Abstract
Bisulfite sequencing, a standard method for DNA methylation profile analysis, is widely used in basic and clinical studies. This method is limited, however, by the time-consuming data analysis processes required to obtain accurate DNA methylation profiles from the raw sequence output of the DNA sequencer, and by the fact that quality checking of the results can be influenced by a researcher's bias. We have developed an interactive and easy-to-use web-based tool, QUMA (quantification tool for methylation analysis), for the bisulfite sequencing analysis of CpG methylation. QUMA includes most of the data-processing functions necessary for the analysis of bisulfite sequences. It also provides a platform for consistent quality control of the analysis. The QUMA web server is available at http://quma.cdb.riken.jp/.
INTRODUCTION
DNA methylation at the C-5 position of cytosine is a major epigenetic silencing mechanism in many eukaryotic organisms. The sequence context of methylated cytosines in the genome depends on the organism. In mammals, the genomes are methylated almost exclusively at CpG dinucleotides, while those of flowering plants are methylated at both CpG and non-CpG sequences. In mammals, CpG methylation plays a pivotal role in genomic imprinting, development, cell growth and survival and gene regulation (1,2). Abnormal DNA methylation profiles are associated with a broad range of human diseases, especially cancers (3,4). Bisulfite sequencing analysis has been the standard method used by biological and medical researchers to detect cytosine methylation profiles in genomic DNA at the single-nucleotide level (5). This method, however, is limited by the laborious and time-consuming data analysis steps required: making the alignment, trimming raw sequences, extracting DNA methylation profiles, excluding low-quality sequences, performing statistical analysis and drawing figures that summarize the methylation patterns. Furthermore, due to the error-prone nature of the bisulfite chemical reaction and subsequent PCR amplification (6–8), a quality check of the aligned sequences is essential. Thus, a user-friendly tool for both data analysis and a quality check of the bisulfite sequencing is desirable.
Several software tools are currently available for bisulfite sequencing analysis (9–14), and they have both advantages and disadvantages. MethTools is a pioneering tool for bisulfite sequencing analysis (9), and comprehensive analysis results are quickly obtained from input data. However, it requires aligned sequences as input data, so that trimming and multiple-alignment of raw sequences are necessary prior to data submission. Similarly, CyMATE (13), a unique mapping tool for both CpG and non-CpG methylation, and CpG PatternFinder (14), a Windows-based program, also require aligned sequences as input data. MethTools and CyMATE receive input data through a web submission form and return their results by Email. MethylMapper (11) is a perl script program for bisulfite sequence analysis that is executable from the command line. It is designed for high-throughput mapping, but not for routine bisulfite sequencing. BiQ Analyzer (10) and CpGviewer (12) are local executable programs with a graphical and interactive user interface, and they accept raw bisulfite sequences as input data. One of the important features of the BiQ analyzer is a quality control function for evaluating bisulfite-unconversion, a typical experimental artifact in bisulfite sequencing, which is based on a comparison with the frequency of unconverted cytosines at non-CpG sites. This function is important in practice when evaluating results from the vertebrate genome, which is methylated primarily at CpG sites. However, the BiQ analyzer requires a significantly longer time to execute the analysis compared with the other programs, especially when a large number of sequences are used. CpGviewer has several useful features, including the ability to accept electropherogram formats as input sequence data, but it does not have the quality check function for bisulfite-unconversion like the BiQ analyzer.
Here, we report an interactive web-based bisulfite sequencing analysis tool called QUMA (quantification tool for methylation analysis) for CpG methylation analysis, intended for routine use by experimental researchers studying organisms with genomes methylated primarily at CpG sites, such as those of vertebrates. QUMA has four major features. First, it is easy-to-use and needs only two types of input: a PCR target genomic sequence and raw bisulfite sequences. With its user-friendly interface, only a few clicks are needed to quickly align, visualize and quantify the bisulfite sequence data in a comprehensive manner. Almost all the displayed data are downloadable. Second, QUMA is an all-in-one tool that includes most of the data-processing functions necessary for the analysis of bisulfite sequences. In addition, many optional parameters are available to change the output style according to the user's preferences. Third, QUMA provides a helpful feature that allows the user to control the quality of aligned sequences easily, by changing the cutoff parameters; if the input data and cutoff parameters are indicated, anyone can reproduce the analysis, by using the QUMA web server. Fourth, QUMA server can be launch locally, on a personal computer connected to a local network, by using a bootable CD. This feature is especially helpful to the researcher who must analyze sensitive data.
