Abstract
The traditional experimental approaches used for changing the flux or the concentration of a particular metabolite of a metabolic pathway have been mostly based on the inhibition or over-expression of the presumed rate-limiting step. However, the attempts to manipulate a metabolic pathway by following such approach have proved to be unsuccessful. Metabolic Control Analysis (MCA) establishes how to determine, quantitatively, the degree of control that a given enzyme exerts on flux and on the concentration of metabolites, thus substituting the intuitive, qualitative concept of rate limiting step. Moreover, MCA helps to understand (i) the underlying mechanisms by which a given enzyme exerts high or low control and (ii) why the control of the pathway is shared by several pathway enzymes and transporters. By applying MCA it is possible to identify the steps that should be modified to achieve a successful alteration of flux or metabolite concentration in pathways of biotechnological (e.g., large scale metabolite production) or clinical relevance (e.g., drug therapy). The different MCA experimental approaches developed for the determination of the flux-control distribution in several pathways are described. Full understanding of the pathway properties when is working under a variety of conditions can help to attain a successful manipulation of flux and metabolite concentration.
1. INTRODUCTION
Is an effort to manipulate the metabolism of an organism worthy and reasonable, knowing that this cellular process has been continuously modified and refined through evolution and natural selection for adapting, in the most convenient manner, to the ongoing environmental conditions? The answer to this question seems obvious when three broad areas of research and development are identified in which manipulation of metabolic pathways is relevant: (a) drug design to treat diseases, (b) genetic engineering of organisms of biotechnological interest, and (c) genetic syndromes therapy.
Historically, drug design was the first area in which modification of metabolism was tried: the primary goal of drug administration is the inhibition of essential metabolic pathways, for example, in a parasite or a tumor cell. Thus, any metabolic pathway can be a potential therapeutic target. In the absence of a solid theoretical background that may build a strategy for the rational design of drugs, the pharmaceutical industry has applied the knowledge of inorganic and organic chemistry for the arbitrary and rather randomized modification of metabolic intermediaries by replacing hydrogen atoms in a model molecule with any other element or compound. This approach has been successful in the battle against many diseases. However, in many other instances such an approach has been unsuccessful.
The era of rational drug design probably started in the 50s when Hans Krebs proposed that, after having an exact description of a metabolic pathway, the “pacemaker” enzyme or “rate-limiting step” had to be identified. This approach certainly decreased the amount of intermediaries to be chemically modified, focusing only on the substrates, products, and allosteric effectors of the “rate-limiting step,” instead of dispersing efforts on all the metabolic pathway intermediates. The experimental approaches used in the identification of the pacemaker, key enzymes, “bottlenecks.” limiting steps, or regulatory enzymes [1, 2] were
inspection of the metabolic pathway architecture: due to cell economy and for reaching the highest efficiency, pathway control must reside in the enzymes localized at the beginning of a pathway or after a branch (teleological approach);
determination of nonequilibrium reactions: those reactions in which the quotient between the mass action ratio (Γ) and its equilibrium constant (Keq) is low, Γ/Keq ≪1 (thermodynamic approach);
identification of the steps with the lowest maximal rates (Vmax) in cellular extracts: the key enzyme of the pathway is the one that has the lowest rate (kinetic approach);
enzymes with sigmoidal kinetics: steps that are susceptible to alteration in their kinetic properties by compounds different from substrates and products and which may coordinate the entire metabolism (NADH/NAD+; NADPH/NADP+, ATP/ADP; acetyl CoA/CoA; Ca2+/Mg2+; high pH/low pH) or at least two metabolic pathways (citrate, Pi, AMP, malonyl-CoA);
crossover theorem. Comparing the intermediary concentrations between a basal and an active steady-state pathway flux, the rate-limiting step in the basal condition will be that for which its substrate concentration diminishes and its product concentration increases when the system changes from the basal to the active state or vice versa (crossover point on a histogram of each intermediary versus its normalized variation in concentration);
the shape of the metabolic flux inhibition curve: a sigmoidal curve on a plot of inhibitor concentration versus flux shows that the sensitive step to the inhibitor exerts no control, that is, there is not proportionality between enzyme activity inhibition and pathway flux inhibition because there is an “excess” of enzyme. On the other hand, a hyperbolic curve indicates that the enzyme susceptible to the inhibitor controls the flux.
2. CONTROLLING SITES IN A METABOLIC PATHWAY
Once a site in a metabolic pathway has been identified with at least one of the criteria described above as “the rate-limiting step,” researchers have frequently concluded that such enzyme or transporter is the only limiting step of the metabolic flux and extend this conclusion to all cell types and to all conditions.
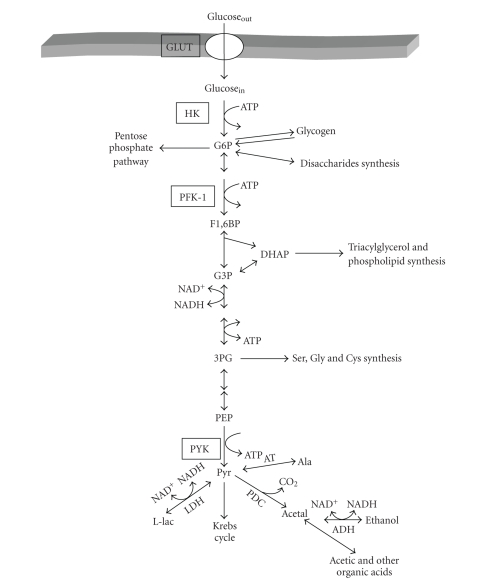
For example, inspection of the glycolytic pathway (teleological approach) suggests that hexokinase (HK) and phosphofructokinase-1 (PFK-1) (which are at the beginning and after a branch of the pathway) are the key steps of glycolysis. However, all studies on glycolysis in the 60s, 70s, and 80s were performed by taking into account only the intracellular reactions from HK to LDH (i.e., without including the glucose transport reaction through the plasma membrane) and by considering glycolysis as a linear pathway without branches. To this regard, it is recalled that the glucose transporter (GLUT) includes a family of proteins and genes that are susceptible of regulation. Thus, if the extracellular glucose is considered as the initial glycolytic substrate, then another potential key step would be GLUT. Hence, if all the branches of the pathway are considered (Figure 1), then according to the teleological approach there will be additional potential rate-limiting sites.
Figure 1.
Glycolytic pathway and principal branches. GLUT, glucose transporter; HK, hexokinase; PFK-1, phosphofructokinase-1; G6P, glucose-6-phosphate; F1,6BP, fructose 1,6 bisphosphate; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde-3-phosphate; 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; pyr, pyruvate; PYK, pyruvate kinase; L-lac, L-lactate; acetal, acetaldehyde; AT, alanine transaminase. S. cerevisiae lacks the LDH gene.
Application of the thermodynamic and kinetic approaches to glycolysis reveals that HK, PFK-1, and pyruvate kinase (PYK) are the rate-limiting steps because in the living cell they catalyze reactions that are far away from equilibrium (Γ/Keq = 10−3–10−4), and they are also the slowest enzymes in the pathway by at least one order of magnitude (they have the lowest Vmax values).
The use of the enzyme cooperativity approach has established that the regulatory steps of glycolysis are (i) PFK-1 and PYK because they are allosteric enzymes and (ii) HK because it is inhibited by its products (G6P and ADP, or AMP as an ADP-analogue). The application of the crossover theorem (approach no. v) to glycolysis has shown a consistent variation in the PFK-1 substrate (F6P) and product (F1,6BP). Up to now, there are few studies on control of glycolysis using the shape of the inhibitor titrating curve (approach no. vi), due to the lack of specific inhibitors for any of the three presumed key steps. An exception is iodoacetate which is indeed a potent inhibitor of GAPDH, but also of other highly reactive cysteine-containing enzymes [3–5]. By using iodoacetate as specific inhibitor, both GAPDH activity and flux showed identical titration curves, leading to the conclusion that GAPDH was the rate-limiting step of glycolysis in Streptococcus lactis and S. cremoris [6] (see, however, Section 3.2; Glycolysis in lactobacteria below).
All together, these results constitute the main reason why many intermediary metabolism researchers, including the authors of biochemistry text books, have proposed HK, PFK-1, and PYK as the rate-limiting steps of glycolysis. In consequence, to vary the glycolytic flux, one of these enzymes has to be modified.
Although the above-described experimental approaches are qualitative, full control has been automatically assigned to the “key” steps because the concept of the rate-limiting step assumes that there is only one single enzyme controlling the metabolic pathway flux (and the concentration of the final product of the pathway) and, in consequence, assigns values of zero to the control exerted by the other enzymes and transporters. However, as analyzed for glycolysis, researchers have commonly “identified” more than one limiting step. In the case of oxidative phosphorylation (OXPHOS), in the 70s and 80s some researchers considered cytochrome c oxidase as the rate-limiting step, whereas others preferred the ATP/ADP translocator or the Krebs cycle Ca2+-sensitive dehydrogenases (for a review, see [7]).
Rephrasing the initial question, which could be the aim of manipulating a metabolic pathway such as glycolysis, knowing its universal distribution in the living organisms? From a clinical standpoint, the inhibition of glycolysis is relevant for the treatment of human parasitic or pathological diseases such as cancer. The glycolytic reactions are almost identical in all organisms; in addition, the enzymes catalyzing these reactions are highly conserved throughout the evolutionary scale (their amino acid sequences are highly similar). In mammals, the genes of the 12 glycolytic enzymes are scattered throughout the genome, generally in different chromosomes, whereas in bacteria many of the glycolytic enzymes are clustered in operons [8]. However, there are organisms (like some human parasites) that contain enzymes with remarkable differences in their biochemical properties (substrate selectivity, catalytic capacity, stability, and oligomeric structure), or in genetic expression regulation in comparison to the human enzymes, which could be considered as drug targets.
Furthermore, some glycolytic products are of commercial interest such as ethanol for wine, beer, and other alcoholic beverages; CO2 for bread manufacturing; and lactic acid and other organic acids for cheese production. Thus, from a biotechnological standpoint, it is convenient to accelerate the pathway flux to diminish the processing time and it is also desirable to increase the concentration of the metabolite to obtain robust commercial products. Here, it is important to emphasize that the metabolic pathways are designed to attain changes in flux with minimal disturbances in the intermediary concentrations. For example, the glycolytic flux in skeletal muscle can increase from rest to an active state by 100 fold, without large changes in metabolites. Then, it is physiologically more common to change a metabolic flux and the production of the final metabolite in the pathway than varying the intermediary concentrations [2]. However, we will see that, by using a suitable approach of metabolic control analysis, it is possible to design strategies to manipulate not only fluxes but also metabolic intermediary concentrations.
3. IN VIVO OVEREXPRESSION EXPERIMENTS OF ENZYMES
3.1. Glycolysis in yeasts
When the yeast Saccharomyces cerevisiae is exposed to high glucose (>2%; 0.11 M), the genes of all glycolytic enzymes are induced (PDC and ENO increase their expression by 20 fold; PGK, PYK, and ADH, 3–10 times; and the others, 2 fold in average) [8–11]. However, when the methodological development of genetic engineering allowed modulating the expression of enzymes within cells, researchers turned to the rate-limiting step concept to manipulate a metabolic pathway to increase flux and/or its intermediates, hypothesizing that the overexpression of only one, or of a few key glycolytic genes, should increase the flux.
