Cell 81 269–278 [DOI] [PubMed] [Google Scholar]
- 54.Mailand, N., and Diffley, J. F. (2005) Cell 122 915–926 [DOI] [PubMed] [Google Scholar]
- 55.Littlepage, L. E., and Ruderman, J. V. (2002) Genes Dev. 16 2274–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D., and Futcher, B. (1998) Mol. Biol. Cell 9 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aitken, A. (2006) Semin. Cancer Biol. 16 162–172 [DOI] [PubMed] [Google Scholar]
- 58.van Hemert, M. J., Steensma, H. Y., and van Heusden, G. P. H. (2001) BioEssays 23 936–946 [DOI] [PubMed] [Google Scholar]
- 59.Hermeking, H., and Benzinger, A. (2006) Semin. Cancer Biol. 16 183–192 [DOI] [PubMed] [Google Scholar]
- 60.Hall, M. C., Torres, M. P., Schroeder, G. K., and Borchers, C. H. (2003) J. Biol. Chem. 278 16698–16705 [DOI] [PubMed] [Google Scholar]