WEBSITE USAGE
Overview
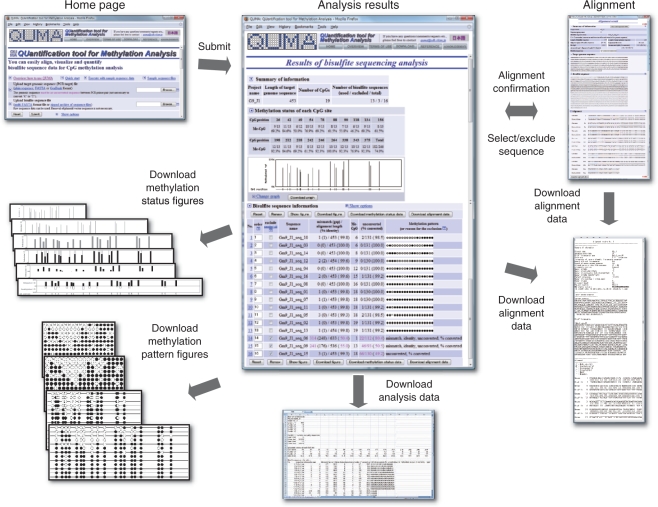
QUMA has useful functions for bisulfite sequencing analysis, including bisulfite sequence alignment, trimming raw bisulfite sequence, checking sequence quality, quantifying the methylation status, visualizing methylation profiles by various diagrams and, if necessary, calculating statistics values. The use of the basic features of QUMA is quite simple; that is, the user selects sequence files and submits them. The bisulfite alignment data, summarized analysis data, diagrams of methylation status and figures of methylation patterns can then be displayed and downloaded (Figure 1). The user's manual with detailed descriptions can be downloaded.
Figure 1.
Flow and output of the QUMA web server. After data submission, an analytical results page is displayed. Methylation status data for each CpG site, a summary of the sequence alignment of the bisulfite sequences, and the methylation pattern of each sequence are shown on this page. The alignment of the genomic sequence with each bisulfite sequence can be shown in a different window. The analysis results data (CSV format, which can be opened from Excel or other spread-sheet software), alignment data (text format), several types of methylation status figures and methylation pattern figures (PNG format) can be downloaded.
Input
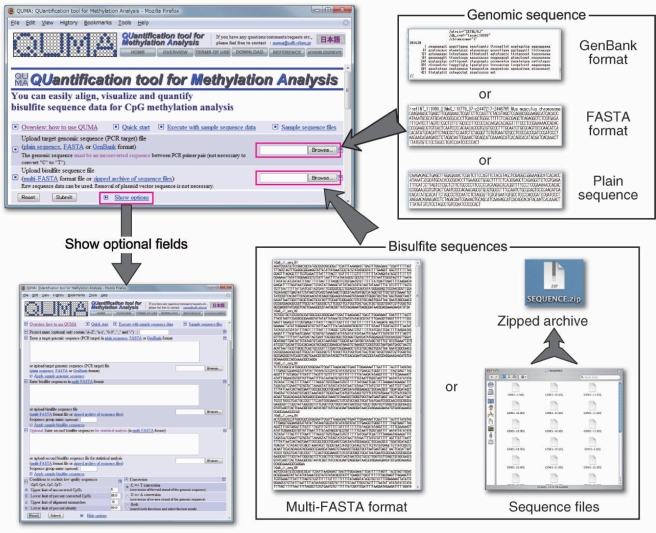
The input data for QUMA consist of a target genomic sequence and raw bisulfite sequences. The home page of the QUMA web server contains ‘file input’ fields for each of these data sets (Figure 2). The acceptable sequence formats for the target genomic sequence file are FASTA, GenBank and plain sequence. The genomic sequence must be unconverted (that is, ‘C’ should not be converted to ‘T’, which mimics bisulfite conversion) in the same strand as the region between the primer pairs for the bisulfite PCR. A set of multiple bisulfite sequences may be loaded either as a multi-FASTA format file or as a zipped archive of text sequence files (FASTA, GenBank and plain sequence). Raw bisulfite sequences can be used as input data and it is not necessary to remove the plasmid vector sequences. Most recent operating systems (Mac OS X 10.3 or later and Windows ME or later) support the creation of a zipped archive by default, and it is easy to create such an archive, which can contain even hundreds of sequence files, on a personal computer. Thus, it may be convenient to use a zipped archive as an input file of multiple bisulfite sequences, especially for those who are unfamiliar with the multi-FASTA format. Optional parameter fields, which are represented as ‘Show options’ by default, allow users to input sequences directly into a text field by ‘copy and paste’, to change the cutoff values for unconverted cytosines at non-CpG sites and alignment mismatch, and to change the direction of the bisulfite conversion of the genomic sequence.