Historically, Heinisch [9] in Germany was the first author to obtain a 3.5 fold overexpression of PFK-1 in S. cerevisiae, but surprisingly he observed that the rate of ethanol production was not modified. Subsequent experiments for increasing the ethanol production rate by overexpressing either each of the presumed limiting steps, or in combination with other glycolytic enzymes (Table 1), have been unsuccessful and, even in some cases, a slight decrease in flux has been attained. For instance, the simultaneous overexpression of seven enzymes of the final section of glycolysis induced only a 21% increase in ethanol production after 2 hours of culture (Table 1) [11]. This was accompanied by a 10–20% decrease in PFK-1 expression, which might have attenuated the flux increase.
Table 1.
Overexpression of glycolytic enzymes in different cell types.
| Cell type | Enzyme | Activity (overexpression fold) | Flux (% Control) | Reference |
|---|---|---|---|---|
| Saccharomyces cerevisiae | HK | 13.9 | 107 | [12] |
| PFK-1 | 3.5, 3.7,5 | 102 | [9, 10, 12] | |
| PYK | 8.6 | 107 | [12] | |
| PDC | 3.7 | 85 | [13] | |
| ADH | 4.8 | 89 | [12] | |
| PFK-1 + PYK | 5.6 + 1.3 | 107 | [12] | |
| GAPDH + PGK + PGAM + ENO + PYK + PDC + ADH | 1.4 + 1.7 + 16 + 4 + 10.4 + 1.08 + 1.4 | 121 | [12] | |
| GAPDH + PGK + PGAM + ENO + PYK + PDC + ADH | 1.5 + 1.4 + 3.4 + 1.5 + 2.5 + 1.1 + 1.2 | 94 | [11, 14] | |
| Escherichiacoli | PFK | 8.7 | 72 | [15] |
| PYK | 2.9, 4.2 | 91,95 | [16] | |
| Lactococcus lactis | GAPDH | 14-210 | 100 | [17] |
| Aspergillus niger | PFK | 3 | 100 | [18] |
| PYK | 5 | 100 | ||
| Chinese hamster ovary | PFK | 2.2, 3.4, 3.7 | 100 | [19] |
Flux to ethanol was for S. cerevisiae and E. coli; flux to citrate was for A. niger; and flux to L-lactate was for hamster.
In yeasts, HK is not product inhibited by G6P or ADP; instead, it is strongly feedback inhibited by trehalose-6-phosphate (Tre6P). This metabolite is synthesized from G1P by Tre6P synthase and Tre6P phosphatase. Deletion of the Tre6P synthase gene does not bring about an increased ethanol production, but it rather induces a defective cellular growth on glucose and fructose and a lowered ethanol production, as a result of a highly active HK that leads to hyperaccumulation of hexose phosphate metabolites (particularly F1,6BP) and fast depletion of ATP, Pi, and downstream metabolites. The explanation for this event is that, in the Tre6P synthase mutants, the rate of glucose phosphorylation exceeds the rate of glycolytic ATP synthesis (named “turbo effect”). Heterologous expression of a Tre6P-insensitive HK does not recover completely the wild-type phenotype. Furthermore, deletion of the Tre6P synthase gene in the Tre6P-insensitive HK strain did affect growth, suggesting other interactions and functions of Tre6P synthase in the control of sugar metabolism, at least in Schizosaccharomyces pombe [20].
Davies and Brindle [10] obtained a 5-fold overexpression of PFK-1 in S. cerevisiae, but the increase in ethanol production was not attained under anaerobic conditions. There was a slight increase in ethanol production in resting cells in aerobic conditions, under which the mitochondrial metabolism contributes to the ATP supply. In all these works, it may be noted that enzyme overexpression indeed affects the concentration of several intermediaries, but this effect has not been further examined.
It is worth noting that the experiments described in Table 1 do not rigorously reproduce the physiological situation, in which overexpression of all the enzymes should be carried out in the proportions found in the organisms. The rationale behind this observation is that overexpression of only one “limiting” step leads to a flux control redistribution, a condition at which other steps now become rate limiting. Thus, the concept of “rate-limiting step” offers no simple answer to the question of increasing the yeast glycolytic flux, and it rather makes this problem to appear as a difficult task to solve. In contrast, it seems that all relevant controlling steps have to be overexpressed, thus reproducing what natural selection has already successfully accomplished.
In addition to S. cerevisiae, overexpression of glycolytic enzymes in other organisms such as E. coli [15, 16], lactobacteria [17], tomato [21], potato [22], and hamster ovary cells [19] has been accomplished, although without increasing flux (Table 1). It is somewhat surprising to note that in the glycolytic enzyme overexpression experiments, the strong inhibitory effect of G6P (or Tre6P in S. cerevisiae), and citrate on HK and PFK-1, respectively, have been neglected. This regulatory mechanism does not disappear in the cells overexpressing the enzymes but, on the contrary, it is exacerbated. Then, what would be the aim of overexpressing HK, PFK-1 or any other allosteric, or strongly product-inhibited enzyme if they will be more inhibited?
A successful experiment of increasing the glycolytic flux was performed in primary cultures of rat hepatocytes [23]. HK and glucokinase (GK) were overexpressed by using adenovirus as carrier. The transformed hepatocytes showed higher activity of 18.7- and 7.1-times for HK and GK, respectively, at 3 mM glucose, and of 6.3- and 7.1-times at 20 mM glucose. However, at 20 mM glucose, the flux to lactate was not modified in HK-transformed cells, just like the experiments described above (Table 1). In contrast, with GK overexpression, a 3-fold increase in flux was achieved. The mechanistic difference is the HK inhibition by G6P (10 mM G6P inhibits HK activity by 90%), whereas GK is not product inhibited.
3.2. Glycolysis in lactobacteria
Lactococcus lactis is used in cheese production. For this purpose, L. lactis ferments lactose to lactic acid by glycolysis. The end products, lactate and H+, are expelled and acidify the external medium which contributes to cheese flavor and texture and inhibits the growth of other bacteria. Similarly to yeast, the lack of carbon source in lactobacteria promotes a metabolic change that leads to the production of formic and acetic acids, ethanol, and, in a lower proportion, L-lactic acid, altering the product quality. Thus, from a commercial point of view, it does not seem important to know what controls the flux to lactate (because its rate of production is adequate), but what controls the branching flux.
To understand the process, and to eventually inhibit the production of secondary acids, Andersen et al. [24] constructed LDH mutants, using a synthetic promoter library for tuning the gene expression. In mutants lacking this enzyme, most of the pyruvate was transformed into acetic and formic acids (Figure 1). In turn, flux to lactate was affected in mutants expressing only 10% or less of wild-type LDH levels, which indicated that LDH exerts no control of the glycolytic flux in wild-type bacteria. Only with a normal content of this enzyme (100%), flux toward secondary acids was prevented. Therefore, the flux to formic and acetic acids is negatively controlled by LDH, and positively by PYK [17, 25]. As in S. cerevisiae, overexpression of PFK-1, PYK, or GAPDH in lactobacteria did not increase the flux to L-lactic acid [17, 25]. Similarly to E. coli glycolysis [26], glycolysis in L. lactis was controlled by the ATP demand when working below its maximum capacity [27, 28], whereas, under high-rate conditions, the glucose and lactate transporters exerted the main flux control [28]. Furthermore, this kind of observations indicates that the flux control may reside outside the pathway [27–29], and it also supports the proposal by Hofmeyr and Cornish-Bowden [30] that the end-product demand (which is usually overlooked in studies of metabolism because these metabolites are frequently not considered as part of the pathway) might be essential in flux control.
3.3. Glutathione and phytochelatin synthesis in plants
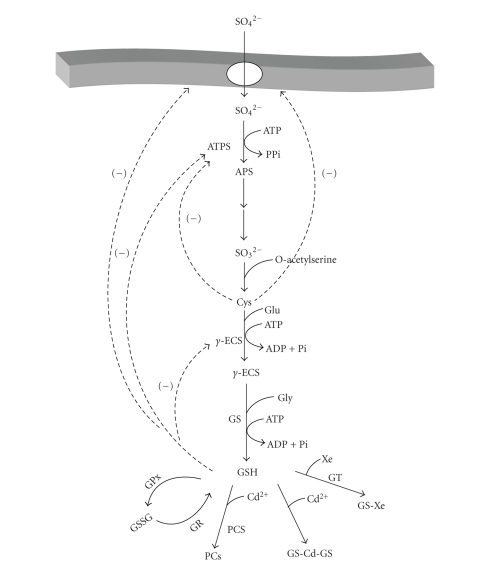
Glutathione (γ-Glu-Cys-Gly; GSH) is the most abundant nonproteinaceous thiol compound (1–10 mM) in almost all living cells. GSH is involved in the oxidative stress processing, xenobiotic detoxification, and, in some plants and yeasts, in the inactivation of toxic heavy metals (for a recent revision see [31]). GSH is synthesized by two enzymes: γ-glutamylcysteine synthetase (γ-ECS) and glutathione synthetase (GS) (Figure 2), which catalyze reactions with high-equilibrium constants (Keq > 1000). Under a low GSH demand (unstressed conditions), the producing block of enzymes has to receive information from the last part of the pathway to (i) avoid the excessive and toxic accumulation of the intermediary γ-EC and (ii) reach a stable steady state [32]. This information transfer is mediated by GSH, which exerts strong competitive inhibition of γ-ECS [33] (Figure 2). GSH and Cys also exert inhibition on the ATP-sulfurylase (ATPS) and on sulfate transporters (Figure 2) (for a review, see [31]). The feedback inhibition of γ-ECS has led several researchers to propose that this enzyme is the rate-limiting step of GSH synthesis [33–35]. Although there are no studies about the pathway's behavior under stressed conditions, which means under a high GSH demand, the proposal that γ-ECS is the key enzyme has been automatically extended to any environmental condition such as heavy metal exposure.
Figure 2.
Sulfur assimilation and glutathione and phytochelatins synthesis in plants ATPS, ATP sulfurylase; APS, adenosine 5′ phosphosulphate; γ-ECS, γ-glutamyl cysteine synthetase; γ-EC, γ-glutamyl cysteine; GS, glutathione synthetase; GSH, reduced glutathione; GSSG, oxidized glutathione; GPx, GSH peroxidase; GR, GSH reductase; PCS, phytochelatin synthase; PCs, phytochelatins; GT, GSH-S-transferases; Xe, xenobiotic; GS-Xe, glutathione-xenobiotic complex. The reactions are not shown stochiometrically. GR uses the cofactor NADPH. The Cd2+-GSH complex formation (cadmium bis-glutathionate) is fast and spontaneous and does not require enzyme catalysis. Modified from [31].
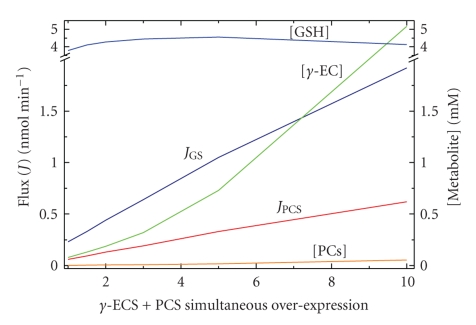
By assuming that γ-ECS is the rate-limiting step, many research groups have tried to increase, in plants and yeasts, the rate of synthesis and the concentration of GSH and phytochelatins (PCs) with the aim of fortifying their heavy metal resistance and storage capacity, mainly toward Cd2+. The development of organisms able to grow in soils and water systems contaminated with heavy metals, which may have the ability of accumulating toxic metal ions, is of biotechnological interest for bioremediation strategies.
With this goal in mind, researchers have then overexpressed γ-ECS and other pathway enzymes, including phytochelatin synthase (PCS) (Table 2). Some of these experiments have been partially successful in increasing GSH levels, although this has been rather marginal with no correlation between enzyme levels and GSH concentration. Unfortunately, these overexpression experiments have not been accompanied by determinations of fluxes or other relevant metabolite concentrations such as PCs or Cys. On the other hand, the overexpression of PCS has surprisingly induced oxidative stress and necrosis instead of increasing Cd2+ accumulation and resistance [36]. This result suggests that, under high GSH demand (i.e., for PCs synthesis and for direct heavy metal sequestration by GSH), the GSH concentration does not suffice for maintaining the other essential GSH functions such as oxidative stress management and xenobiotic detoxification.
Table 2.
GSH and phytochelatin synthesis enzymes overexpression in plants and yeasts.
| Overexpressed enzyme (activity fold) | Organism (experimental condition) | Metabolite (increment fold) | Reference |
|---|---|---|---|
| ATP sulfurylase (2.1) | Brassica juncea | 2.1 [GSH] | [37] |
| ATP sulfurylase (4.8) | Tobacco (unstressed) | 1.3 [SO42−] | [38] |
| O-acetyl-serine thiol-lyase (2.5) | Tobacco (unstressed) | 2 [Cys] | [39] |
| 0 [GSH] | |||
| Serine acetyl transferase (>10) | Potato chloroplasts (unstressed) | 2 [Cys] | [40] |
| 0 [GSH] | |||
| E. coli GS (90) | Populus tremula (unstressed) | 0 [GSH] | [34] |
| GS (3) | S. cerevisiae (unstressed) | 0 [GSH] | [41] |
| E. coliγ-ECS (>2) | Brassica juncea (unstressed) | 0 [GSH] | [35] |
| B. juncea (+100 μM Cd2+) | 4 [GSH](a) | ||
| γ-ECS (2.1) | S. cerevisiae (unstressed)) | 1.3 [GSH] | [42] |
| E. coliγ-ECS (50) | Populus tremula (unstressed) | 4.6 [GSH] | [34] |
| E. coliγ-ECS (4.9) | Brassica juncea (unstressed) B. juncea (+200 μM Cd2+) | 3.5 [GSH](b) | [43] |
| 1.5 [GSH](b) | |||
| E. coliγ-ECS (40) | Tobacco (unstressed) | >4 [GSH] | [44] |
| γ-ECS (9.1) + GS (18) | S. cerevisiae (unstressed) | 1.8 [GSH] | [45] |
| PCS (>2) | Arabidopsis thaliana (+85 μM Cd2+) | 0 [GSH] | [36] |
| Vacuolar transporter of PC-Cd complexes (>2) | S. pombe | Higher Cd2+ | [46] |
| resistance |
(a)The increase was only in roots with no effect on shoots. (b)The increase was only in shoots with no effect on roots.
Another problem in the study of GSH biosynthesis for its eventual manipulation is that the pathway has been analyzed considering only the GSH-synthetic reactions without taking into account the GSH-consuming reactions (Figure 2), [31]. The analysis of an incomplete pathway leads to misleading conclusions about the control of flux. Metabolic modeling has shown that only with the incorporation of the consuming reactions of the pathway end products, a true steady state can be established [30]. In conclusion, without a solid theoretical framework, the overexpression of only one enzyme (the “rate-limiting step”), or of many arbitrarily selected enzymes (Tables 1 and 2), the problem of increasing the flux or metabolite concentrations cannot be solved.
3.4. Overexpression of proteins from other metabolic pathways
There are some successful examples of the genetic engineering approach to manipulate metabolism:
overexpression (approx. 23 fold) of the five genes of the tryptophan synthesis pathway in S. cerevisiae, to increase (9-fold) flux [47];
increase in amino acids (Trp, Ile, Lys, Val, Thr) and trehalose production in Corynebacterium glutamicum, in which some proteins of each metabolic pathway are simultaneously overexpressed, but some of them with mutations that confer insensitivity to feedback inhibition [48–53]. In these transformed bacteria, the end products are indeed overproduced and their excretion is accelerated;
overexpression of PFK and PyK to increase ethanol production by 35% in E. coli, although lactic acid formation was not modified [16];
mannitol 1-phosphate dehydrogenase and mannitol 1-phosphatase overexpression to increase mannitol production by 27–50% in LDH-deficient Lactococcus lactis [54];
increase in sorbitol production (5 fold) in LDH-deficient Lactobacillus plantarum through the overexpression of sorbitol 6-phosphate dehydrogenase (activity up to 250 fold in mutants versus wild type) [55];
overexpression of PFK (14 fold) or LDH (3.5 times) to increase 2-3 times the homolactic fermentation flux in Lactococcus lactis growing on maltose, and in parallel decrease fluxes toward secondary acids and ethanol [56].
4. DOWNREGULATION OF ENZYMES TO MANIPULATE METABOLISM
4.1. Glycolysis in tumor cells
Glycolysis is enhanced in human and animal cancer cells (reviewed in [57]). Several glycolytic enzymes are overexpressed in at least 70% of human cancers [58]. Except for glucose transporter 1 (GLUT-1), the other 11 glycolytic enzymes (HK to LDH) are overexpressed in brain and nervous system cancers. Prostate and lymphatic nodule cancers (Hodgkin and non-Hodgkin lymphomas; myelomas) overexpress 10 glycolytic enzymes (except for HK; in prostate cancer GLUT1 is also overexpressed). There is a second group of cancers that overexpresses 6–8 glycolytic genes (skin, kidney, stomach, testicles, lung, liver, placenta, pancreas, uterus, ovary, eye, head and neck, and mammary gland). A third group includes those cancers overexpressing 1 or 2 glycolytic genes (bone, bone marrow, cervix, and cartilage) [58].
In animals, gene expression of glycolytic enzymes is regulated (both coordinately and individually) under hypoxic conditions by hypoxia-responsive transcription factors such as HIF-1α (hypoxia-inducible factor 1α), SP family factors, AP-1, and possibly MRE (metal response elements) [8, 59–61]. HIF-1α is probably the principal coordinator in gene induction. There are binding sites (consensus sequence ACGT) for HIF-1α in the promoters of genes for HK [62], PFK-1, ALDO, GAPDH, PGK, ENO, PYK, and LDH (reviewed in [8]). TPI and perhaps HPI and PGAM are also induced by hypoxia, but it is not clear whether HIF-1α mediates this induction [8], and whether this factor regulates other metabolic pathways associated with glucose catabolism. For example, although glycogen phosphorylase is overexpressed under hypoxia in human tissues [63], the role of HIF-1 has not been demonstrated.
If direct manipulation of pathway genes becomes difficult, then the overexpression or repression of transcription factors such as HIF-1α, AP1, and MREs might solve the problem of changing flux, although overexpression of transcription factors may also be difficult due to the numerous upstream and downstream factors involved.
4.2. Glycolysis in Trypanosoma brucei
The kinetoplastid parasites Trypanosoma cruzi, Trypanosoma brucei, and Leishmania are the causative agents of Chagas disease, African trypanosomiasis, and leishmaniasis, respectively. The available drugs to treat these diseases are highly toxic for humans. Moreover, the parasites may become resistant, and hence the search for new drugs and drug targets is relevant for solving these public health problems.
In these parasites, the metabolism is organized in a peculiar way; they have a subcellular structure called glycosome in which several metabolic pathways take place: gluconeogenesis, reactions of the pentose phosphate pathway, purine salvage and pyrimidine biosynthesis, β-oxidation of fatty acids, fatty acid elongation, biosynthesis of ether lipids, and the first seven steps of glycolysis. In fact, approximately 90% of glycosome enzyme content corresponds to glycolytic enzymes [64]. Glycosomal glycolytic enzymes have unique structural, kinetic, and regulatory features not found in their human counterparts, and therefore have been the subject of extensive biochemical studies to use them as drug targets [65]. The rationale behind this is to synthesize inhibitors that affect mainly the parasitic enzymes with relatively low effect on the human enzymes since the infective parasite stages rely mostly on glycolysis for ATP supply.
There are reports on the design of presumed specific inhibitors for some of the T. brucei glycolytic enzymes: GLUT (bromoacetyl-2-glucose) [66], HK, HPI, PFK, ALDO, TPI, GAPDH, PGK, PYK, and glycerol-3-phosphate dehydrogenase [67]. Although the purified enzymes display very low Ki values for these inhibitors and some of them inhibit parasite growth or infective capabilities, their effect on inhibiting the glycolytic flux has not been explored. Therefore, it is not yet possible to directly ascribe the effects seen in parasite culture with the in vitro effects on the isolated enzymes. To identify the best drug targets, determination of the flux control steps of glycolysis in T. brucei has been recently initiated [68].
4.3. Trypanothione synthesis in kinetoplastid parasites
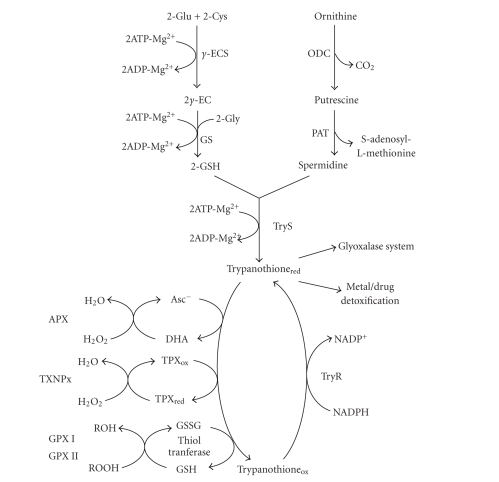
Trypanothione (TSH2) is a reducing agent present in trypanosomatids that is synthesized from one spermidine and two GSH molecules by TSH2 synthetase (TryS) (Figure 3). This metabolite and its reducing enzyme, TSH2 reductase (TryR), replace the antioxidant and metabolic functions of the more common GSH/GSH reductase system present in mammals. In fact, most of the antioxidant metabolism of these parasites depend on TSH2 (Figure 3) [69, 70]. Thus, the enzymes of this metabolic pathway have been proposed as drug targets for killing the parasites.
Figure 3.
Trypanothione synthesis in trypanosomatids. The trypanothione producing enzymes are γ-ECS, GS, ODC, aminopropyl transferase (PAT), and TryS. The trypanothione consuming enzymes are ascorbate peroxidase (APX); tryparedoxin peroxidases (TXNPx); trypanothione-glutathione thiol transferase (thiol transferase); and glutathione peroxidases I (GPX I) and II (GPX II). The regenerating enzyme is TryR. APX, thiol transferase, and GPX II have only been described in T. cruzi. This last parasite lacks ODC activity, but it has developed high-affinity transporters for putrescine, cadaverine, and spermidine [71].
Several studies have focused in assessing TryR as drug target. Diminution in its gene transcription yields a loss of activity between 56–90%, depending on the genetic technique [72–75]. In knockdown T. brucei cells (i.e., when TryR activity has diminished to less than 10% of the wild-type level), the parasites show growth diminution and higher sensitivity to H2O2 in culture and loss of infectiveness in mice. However, TSH2 and thiol compound contents were not affected [75]. TryR downregulation by >85% in Leishmania species causes inability to survive under oxidative stress inside macrophages [72–74]. In contrast, when TryR is 14- and 10 fold overexpressed in Leishmania and T. cruzi, respectively, there are no significant differences in H2O2 susceptibility between control and transfected cells; both types of cells are also equally resistant to the oxidative stress-inducers gentian violet, and nitrofurans [76]. Intriguingly, the cellular levels of TSH2, GSH, and glutathionyl-spermidine, determined in both types of experiments (TryR suppression and overexpression) were similar in control and transformed cells.