Figure 2.
The home page and input data formats of the QUMA web server. QUMA needs two types of input files: a PCR target genomic sequence and bisulfite sequences. Acceptable file formats are plain sequence, FASTA or GenBank format for the genomic sequence, and a multi-FASTA format or a zipped archive of sequence files for bisulfite sequences. Optional fields can be seen from the ‘Show options’ link.
Output
After submitting the input data, bisulfite alignment, sequence trimming, exclusion of problematic sequences and methylation status analysis are performed by the QUMA web server. A typical run takes a few seconds to process 30 bisulfite sequences and the analysis result page is then displayed (Figure 1). The positions of CpG sites and methylation status of each CpG site are shown both in a table and as a diagram, which can be switched to several different formats. The summarized information on sequence alignments and the methylation pattern of each bisulfite sequence are also indicated in a table. A detailed bisulfite alignment between the target genomic sequence and each bisulfite sequence can be displayed from links. DNA methylation patterns can be displayed at the click of a button in several types of black/white circle-style figures, which are frequently used to represent methylation patterns. Optional parameters allow users to change the disposition and resolution of the black/white circle-style figures. Almost all the data shown in the web pages are downloadable in standard file formats (text for alignment data, CSV for analysis data and PNG for graphics), which can be opened by many applications. The user can set the preferences for sorting the order of bisulfite sequence information in the table or use several parameters that can be selected in optional fields.
Quality control
To control the quality of the analysis results, we introduced two types of criteria for detecting problematic sequences: a sequencing quality check and a bisulfite conversion quality check.
Due to the low complexity of sequences after bisulfite conversion, sequencing errors sometimes occur during the bisulfite sequencing analysis. To control for sequence quality, we use the identity and number of mismatches in the local pairwise alignment of the bisulfite sequence and the target genomic sequence as parameters. We adopted a 90% lower limit for identity and 10 as the upper limit of mismatches as default values.
Incomplete bisulfite conversion of cytosines is a frequently encountered problem in bisulfite sequencing analysis (6–8). Although low-level non-CpG methylation does occur in the mammalian genome (15), the presence of a certain number of unconverted cytosines in a single bisulfite sequence clone has been a practical indicator of incomplete bisulfite conversion in analyzing mostly CpG-methylated genome of vertebrates (16). Therefore, to control for the quality of bisulfite conversion, we adopted the conversion efficiency of cytosines at non-CpG sites and the number of unconverted cytosines at non-CpG sites as parameters. We set 95% as the lower limit for the conversion efficiency and 5 for the upper limit of the number of unconverted cytosines as default values. The default values of the parameters were determined based on our experience and usually work well for analyzing mammalian CpG methylation.
QUMA shows these parameters for all bisulfite sequences in a summary table, and displays a diagram of DNA methylation patterns excluding sequences that do not match the above criteria. Users can check the sites of the mismatches and unconverted non-CpG cytosines in detailed alignments of these sequences, decide on their inclusion or exclusion, and, if necessary, change the cutoff values of the parameters through QUMA's interactive interface. This option is useful in analyzing polymorphism-containing sequences (e.g. polymorphic alleles of imprinted genes and repetitive sequences) or sequences from organisms with genomes that contain significant amounts of non-CpG methylation.