Other studies have proposed TryS as an alternative drug target. Knockdown of TryS by siRNA in procyclic T. brucei causes (i) viability impairment and arrest of proliferation when TSH2 levels decrease to 15% of the wild-type level, (ii) increased sensitivity to H2O2 and alkyl hydroperoxides, (iii) damage to the plasma membrane, and (iv) diminution of the TSH2 content and accumulation of GSH and glutathionyl-spermidine [77]. A similar metabolite variation (lower TSH2; higher GSH) was attained with a TryS knockdown induced by siRNA in the bloodstream form of T. brucei [78]. This TryS knockdown also induced an increased sensitivity to different compounds that affect TSH2 metabolism such as arsenicals, melarsen oxide, trivalent antimonials, and nifurtimox [78]. Indeed, western blot analysis showed, in addition to the expected (10-fold) decrease in TryS protein, a 2-3-folds increase in γ-ECS and TryR. The changes in expression of other enzymes suggest unveiled compensatory or pleiotropic effects on TSH2 metabolism.
Other researchers have selected γ-glutamylcysteine synthetase (γ-ECS), the presumed rate-limiting step of GSH synthesis, as an alternative drug target of TSH2 synthesis in T. brucei (Figure 3). Knockdown of γ-ECS gene in the parasite induces cell death and depletion of GSH and TSH2 only after 80% decrease in the enzyme content [79]. The γ-ECS knockdown cells are rescued from death by adding external GSH, which elevates the cellular GSH and TSH2 levels [79].
Glutathione synthetase (GS) has not been manipulated in trypanosomatids, or in any other organism, perhaps because it has been considered as a nonrate-limiting step of GSH and TSH2 biosynthesis. However, DNA microarray analysis of antimonite-resistant Leishmania tarentolae shows increased transcription of γ-ECS, GS, and P-glycoprotein A RNAs [80]. Although it was not evaluated whether increase in gene transcription correlated with an increase in enzyme activity, it may be possible that under high GSH demand (i.e., under oxidative stress conditions) GS might exert control of TSH2 synthesis. On the other hand, ornithine decarboxylase (ODC) overexpression in T. brucei (the presumed limiting step of spermidine synthesis) causes no change in TSH2 levels [81]. Therefore, ODC does not seem to be a controlling step of TSH2 synthesis.
Although almost full inhibition (>80%) of gene transcription or activity of any of these enzymes results in parasite death, the question remains of how TSH2 metabolism is affected when the enzymes are less inhibited. For example, in the therapeutic treatment of patients it is certain that drugs have to be administered for long periods of time. If the parasites are not completely cleared from the patient, disease recurrence and generation of drug-resistant parasites are possible. The results described above indicate that each enzyme by itself has low control on TSH2 synthesis and concentration; therefore, highly specific and very potent inhibitors have to be designed in order to attain the required full activity blockade to affect TSH2 metabolism in these parasites.
5. THEORY OF METABOLIC CONTROL ANALYSIS
The metabolic control analysis (MCA) was initially developed by Kacser and Burns in Scotland [82, 83] and by Heinrich and Rapoport in East Germany [84, 85]. This analysis establishes a theoretical framework that explains the results observed with the enzyme overexpression and downregulation experiments. In addition, it helps to identify and design experimental strategies for the manipulation of a given process in an organism (heavy metal hyperaccumulation; increased production of ethanol, CO2, lactate or acetate; or inhibition of a metabolic pathway flux with therapeutic purposes). MCA rationalizes the quantitative determination of the degree of control that a given enzyme exerts on flux and on the concentration of metabolites. Different experimental approaches have been developed to detect and direct what has to be done and measured, in order to identify and understand why an enzyme exerts a significant or a negligible control on flux and metabolite concentration in a metabolic pathway. Thus, the application of this analysis avoids the “trial and error” experiments for identifying and manipulating the conceptually wrong “rate-limiting step.”
To understand how a metabolic pathway is controlled and could be manipulated, its control structure has to be evaluated. The control structure of a pathway is constituted by the flux control coefficient (CviJ), which is the degree of control that the rate (v) of a given enzyme i exerts on flux J; the concentration control coefficient (CviX), which is the degree of control that a given enzyme i exerts on the concentration of a metabolite (X); and the elasticity coefficients. The control coefficients are systemic properties of the pathway that are mechanistically determined by the elasticity coefficients (εXvi), which are defined as the degree of sensitivity of a given enzyme vi (i.e., the enzyme's ability to change its rate) when any of its ligands (X: substrate, products or allosteric modulators) is varied.
The flux control coefficient is defined as
| (1) |
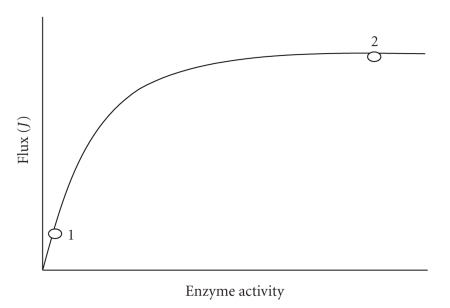
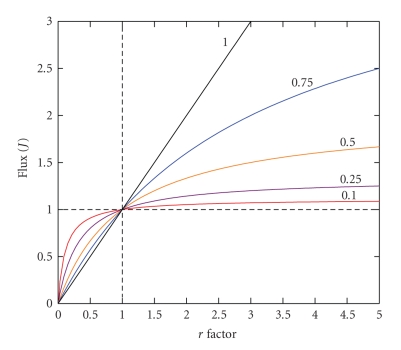
in which the expression dJ/dvi describes the variation in flux (J) when an infinitesimal change is done in the enzyme i concentration or activity. In practice, the infinitesimal changes in vi are undetectable, and hence measurable noninfinitesimal changes are undertaken. If a small change in vi promotes a significant variation in J, then this enzyme exerts an elevated flux control (Figure 4, position 1). In contrast, if a rather small or negligible change in flux is observed when vi is greatly varied, then the enzyme does not exert significant flux control (Figure 4, position 2). To obtain dimensionless and normalized values of CviJ the scaling factor vio/Jo is applied, which represents the ratio between the initial values from which the slope dJ/dvi is calculated. If all CviJ of the pathway enzymes and transporters are added up, the sum comes to one (summation theorem).
Figure 4.
Experimental determination of flux control coefficient.
The MCA clearly distinguishes between the control exerted by a given enzyme on flux (flux control coefficient) and on the metabolite concentration (concentration control coefficient). Thus, an enzyme can have significant control on a metabolite concentration but not on the pathway flux. This distinction is important for biotechnology purposes. On one hand, the use of the rate-limiting step concept for manipulating metabolic pathways does not make such differentiation, which probably has contributed to the many unsuccessful experiments reported in the literature; on the other hand, it should be clearly defined whether the aim of the project is to increase flux and/or a metabolite concentration since MCA establishes for each aim a different experimental design.
To determine the flux control coefficient of a given enzyme, small variations in the enzyme content, or preferentially, in activity are required, without altering the rest of the pathway, and then the changes in flux are determined. The experimental points are plotted as shown in Figure 4 to calculate the slope at the reference point vio/Jo. This experiment, apparently easy to perform, has demanded great intellectual and experimental effort. Several experimental strategies have been developed to determine CviJ:
formation of heterokarionts and heterocygots (classical genetics),
titration of flux with specific inhibitors,
elasticity analysis,
mathematical modeling (in silico biology),
in vitro reconstitution of metabolic pathways,
genetic engineering to manipulate in vivo protein levels.
5.1. Classical mendelian genetics
The arginine biosynthesis in Neurospora crassa was the first metabolic pathway in which flux control coefficients were experimentally determined by Kacser's laboratory [86]. This fungus forms multinucleated mycelia that facilitate the generation of polyploid cells. By mixing different ratios of spores containing genes encoding wild (active) and mutant (inactive) enzymes of this pathway, it was possible to generate heterokaryont mycelia with different content, and activity, of four pathway enzymes. The authors built plots of enzyme activity versus flux (see Figure 4) for acetyl-ornithine aminotransferase, ornithine transcarbamoylase, arginine-succinate synthetase, and arginine-succinate lyase. All the experimental points of these heterokaryonts localized near to position 2 of Figure 4 with CviJarg = 0.02–0.2 (flux control by these enzymes was only 2–20%), which indicated that none of these enzymes exerted significant control on arginine synthesis. The authors did not determine the remaining flux control (75%), which might reside in carbamoyl-phosphate synthetase I (this mitochondrial ammonium-dependent isoform can be bound to the mitochondrial inner membrane or form complexes with ornithine transcarbamoylase [87, 88]) and in mitochondrial citruline/ornithine transporter, both of which have been proposed as limiting steps, or might be in the arginine demand for protein synthesis.
Organisms with many alleles of one enzyme may form homo-and heterozygotes expressing different activity levels. Drosophila melanogaster has three ADH alleles encoding for isoforms with different Vmax. When three natural homozygotes, a null mutant, and some heterozygotes were generated, different ADH activities were attained but the ethanol consuming rate did not change (Figure 4, position 2). It was concluded that the ADH flux control was near zero [89].
5.2. Titration of flux with inhibitors (control of oxidative phosphorylation)
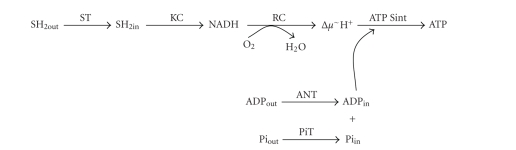
Oxidative phosphorylation (OXPHOS) is the only pathway for which specific and potent inhibitors for many enzymes and transporters are available. OXPHOS is divided in two segments (Figure 5): the oxidative system (OS) formed by substrate transporters (pyruvate, 2-oxoglutarate, glutamate, glutamate/aspartate, dicarboxylates), Krebs cycle enzymes, and the respiratory chain complexes; and the phosphorylating system (PS) constituted by the ATP/ADP (ANT) and Pi (PiT) transporters, and ATP synthase. The proton electrochemical gradient (Δμ−H+) connects the two systems.
Figure 5.
Mitochondrial oxidative phosphorylation. ST, oxidizable substrate transporter; KC, Krebs cycle; RC, respiratory chain; (Δμ∼H+), proton electrochemical gradient; ANT, adenine nucleotide translocator; PiT, phosphate transporter; ATP Sint, ATP synthase.
When the flux (ATP synthesis) is titrated by adding increasing concentrations of each specific inhibitor, plots are generated in which the enzyme activity is progressively diminished by increasing inhibitor concentration. Hence, the CviJ value depends on the type of inhibitor used
- for irreversible inhibition,
(2) - for simple noncompetitive inhibition,
(3) - for simple competitive inhibition,
(4)
where Jo is the pathway flux in the absence of inhibitor; Imax, minimal inhibitor concentration to reach maximal flux inhibition; K i, inhibition constant; S, substrate concentration; Km, Michaelis-Menten constant; and d J/d I, initial slope ([I] = 0) of inhibition titration curve.
To estimate flux control coefficients from inhibitor titration of ADP-stimulated (state 3) respiratory rates (i.e., mitochondrial O2 consumption coupled to ATP synthesis), (2) for irreversible inhibitors was used because researchers assumed that mitochondrial inhibitors such as rotenone, antimycin, carboxyatractyloside, and oligomycin were “pseudoirreversible,” due to the enzyme's high affinity for them. However, under this assumption flux control coefficients were usually overestimated [90, 91]. To solve this problem, Gellerich et al. [92] developed (5) for noncompetitive tightly-bound inhibitors and, by using nonlinear regression analysis, it was possible to include all experimental points from the titration curve thus increasing accuracy in calculating CviJ:
| (5) |
in which Jo and Ji are the respiration fluxes in the noninhibited (E = Eo) and inhibited (E = 0) states; K d is the dissociation constant of the enzyme-inhibitor complex, and n is an empirical component that expresses the relationship between substrate concentration and the reaction catalyzed by the enzyme E.
The analysis of data in Table 3 shows that OXPHOS is not controlled by only one limiting step, but the flux control is rather distributed among several enzymes and transporters. It is worth noting that the value of the flux control coefficient depends on the content of enzyme or transporter, which varies from tissue to tissue. Perhaps the ATP/ADP translocase in AS-30D hepatoma mitochondria might reach the status of being the “OXPHOS limiting step” with a CANTJOxPhos = 0.70, or the Pi transporter in kidney mitochondria [93], or the ATP/ADP translocase and the respiratory chain complex 3 in liver mitochondria [94], but it should be noted that other steps also exert significant control (Table 3). Although the distribution of control varies between tissues, the flux control mainly resides in the PS of organs with high ATP demand such as the heart (CPT+ANT+ATPsynthaseJOxPhos = CPSJOxPhos = 0.73), kidney (CPSJOxPhos = 0.75; COSJOxPhos = 0.31), and fast-growing tumors (CPSJOxPhos = 0.98). In contrast, in the liver (COSJOxPhos = 0.80; CPSJOxPhos = 0.65) and brain (COSJOxPhos = 0.35; CPSJOxPhos = 0.41), the control is shared by both systems.
Table 3.
Control distribution of oxidative phosphorylation.
| Enzyme | CviJATP | Rat organ mitochondria | Specific inhibitor | Inhibition mechanism | Reference |
|---|---|---|---|---|---|
| NADH-CoQ-oxidoreductase (Site 1 of energy conservation or Complex I of respiratory chain) | 0.15 | Heart (0.5 mM pyr + 0.2 μM Ca2+) | Rotenone | Noncompetitive tightly bound | [93] |
| 0.26 | Heart (10 mM pyr + 10 mM mal) | [95] | |||
| 0.31 | Kidney (0.5 mM pyr + 0.2 μM Ca2+) | [93] | |||
| 0.06 | Kidney (10 mM pyr + 10 mM mal) | [95] | |||
| 0.06–0.10 | Brain (0.05 mM pyr + 0.4 μM Ca2+) | [91] | |||
| 0.25 | Brain (10 mM pyr + 10 mM mal) | [95] | |||
| 0 | Tumor (10 mM glut + 3 mM mal) | [96] | |||
| 0.27 | Liver (10 mM pyr + 10 mM mal) | [95] | |||
| 0.13 | Skeletal muscle (10 mM pyr + 10 mM mal) | [95] | |||
| CoQ.cytochrome c oxidoreductase (Site 2 of energy conservation or Complex III of respiratory chain) | 0.01 | Heart | Antimycin | Noncompetitive tightly bound | [93] |
| 0.19 | Heart | [95] | |||
| 0.02 | Kidney | [95] | |||
| 0.05–0.11 | Brain | [91] | |||
| 0.02 | Brain | [95] | |||
| 0 | Tumor | [96] | |||
| 0.43 | Liver (5 mM Succ + 1 μM Ca2+) | [94] | |||
| 0.07 | Liver | [95] | |||
| 0.22 | Skeletal muscle | [95] | |||
| Cytochrome c oxidase (Site 3 of energy conservation or Complex IV of respiratory chain) | 0.11 | Heart | Cyanide or azide | Noncompetitive simple | [93] |
| 0.13 | Heart | [95] | |||
| 0.04 | Kidney | [95] | |||
| 0.02–0.07 | Brain | [91] | |||
| 0.02 | Brain | [95] | |||
| 0.04 | Tumor | [96] | |||
| 0.23 | Liver | [94] | |||
| 0.03 | Liver | [95] | |||
| 0.20 | Skeletal muscle | [95] | |||
| ATP/ADP transporter (adenine-nucleotides or ATP/ADP transporter, carrier or exchanger) | 0.24 | Heart | Carboxy-atractyloside (CAT) | Noncompetitive tightly bound | [93] |
| 0.04 | Heart | [95] | |||
| 0 | Kidney | [93] | |||
| 0.07 | Kidney | [95] | |||
| 0.08 | Brain | [91] | |||
| 0.08 | Brain | [95] | |||
| 0.60–0.70 | Tumor | [96] | |||
| 0.48 | Liver | [93] | |||
| 0.01 | Liver | [93] | |||
| 0.37 | Skeletal muscle (10 mM Glut + 3 mM mal) | [97] | |||
| 0.08 | Skeletal muscle | [95] | |||
| ATP synthase | 0.34 | Heart | Oligomycin | Noncompetitive tightly bound | [93] |
| 0.12 | Heart | [95] | |||
| 0.32 | Kidney | [93] | |||
| 0.27 | Kidney | [95] | |||
| 0.09–0.20 | Brain | [91] | |||
| 0.26 | Brain | [95] | |||
| 0.28 | Tumor | [96] | |||
| 0.05 | Liver | [94] | |||
| 0.20 | Liver | [95] | |||
| 0.10 | Skeletal muscle | [97] | |||
| 0.10 | Skeletal muscle | [95] | |||
| Pi transporter | 0.15 | Heart | Mersalyl | Noncompetitive simple | [93] |
| 0.14 | Heart | [95] | |||
| 0.43 | Kidney | [93] | |||
| 0.28 | Kidney | [95] | |||
| 0.13 | Brain | [91] | |||
| 0.26 | Brain | [95] | |||
| 0 | Tumor | [96] | |||
| 0.05–0.12 | Liver | [94] | |||
| 0.26 | Liver | [95] | |||
| 0.15 | Skeletal muscle | [97] | |||
| 0.08 | Skeletal muscle | [95] | |||
| Pyruvate transporter | 0.15 | Heart | α-cyano-4-hydroxy-cinnamate | Noncompetitive simple | [95] |
| 0.03 | Kidney | [95] | |||
| 0.08 | Brain | [91] | |||
| 0.26 | Brain | [95] | |||
| 0.21 | Liver | [95] | |||
| 0.20 | Skeletal muscle | [95] | |||
| Dicarboxylates transporter | 0.05–0.14 | Liver | Malate or butyl-malonate | Competitive simple | [94] |
| External ATPase | 0.40 | Skeletal muscle | Purified ATPase addition | [94] | |
The situation in skeletal muscle appears controversial. Wisniewski et al. [97] determined that the OXPHOS control was shared by the PS (CPSJOxPhos = 0.62) and the ATP demand (purified ATPase). In turn, Rossignol et al. [95] concluded that the OS exerted the main control (COSJOxPhos = 0.68), but these authors apparently used low-quality mitochondria (low respiratory control values that lead to low rates of ATP synthesis associated with high rates of respiration) that were not incubated under near physiological conditions (10 mM pyruvate, 10 mM malate, 10 mM Pi, pH 7.4 in Tris buffer), and the authors incorrectly assumed that rotenone and antimycin were irreversible inhibitors. It is notorious that in all works shown in Table 3 at least one of these mistakes is evident.
There are some inhibitors for enzymes and transporters from other pathways, but they are not quite specific and may affect other sites. Due to the fact that there are no inhibitors for every step in these pathways, only one flux control coefficient has been determined by inhibitor titration. Examples of these inhibitors are 6-chloro-6-deoxyglucose for glucose transporters in bacteria, 2-deoxyglucose for HPI, iodoacetate for GAPDH [6], 1,4-dideoxy-1,4-imino-D-arabinitol for glycogen phosphorylase [98], oxalate and oxamate for LDH, 6-amino nicotinamide for the phosphate pentose pathway [99], amino-oxyacetate for aminotransferases and kirureninase (tryptophan synthesis), norvaline for ornithine transcarbamylase, mercaptopycolinate for PEP carboxykinase, acetazolamide for carbonic anhydrase, and isobutyramide for ADH (compiled by Fell [2]).
Potential uses of the experimental approach —
Mitochondrial pathologies are a heterogeneous group of metabolic perturbations characterized by morphological abnormalities and/or OXPHOS dysfunction [100]. Mitochondrial DNA analysis has revealed specific mutations for some mitochondriopathies. Although the specific OXPHOS mutations causing the disease may appear in all tissues, the functioning of only some of them is altered. The organ's sensitivity might be related to the different flux control coefficients of the mutated enzyme in the different tissues (Table 3) and to their ATP supply dependence from OXPHOS versus glycolysis.
MCA allows for the analysis of a metabolic flux or intermediate concentration by focusing either on one step or by grouping enzymes in blocks or in pathways. Thus, a comparative analysis of OXPHOS control distribution reveals that heart, kidney, some fast growing tumors (rat AS-30D hepatoma, mouse fibrosarcoma, human breast, lung, thyroid carcinoma, melanoma) [101], and perhaps skeletal muscle are more susceptible to mitochondrial mutations in ATP synthase, which is the only PS site with subunits encoded in the mitochondrial genome. On the other side, liver and brain might be more susceptible to mitochondrial mutations of the respiratory chain enzymes (see Table 3). Considering that the brain is a fully aerobic organ [102], whereas the liver depends on both OXPHOS (70–80%) and glycolysis (20–30%) for ATP supply [103], then it can be postulated that the brain is more sensitive to mutations in the mitochondrial genome than the liver because subunits of complexes I, III, and IV are encoded by the mitochondrial genome.
Titration of flux with specific inhibitors to determine the flux control coefficients of OXPHOS has been applied to intact tumor cells [90]. The results showed that the flux control resided mainly in site 1 of the respiratory chain (CSitelJOxPhos = 0.30), whereas the other evaluated sites exerted a marginal control [90]. This observation could have therapeutic application if site 1 does not exert control in healthy cells, leading to less severe side effects.
The use of inhibitors in intact cells to determine control coefficients might pose two problems: hydrophilic inhibitors such as carboxyatractyloside (for ANT) and α-cyano-4-hydroxy-cinammate (for pyruvate transporter) cannot readily enter the cell due to the presence of the plasma membrane barrier; the other problem is that hydrophobic but slow inhibitors, such as oligomycin, require long incubation times to ensure the interaction with the specific sites. These problems can be solved by incubating the cells for long periods of time and taking care of cell viability, for instance, AS-30D hepatoma cells are fairly resistant to this mechanical manipulation as they maintain high viability after a lengthy incubation under smooth orbital agitation of 1 h at 37°C [90].