Statistical analysis
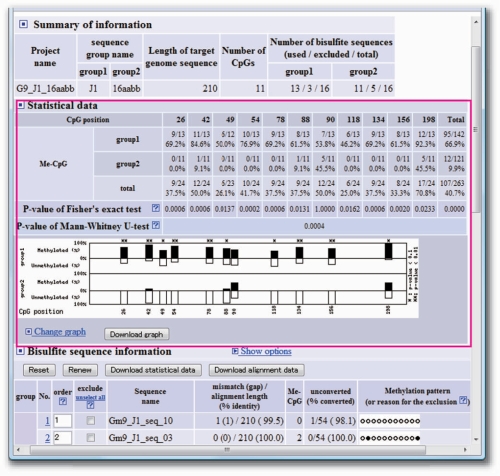
In cases in which two groups of bisulfite sequences are fed into the optional fields on the home page of the QUMA web server (Figure 2), QUMA performs a statistical analysis between the methylation profiles of the two groups. In addition to the standard analysis results, statistical significances (P-values) and a diagram of comparative methylation status are shown (Figure 3). The statistical significance of the difference between two bisulfite sequence groups at each CpG site is evaluated with Fisher's exact test (17), while that of the entire set of CpG sites is evaluated with the Mann–Whitney U-test (18). For a statistical analysis at each CpG site, the two-tailed P-value of Fisher's exact test is calculated from the 2 × 2 tables at each CpG site. This P-value is used to show the independence of CpG methylation between two groups at a given CpG site. For statistical analysis of the entire set of CpG sites, the two-tailed P-value of the Mann–Whitney U-test is determined from ranks of the ratio of methylated CpGs to all the CpGs at each bisulfite sequence. This P-value indicates the independence of distribution of the ratio of CpG methylation to all CpGs.
Figure 3.
An example of statistical analysis output at the QUMA web server. In the statistical data section (magenta rectangle), the position of CpG sites, methylation status of each CpG site, and statistical significance (P-values) of differences between two bisulfite sequence groups are shown. A diagram representing the comparative methylation status can be displayed in several different formats.
One limitation is that the statistical analysis by both tests does not take the CpG methylation pattern into account.
IMPLEMENTATION
QUMA is implemented in HTML, JavaScript and Perl-CGI script. The QUMA web server is a Linux server (2.66 GHz Intel Core2Quad Processor, 8 GB RAM) running on CentOS 5. The QUMA web server limits the maximum number of bisulfite sequences per request to 400 to prevent the server from crashing during periods of high use. This should take <60 s (35 s for 450 bp genomic sequence and 384 bisulfite sequences that contain a total of 370 kb). The download version does not limit the maximum number of sequences per analysis request.
To align bisulfite sequences with a genomic sequence, we took a pairwise-alignment approach for QUMA, instead of the multiple-alignment approach used by some other programs (6,10,13,14). QUMA aligns a target genomic sequence with each bisulfite sequence, and therefore the alignment of a certain bisulfite sequence is independent of other bisulfite sequences. However, an alignment between the target sequence and a given bisulfite sequence in a multiple-alignment could be affected by the qualities of other bisulfite sequences. In addition, a pairwise-alignment approach is suitable for prompt interactive changes to the inclusion or exclusion of bisulfite sequences during a quality-check process, because of the independence of each bisulfite sequence result. An output style of pairwise alignment may be suitable for checking mismatches and unconverted cytosines in individual bisulfite sequence clones.
To perform pairwise alignment, we adopted the needle program of the EMBOSS package (19) because of its accurate production of Needleman–Wunsch pairwise alignments (20). A modified score matrix was used for the needle, so that ‘C’ bases in the genomic sequence may match with both ‘C’ and ‘T’ bases in the bisulfite sequence, since unmethylated cytosine is detected as ‘T’ in bisulfite sequencing.