5.3. Elasticity analysis
MCA defines the elasticity coefficients as
| (6) |
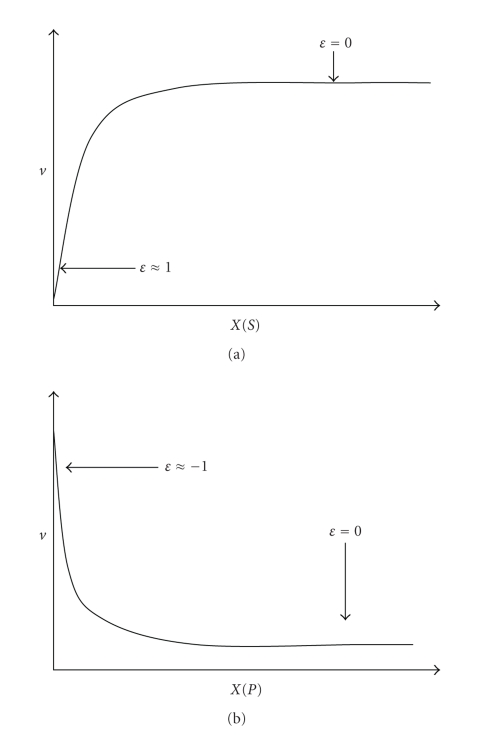
which is a dimensionless number that show the rate variation v of a given enzyme or transporter i when the concentration of a ligand X (substrate S, product P or allosteric modulator) is varied in infinitesimal proportions. The elasticity coefficients are positive for those metabolites that increase the enzyme or transporter rate (substrate or activator), and they are negative for the metabolites that decrease the enzyme or transporter rates (product or inhibitor). An enzyme working, under a steady-state metabolic flux, at saturating conditions of S or P, is no longer sensitive to changes in these metabolites. Thus, its elasticity is close to zero (Figure 6, εXvi = 0). In turn, an enzyme working at S or P concentrations well below the Michaelis constant (KmS or KmP) is expected to be highly sensitive to small variations in these metabolites (Figure 6, εXvi = 1).
Figure 6.
Elasticity coefficients.
The elasticities are intrinsically linked to the actual enzyme kinetics. If the kinetic parameters of an enzyme are known (Vmf, Vmr, KmS, and KmP), then the enzyme elasticity for any given metabolite concentration may be calculated as shown in the following equations.
For substrate,
| (7) |
and for product,
| (8) |
in which Γ is the mass action ratio, and Keq is the equilibrium constant preferentially determined under physiological conditions.
An enzyme with low elasticity cannot increase (or decrease) its rate despite large variations in S (or P) concentration; in consequence, such enzyme exerts a high flux control. In turn, an enzyme with a high elasticity can adjust its rate to the variation in S or P concentrations, and thus it does not interfere with the metabolic flux, exerting a low flux control. This inverse relationship between the elasticity and the flux control coefficients is expressed in a formal equation denominated connectivity theorem. A metabolic pathway can be divided in two blocks around an intermediary X: the producing (synthetic, supply) and the consuming (demand) enzyme blocks of X are i1 and i2, respectively. Thus, the connectivity theorem for this two-block system is
| (9) |
The negative sign of the right part of the equation cancels with εXvi1, which is negative because X is a product of enzyme block i1 (Figure 6).
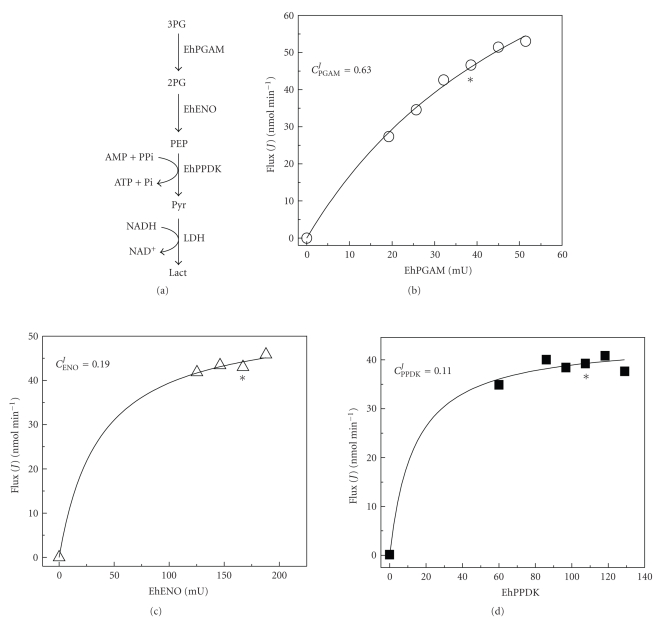
To obtain the flux control coefficients, this approach requires experimental determination of the elasticity coefficients. How can this be done? Many strategies have been designed [90, 103–108], but the most used and probably more trustworthy is that in which the initial pathway metabolite (So) concentration is varied to increase the X concentration (any intermediary in the pathway), and measuring in parallel the variation in flux. Under steady-state conditions, the flux rate is equal to the rate of end-product formation (i.e., lactate or alcohol for glycolysis; oxygen consumption for OXPHOS) and to the rate of any partial reaction. Then, plots of X versus flux (Figure 7) are generated. The slope, calculated at the reference coordinate (Xo, Jo) that is equivalent to (So, vio), yields the elasticity coefficient of the consuming block of X. In another set of experiments, an inhibitor is added to block one or more enzymes after X. The X concentration and flux are determined and plotted as shown in Figure 7, from which the elasticity coefficient of the producing block is calculated.
Figure 7.
Experimental determination of the elasticity coefficients for substrates and products.
The flux control coefficients are determined by using the connectivity theorem and considering that the sum of the control coefficients comes to 1, C1 + C2 = 1 (summation theorem):
| (10) |
This method for determining CviJ using the elasticities of the two blocks was called double modulation by Kacser and Burns [83]. Years later, Brand and his group [103, 104] renamed this method as top-down approach. By applying the procedure shown in Figure 7 and using (10) for different metabolites along the metabolic pathway, it is possible to identify those sites that exert a higher control (which may be the sites for therapeutic use or biotechnological manipulation) and those that exert a negligible control under a given physiological or pathological situation.
Elasticity analysis has been used to evaluate the OXPHOS control distribution in tumor cells [90]. Almost all studies on this subject have been carried out with isolated mitochondria incubated in sucrose-based medium at 25 or 30°C or with the more physiological KCl-based medium but still at 30°C (Table 3). Furthermore, these studies did not consider that the product, ATP, never accumulates in the living cells, which does occur in experiments with isolated mitochondria. Under such a condition, a steady state in ATP production can never be reached as in living cells. In other words, the distribution of control in mitochondria (Table 3) has been determined in the absence of an ATP-consuming system. A remarkable exception to this incomplete experimental design was the work done by Wanders et al. [105], in which isolated liver mitochondria were incubated with two different ATP-consuming systems (or ADP-regenerating systems): HK + glucose and creatine kinase (CK) + creatine. Under this more physiological setting, the OXPHOS flux control distributed between ANT and the ATP-consuming system; however, flux control by the other pathway components was not examined. Therefore, to accurately evaluate OXPHOS control distribution, mitochondria should be incubated in the presence of an ATP-consuming system or in their natural environment (i.e., inside the cell).
The rate of OXPHOS in intact cells is determined from the rate of oligomycin-sensitive respiration: in the steady state, the enzyme rates are the same and constant; in branched pathways the sum of the branched fluxes equals the flux that supplies the branches. The global elasticity of the ATP-consuming processes (e.g., synthesis of protein, nucleic acid, and other biomolecules, as well as ion ATPases to maintain the ionic gradients, mechanical activity such as muscular contraction or flagellum and cilium movement, and secretion of hormones, digestive enzymes and neurotransmitters) is estimated by inhibiting flux with low concentrations of oligomycin or a respiratory chain inhibitor. To determine the elasticity of the ATP-producing block, flux, and [ATP] are varied with streptomycin, an inhibitor of protein synthesis (Figure 7). The elasticity coefficients are calculated from the initial coordinate slopes (without inhibitors) of each titration. With this procedure, it has been determined that the ATP-consuming block exerts a significant flux control of 34% [90]. Remarkably, this flux control value obtained in cells is quite similar to the flux control coefficients of the ATP-consuming system (HK or CK) reported by Wanders et al. [105] with isolated mitochondria.
Elasticity analysis by enzyme blocks allows the inclusion of the end-product demand as another pathway block. The conclusions obtained from this analysis have formulated the supply-demand theory [30], which proposes that when flux is controlled by one block (demand), the concentration of the end-product is determined by the other block (supply). The ratio of elasticities determines the distribution of flux control between supply and demand blocks. For instance, if εXSupply > εXDemand (i.e., demand becomes saturated by the end-product X, and hence its elasticity is near zero), then the demand block exerts the main flux control. For concentration control, at larger εXDemand − εXSupply, smaller absolute values of both CSupplyX and CDemandX are attained; hence, under demand saturation, the supply elasticity fully governs the magnitude of the variation in the end-product concentration. On the other hand, when demand increases, it loses flux control and induces a diminution in the end-product concentration. In turn, supply gains flux control and loses concentration control. In the presence of feed-back inhibition, the system can maintain the end-product concentration orders of magnitude away from equilibrium (at a concentration around the K0.5 of the allosteric enzyme).
As mentioned before, the demand is not usually included in the pathway because it is erroneously thought that it is not part of it. But then, is it valid to analyze the control of a metabolite synthesis if its demand is not considered? When the demand block is not included, it is assumed that the metabolic pathway produces a metabolite at the same rate regardless whether the metabolite demand is high or low. This reasoning is incorrect because a metabolic pathway indeed responds to changes in the metabolite demand and, more importantly, a pathway without end-products consumption reactions is unable to reach a steady state.
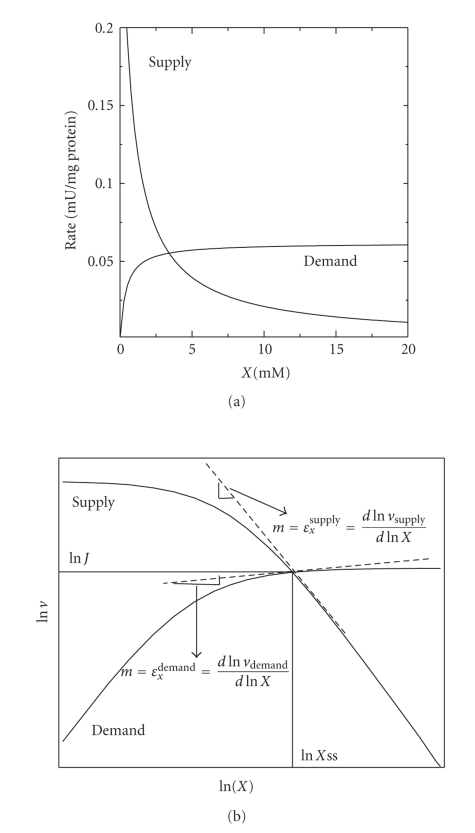
Therefore, a metabolic pathway can be divided in supply and demand blocks. The intermediary X linking the two blocks is one of the end-products of the producing block (e.g., pyruvate or lactate or ethanol, and ATP for glycolysis). The variation in rate of the two blocks in response to a variation in X can be theoretical or experimentally determined (Figure 8(a)). It is worth noting that, for this supply-demand approach, it is not necessary to know the kinetics of each pathway enzyme because the rate response of each block reflects the global kinetics of all participating enzymes. When the X concentration is increased, the rate of the supply block decreases (i) because X is its product and (ii) because usually an enzyme within this block receives information from the final part of the pathway, decreasing its rate through feedback inhibition. In turn, the rate of the demand block increases as X is its substrate.
Figure 8.
(a) Kinetics of the synthesis (supply) and consuming (demand) blocks of the intermediary X. The kinetic parameters are from enzymes in tobacco glutathione (GSH) synthesis. X represents the intermediary concentration, in this case GSH. (b) Rate plots of the supply and demand blocks in a natural logarithmic scale.