The software source code of the QUMA is freely available on the download section of the QUMA web server (http://quma.cdb.riken.jp/) under the GNU General Public License (GPL). Also, a CD-image file of a ‘LiveCD’ Linux version of QUMA is freely available at the download section. LiveCD is a computer operating system that easily boots a personal computer from a single CD without installation, set-up or changes to the hard disk. After rebooting and ejecting the CD, a computer can be rebooted from its original operating system. The LiveCD version of QUMA enables the user to launch the QUMA server easily on a personal computer connected to a local network, and the server will be accessible from other personal computers connected to the local network. This option may be useful for analyzing sensitive data without submitting sequences to a site outside the user's institution.
CONCLUSION
QUMA is an easy-to-use, all-in-one, interactive web-based tool for the bisulfite-sequencing analysis of CpG methylation that aligns and trims raw sequences, analyzes CpG methylation profiles, performs statistical comparisons, checks the quality of sequencing data and displays the results. QUMA is designed to be useful and understandable for experimental researchers, even if they are unfamiliar with bisulfite sequencing analysis. This tool may improve the speed of the entire analysis of bisulfite sequencing data, and may help standardize the analysis, communication and quality of results across different computing environments. This website is free and open to all users and there is no login requirement.
ACKNOWLEDGEMENTS
We thank Akiko Yamagiwa for sample bisulfite sequences of the mouse Gm9 region (21), Morito Sakaue, Masahumi Kawaguchi and Yoko Dote for useful comments, Hazuki S. Hiraga for checking the website, and the Information Networks Office of RIKEN Kobe Institute for helpful suggestions in setting up the Internet connection for the server. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan to M. Okano. Funding to pay the Open Access publication charges for this article was provided by RIKEN.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 2.Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr. Opin. Cell. Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:e65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajkova P, el-Maarri O, Engemann S, Oswald J, Olek A, Walter J. DNA-methylation analysis by the bisulfite-assisted genomic sequencing method. Methods Mol. Biol. 2002;200:143–154. doi: 10.1385/1-59259-182-5:143. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Wylie RC, Hansen NJ, Andrews LG, Tollefsbol TO. Profiling DNA methylation by bisulfite genomic sequencing: problems and solutions. Methods Mol. Biol. 2004;287:169–179. doi: 10.1385/1-59259-828-5:169. [DOI] [PubMed] [Google Scholar]
- 9.Grunau C, Schattevoy R, Mache N, Rosenthal A. MethTools—a toolbox to visualize and analyze DNA methylation data. Nucleic Acids Res. 2000;28:1053–1058. doi: 10.1093/nar/28.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 11.Ordway JM, Bedell JA, Citek RW, Nunberg AN, Jeddeloh JA. MethylMapper: a method for high-throughput, multilocus bisulfite sequence analysis and reporting. BioTechniques. 2005;39:464–472. doi: 10.2144/000112035. [DOI] [PubMed] [Google Scholar]
- 12.Carr IM, Valleley EM, Cordery SF, Markham AF, Bonthron DT. Sequence analysis and editing for bisulphite genomic sequencing projects. Nucleic Acids Res. 2007;35:e79. doi: 10.1093/nar/gkm330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- 14.Xu YH, Manoharan HT, Pitot HC. CpG PatternFinder: a Windows-based utility program for easy and rapid identification of the CpG methylation status of DNA. BioTechniques. 2007;43:334–342. doi: 10.2144/000112537. [DOI] [PubMed] [Google Scholar]
- 15.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 17.Alan A. 2nd edn. Hoboken, NJ: Wiley; 2002. Categorical Data Analysis. [Google Scholar]
- 18.Ewens W, Grant G. New York, NY: Springer; 2001. Statistical Methods in Bioinformatics: An Introduction (Statistics for Biology and Health) [Google Scholar]
- 19.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 20.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Oda M, Yamagiwa A, Yamamoto S, Nakayama T, Tsumura A, Sasaki H, Nakao K, Li E, Okano M. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 2006;20:3382–3394. doi: 10.1101/gad.1470906. [DOI] [PMC free article] [PubMed] [Google Scholar]