To better visualize the effect of large rate changes, the kinetics of both blocks are plotted in a logarithmic scale. Figure 8(b) shows the kinetics described in Figure 8(a) converted to natural logarithm. The intersection point between kinetic curves, at which the supply and demand rates are identical, represents the pathway steady-state flux (in the Y axis) and end-product concentration (in the X axis). Since the elasticity is also defined as εXvi = dlnvi/dlnX, the slope at the intersection point represents the elasticity of each block towards the intermediary X. Here, the use of the scalar factor is not necessary because it is included in the logarithmic equation. With the elasticity coefficients calculated from plots like those shown in Figure 8, and the connectivity theorem, the flux control coefficient of each block is determined. The example in Figure 8(b) shows that the demand exerts a high flux control (and has low elasticity) and the supply block exerts low control (and has high elasticity).
The fact that the demand may exert a high flux control in metabolite pathways has at least three important implications: (a) the supply block responds to variations in the demand (high elasticity); (b) the demand block has information transfer mechanisms towards the supply block that avoid the unrestricted intermediary accumulation under a low demand, particularly when the supply block has reactions with large Keq (>100; ΔG°′ > 3 Kcal mol−1 at 37°C); and (c) if the main flux control resides in the demand block, then the supply block may only exert control on the intermediary concentration but not on the flux [30, 32]. This last conclusion explains why it is incorrect to consider that an enzyme that controls flux must also control the intermediary concentration.
Regulatory mechanisms of enzyme activity are modulation of protein concentration by synthesis and degradation, as well as covalent modification and variation in the substrate or product concentrations (which are components of the pathway). In addition, another regulatory mechanism is the modulation by molecules that are not part of the pathway, that is, through allosteric interaction with cooperative (sigmoidal kinetics) or noncooperative enzymes (hyperbolic kinetics) (e.g., Ca2+ activates some Krebs cycle dehydrogenases; citrate inhibits PFK-1; malonyl-CoA inhibits the mitochondrial transporter of acyl-carnitine/carnitine; or the initial substrate of a pathway that has not entered the system). For these last cases, Kacser and Burns [83] proposed the use of the response coefficient R which is defined by the following expression:
| (11) |
where M is the external modulator of the i enzyme. The response coefficient is dJ/dM•Mo/Jo. If the elasticity of the sensitive enzyme toward the external effector is also determined, then it is possible to calculate CviJ by using (11). Unfortunately, due to the experimental complexity for determining the elasticity coefficient, this coefficient is often calculated in a theoretical way by using the respective rate equation (Michaelis-Menten or Hill equations) and the kinetic parameters Km and Vmax determined by someone else under optimal assay conditions, which are commonly far away from the physiological ones. Therefore, for this theoretical determination of elasticity only the value of the external modulator concentration is required. It is convenient to emphasize that the determination of the flux control coefficients becomes more reliable when they are calculated from several experimental points (Figure 7), instead of only one, as occurs with the theoretical elasticity analysis.
Groen et al. [106] determined the flux control distribution of gluconeogenesis from lactate in hepatocytes by using both theoretical and experimental elasticity analysis and the response coefficient. These authors concluded that gluconeogenesis stimulated by glucagon was controlled by the pyruvate carboxylase (CPCJglucose = 0.83); in the absence of this hormone, the control was shared by PC, PYK, ENO-PGK segment, and TPI-fructose-1,6-biphosphatase segment [106].
Elasticity analysis has been applied to elucidate the flux control of ATP-producing pathways in fast-growing tumor cells. For OXPHOS, this approach showed that respiratory chain complex I and the ATP-consuming pathways were the enzymes with higher control (CviJ = 0.7) [90]. For glycolysis, the main flux control (CviJ = 0.71) resided in GLUT + HK reactions because HK is strongly inhibited by its product G6P despite extensive enzyme overexpression [107]. Examples of elasticity analysis on other pathways are photosynthesis [108], ketogenesis [109], serine [110] and threonine synthesis in E. coli [111], glycolysis in yeast [112], glucose transport in yeast [113], DNA supercoiling [114], glycogen synthesis in muscle [115], and galactose synthesis in yeast [116].
In conclusion, the elasticity analysis is the most frequently used method for determining flux control coefficients because it does not need a group of specific inhibitors for all the enzymes and transporters of the pathway, neither does it require knowledge of the inhibitory mechanisms or kinetic constants. It is only necessary to produce a variation in the intermediary concentration X by using an inhibitor of either block or by directly varying the X concentration.
5.4. Pathway modeling
In agreement with Fell [2], it seems impossible for a researcher to analyze one by one the rate equation of each enzyme in a metabolic pathway to predict and explain the system behavior as a whole. To deal with this problem, in the last three decades some scientists have constructed mathematical models for some metabolic pathways using several software programs. Thus, the specific variation of a single enzyme activity without altering the rest of the pathway (Figure 4), which has been an experimentally difficult task for applying MCA, becomes easier to achieve with reliable computing models. The term “in silico biology” has been coined for this approach.
There are two basic types of modeling: (a) structural modeling and (b) kinetic modeling. The former is related to the pathway chemical reaction structure and does not involve kinetic information. The use of reactions is based on their stoichiometries. The information obtained with structural modeling is the description of the following:
the exact determination of which reactions and metabolites interact among them;
the conservation reactions. There are metabolites for which their sum is always constant or conserved (e.g., NADH + NAD+; NADPH + NADP+; ubiquinol + ubiquinone; ATP + ADP + AMP; CoA + acetyl-CoA). The identification of conserved metabolites might not be obvious;
enzyme groups catalyzing reactions in a given relationship with another group of enzymes;
elemental modules, which are defined as the minimal number of enzymes required to reach a steady state, which can be isolated from the system (for a review about structural modeling; see [117]).
Kinetic modeling is more frequently used. In addition to an appropriate computing program, this approach requires the knowledge of the stoichiometries, rate equations, and Keq values of each reaction in the pathway (or the Vmax in the forward and reverse reactions), as well as the intermediary concentrations reached under a given steady state. Some currently used softwares are Copasi (http://www.copasi.org/tiki-index.php) based on Gepasi (http://www.gepasi.org/; [118]); Metamodel [119]; WinScamp [120] and Jarnac [121] (both available at http://www.sys-bio.org/); and PySCeS (http://pysces.sourcesforge.net/; [122]). For other programs and links, go to http://sbml.org/index.psp. To reach a steady-state flux, it is necessary to fix the initial metabolite concentration to a constant value and the irreversible and constant removal of the end products. Except for the final reactions in which their products have to be removed from the system, all pathway reactions have to be considered as reversible, notwithstanding whether they have large Keq (if there is an irreversible reaction under physiological conditions, then a reversible rate equation that includes the Keq suffices to maintain the reaction as practically irreversible). Care should be taken to include the enzyme's sensitivity toward its products because this property is related with the enzyme elasticity and hence with its flux control; omission of this parameter may very likely lead to erroneous conclusions.
It should be pointed out that the purpose of kinetic modeling is not merely to replicate experimental data but also to explain them [117]. Thus, pathway modeling is a powerful tool that allows for (i) the detection of those properties of the pathway that are not so obvious to visualize when the individual kinetic characteristics of the participating enzymes are examined; and (ii) the understanding of the biochemical mechanisms involved in flux and intermediary concentration control. Modeling requires the consideration of all reported experimental data and interactions that have been described for the components of a specific pathway, thus allowing for the integration of disperse data, discarding irrelevant facts [84]. Although all models are oversimplifications of complex cellular processes, they are useful for the deduction of essential relationships, for the design of experimental strategies that evaluate the control of a metabolic pathway, and for the detection of incompatibilities in the kinetic parameters of the participating enzymes and transporters, which may prompt the experimental revision of the most critical uncertainties.
With the model initially constructed, the simulation results do not usually concur with the experimental results; in consequence, the model normally requires refinement, a point at which the researcher's thinking and knowledge of biology plays a fundamental role in modifying the structure and parameters of the model. The discrepancies observed between modeling and experimentation unequivocally pinpoint what elements or factors have to be re-evaluated or incorporated so that the model approximates more closely reality (i.e., experimental data). The comparison of the experimentally obtained intermediary concentrations and fluxes with those obtained by simulation is an appropriate validating index of the model; this index indicates whether the model approximation to the physiological situation is acceptable or whether re-evaluation of the kinetic properties of some enzymes and transporters and/or incorporation of other reactions or factors is required.
A reason to why the results obtained by modeling may substantially differ from the experimental results is that the kinetic parameters of the pathway enzyme and transporters and the Keq values used were determined by different research groups, under different experimental conditions and in different cell types. Moreover, enzyme kinetic assays are carried out at low, diluted enzyme concentrations (thus discarding or ignoring relevant protein-protein interactions), and at optimal (but not physiological) pH and “room temperature” (which may be far away from the physiological values). In addition, no experimental information is usually available regarding the reactions reversibility and the product inhibition of the enzymes and transporters (particularly for physiological irreversible reactions, i.e., reactions with large Keq). With worrisome frequency, the researcher has to adjust the experimentally determined Vm and Km values to achieve a model behavior that acceptably resembles that observed in the biological system. Apparently, this type of limitations as well as the sometimes overwhelming amount of kinetic data necessary for the construction of a kinetic model has restricted the number of reliable models that can be used for the prediction of the pathway control structure.
Once the kinetic model stability, robustness, structural and dynamic properties have been evaluated, and experimentally validated, the model may become a virtual laboratory in which any parameter or component can be modified or replaced and any aspect of the pathway behavior can be explored within a wide diversity of circumstances or limits [117]. At this stage, the model is suitable for examining the pathway regulatory properties and control structure.
Glycolysis in S. bayanus, S. cerevisiae [113, 123, 124], and Trypanosoma brucei [125, 126] is the metabolic pathway that has been more extensively modeled. Both cell types have a very active glycolysis and are fully dependent on this metabolic pathway for ATP supply, under anaerobiosis and aerobiosis, respectively. One advantage of modeling glycolysis in these cell types is that most of the kinetic parameters used have been experimentally determined by the same groups under the same experimental conditions. However, the kinetics of the reverse reactions has not been determined and thus these authors used KmP and Keq values reported by others and obtained in other cell types under rather different experimental conditions, or they were adjusted to improve model fitting.
Nevertheless, the simulation results yielded relevant information on the control of the glycolytic flux. In both cases, the enzymes traditionally considered the rate-limiting steps, HK, ATP-PFK-1, and PYK did not contribute to the flux control, whereas the main control resided in GLUT (54% in the parasite and 85–100% in yeast). Under some conditions, HK may exert some control (15%) in S. cerevisiae and some nonallosteric enzymes such as ALDO, GAPDH, and PGK may also exert some flux control in T. brucei.
MCA through kinetic modeling has been applied to several pathways:
glycolysis in erythrocytes [84] in which flux control distributes between HK (71%) and PFK-1 (29%);
carbohydrate metabolism during differentiation in Dictyostelium discoideum [127] with cellulose synthase (86%) as the main controlling step;
sucrose accumulation in sugar cane with HK, invertase, fructose uptake, glucose uptake, and vacuolar sucrose transporter having the most significant flux control [128];
glycerol synthesis in S. cerevisiae with GAPDH (85%) as the main control step [129];
penicillin synthesis in Penicillium chrysogenum controlled (75–98%) either by d-(a-aminoadipyl) cysteinylvaline synthetase (short incubation times <30 hour) or isopenicillin N. synthetase (long incubation times > 100 h) [130];
Calvin cycle [131] controlled by GAPDH (50%) and sedoheptulose-1,7-bisphosphatase (50%);
threonine synthesis in E. coli controlled by homoserine dehydrogenase (46%), aspartate kinase (28%), and aspartate semialdehyde dehydrogenase (25%) [111];
lysine production in Corynebacterium glutamicum mainly controlled by aspartate kinase and permease [132];
nonoxidative pentose pathway in erythrocytes mainly controlled by transketolase (74%) [133];
EGF-induced MAPK signaling in tumor cells controlled by Ras-activation by EGF (21%), Ras dephosphorylation (43%), ERK phosphorylation by MEK (44%), and MEK phosphorylation by RAS (143%) [13];
Aspergillus niger arabinose utilization with flux control shared by arabinose reductase (68%), arabitol dehydrogenase (17%), and xylulose reductase (14%) [134];
glycolysis in L. lactis in which several end products are generated (lactate, organic acids, ethanol, acetoin) [135]. Model predictions indicated that flux toward diacetyl and acetoin (important flavor compounds) was mainly controlled by LDH but not by acetolactate synthetase, the first enzyme of this branch.
We modeled the GSH and PCs biosynthesis (Figure 2) to determine and understand the control structure of the pathway and thus be able to identify potential sites for genetic engineering manipulation that might lead to the generation of improved species in heavy metal resistance and accumulation. Two models were constructed, one for higher plants and the other for yeast, both exposed to high concentrations of Cd2+ [136]. Due to the similarity in the results, only the plant results are analyzed below.
An interesting conclusion from the GSH-PCs synthesis modeling is that control of flux (and GSH concentration) is shared between the GSH supply and demand under both unstressed and Cd2+ exposure conditions (Table 4). This observation strongly differs from the idea that γ-ECS is the rate-limiting step [33–35]. For many researchers, the concept of γ-ECS being the key controlling step has seemed to be correct because (a) γ-ECS receives information from the final part of the pathway, as it is potently inhibited by GSH, the pathway end-product; and (b) γ-ECS is localized in the first part of the pathway (Figure 2). In addition, GS is usually more abundant and efficient than γ-ECS [137].
Table 4.
Control of GSH and PC synthesis in plants exposed to Cd2+.
| Enzyme | 1x γ-ECS + PCS | 2.5x γ-ECS + PCS | ||||||
|---|---|---|---|---|---|---|---|---|
| CviJGSH | CviJPC | CviGSH | CviPC | CviJGSH | CviJPC | CviGSH | CviPC | |
| γ-ECS | 0.58 | 0.60 | 0.68 | 0.76 | 0.45 | 0.61 | 0.70 | 0.60 |
| GS | <0.01 | <0.01 | 0.01 | 0.01 | 0.19 | <0.01 | <0.01 | 0.97 |
| GS-transferase | 0.01 | −0.06 | −0.07 | −0.07 | <0.01 | <0.01 | < −0.01 | −0.05 |
| PCS | 0.40 | 0.44 | −0.63 | −0.56 | 0.33 | 0.44 | −0.62 | 0.57 |
| vacuole PC-Cd transporter | <0.01 | <0.01 | <0.01 | −1.2 | <0.01 | <0.01 | <0.01 | −2.1 |
CviJGSH, control coefficient of enzyme i in GSH synthesis; CviJPC, control coefficient of enzyme Ei on PCs synthesis; CviGSH, control coefficient of enzyme i on GSH concentration; CviPC, control coefficient of enzyme i on PCs concentration. An enzyme with a negative flux control indicates that it is localized in a branch, turning aside the principal flux; an enzyme with a negative concentration control indicates that an increase in its activity decreases metabolite concentration.
However, in most of the studies on the control of GSH synthesis, the GSH demand has not been considered. The GSH synthesis modeling shows that under a physiological feedback inhibition of γ-ECS by GSH a small increase in demand increases flux because the GSH concentration decreases and the γ-ECS inhibition attenuates. In contrast, if the demand remains constant, then an increase in γ-ECS activity or content (by overexpression) does not increase flux because the GSH inhibition is still there and operates on both new and old enzymes. The same pattern is also observed when HK is overexpressed to increase glycolytic flux since it is still inhibited by G6P (see Section 3). On the other hand, γ-ECS indeed exerts significant concentration control on GSH, which means that a γ-ECS increase results in higher GSH concentration (Table 4). This last observation demonstrates that an enzyme controlling a metabolite concentration does not necessarily control the flux.
Cd2+ exposure promotes a high GSH demand because significant oxidative stress surges, thus causing oxidation of GSH through GSH peroxidases, and because GSH and PCs are used for sequestering the toxic metal ion; hence, a higher GSH consuming rate sets up. Under this condition, modeling predicted that control was almost equally shared between the supply and demand blocks, but particularly between γ-ECS and PCS (see Figure 2). Modeling was also able to explain why PCS overexpression can have toxic effects on the cell [36]. An increase in the GSH demand (PCS overexpression) under high-demand conditions (Cd2+ stress) leads to GSH depletion that severely compromises other processes such as the oxidative stress control and xenobiotic detoxification.
The conclusions drawn by this model led us to propose that, to significantly increase the Cd2+ resistance and accumulation, γ-ECS and PCS should be simultaneously overexpressed (Table 4; Figure 9). This particular manipulation promotes an increase in the rate of GSH and PCs synthesis (determined by the high-to-low transition of their flux control coefficients) and in the GSH and PCs concentrations (determined by their high concentration control coefficients). The model predicts that a 2-fold increase in the simultaneous overexpression of γ-ECS and PCS brings about a 1.9–2.4-fold increase in flux to GSH (JGS) and PCs (JPCS) and in PCs concentration (Figure 9); a 5-fold overexpression further increases by 4.5–8.1 times the fluxes and PCs concentration.
Figure 9.
Modeled simultaneous overexpression of two controlling enzymes, one in the supply (γ-glutamylcisteine synthetase, γ-ECS) and the other in the demand branch (phytochelatin synthase, PCS), of the glutathione and phytochelatins synthesis pathway in plants.
This proposed enzyme overexpression should not exceed the GS and the complex PC-Cd (or GS-Cd-GS) vacuolar transporters' maximal activities, in order to keep the cell away from a severe oxidative stress caused by GSH depletion or γ-EC accumulation. Indeed, the concentration of GSH was maintained high and constant although γ-EC accumulated with the simultaneous overexpression (Figure 9). Furthermore, this enzyme manipulation should avoid the increase of the PC-Cd and GS-Cd-GS complexes in cytosol to toxic levels. In other words, excessive enzyme overexpression should be avoided, unless this is accompanied by compensating overexpression of consuming enzymes (GS for γ-ECS overexpression and PCs vacuolar transporters for that of PCS). In yeasts and plants, Cd2+ is ultimately inactivated by the additional interaction with S2− and the subsequent formation of stable high molecular weight complexes with PCs, Cd2+, S2−, and GSH [138, 139]. In parallel to the γ-ECS and PCS overexpression, moderate repression of GSH-S-transferases, which compete for the available GSH (Figure 2), may also promote an increase in GSH concentration and PCs formation flux [136].
MCA is based on infinitesimal changes in an enzyme or metabolite concentration. In contrast, gene overexpression induces large changes in activity; hence, further theoretical background has been developed for predicting the effect on flux and metabolite concentrations induced by large enzyme changes. Such a theoretical background was initially developed by Small and Kacser [140], who depicted (12) based on the flux control coefficients to predict the effect promoted by large changes in enzyme activity:
| (12) |
in which f is the amplification factor (the flux increase), and r represents how many times the enzyme is overexpressed. To predict the flux changes, promoted by identical overexpression of two enzymes (same r value) with different CviJ, the equation is
| (13) |
Figure 10 shows the effect on flux when one or more enzymes with different CviJ are changed by the same r factor. If the sum of CviJ of one or more enzymes is less than 0.25, the impact on flux is discrete when the expression increases 5 folds (which is the most common variation in the overexpression experiments analyzed in Section 2). But for a 3-fold overexpression of a group of enzymes, for which their sum of CviJ is more than 0.5, then a significant flux change is achieved. If the sum of CviJ is 1, the flux varies in a linear proportion with the degree of overexpression. It has to be remarked, however, that the predicted change in flux (Figure 10) will be valid until certain degree, the limits of which being determined by the other pathway enzymes that should stay as noncontrolling steps.
Figure 10.
Effect on flux when one or more enzymatic activities with different control coefficients are varied. This figure represents an enzyme or group of enzymes in which their CviJ sum is indicated in parenthesis and is modified by the same r factor. Number 1 represents the reference control, thus if r < 1, there is suppression, whereas r > 1 represents overexpression.
Figure 10 also shows the effect on flux of decreasing an enzyme activity (third quadrant). This segment plot is useful when inhibition of pathway flux is being pursued for therapeutic purposes or for understanding the molecular basis of the genetic dominance and recessivity. Like in the enzyme overexpression experiment, only a significant effect on flux is achieved when the enzymes with high CviJ values are inhibited. For an enzyme or group of enzymes with CviJ of 0.25, greater than 80% inhibition has to be attained to decrease 50% the pathway flux. In this context, it seems feasible to explain why knockdown of enzymes involved in TSH2 synthesis has to be almost total to detect an effect on TSH2 content or to alter functional or pathogenic properties of the parasites (Section 4.3). The knockdown or knockout experiments in trypanosomatids suggest that γ-ECS, TryS, and TryR most probably have low flux control and concentration-control coefficients since their contents or activities have to be reduced >80% of the normal levels to reach changes in intermediary levels or in oxidative stress handling.
Contrary to the several unsuccessful overexpression experiments carried out to increase the flux or metabolites of a metabolic pathway, modeling may allow for a more focused and appropriate design of experimental strategies of genetic engineering to increase flux or a given metabolite, and for selecting drug targets to decrease flux or metabolite concentration. For these predictions, modeling considers that overexpression of a controlling enzyme or transporter may promote flux or metabolite control redistributions. Thus, a low-control step may become a controlling point when overexpressing another step and, in consequence, the prediction shown in Figure 10 based on (11) and (12) may be inaccurate. By considering the whole pathway components, modeling is also a powerful tool for predicting the effects on flux and metabolite concentration of varying an enzyme activity (by overexpression or drug inhibition).
Model predictions to inhibit a pathway flux —
Kinetic modeling has been used to identify the flux controlling steps in Trypanosoma brucei glycolysis for drug targeting purposes. Interestingly, modeling has predicted controlling steps for the parasite pathway different from those described for glycolysis in human host cells [125, 126].
Entamoeba histolytica is the causal agent of human amebiasis. The parasite lacks functional mitochondria and has neither Krebs cycle nor OXPHOS enzyme activities. Therefore, substrate level phosphorylation by glycolysis is the only way to generate ATP for cellular work [141]. An important difference in amebal glycolysis in comparison to glycolysis in human cells is that it contains the pyrophosphate (PPi)-dependent enzymes phosphofructokinase (PPi-PFK) and pyruvate phosphate dikinase (PPDK), which replace the highly modulated ATP-PFK and PYK present in human cells. Moreover, both have been proposed as drug targets by using PPi analogues (bisphosphonates) [141].
We recently described the construction of a kinetic model of E. histolytica glycolysis to determine the control distribution of this energetically important pathway in the parasite [142]. The model was constructed using the Gepasi software and was based on the kinetic parameters determined in the purified recombinant enzymes [143], as well as the enzyme activities, fluxes, and metabolite concentrations found in the parasite. The results of the metabolic control analysis indicated that HK and PGAM are the main flux control steps of the pathway (73 and 65%, resp.) and perhaps GLUT. In contrast, the PPi-PFK and PPDK displayed low flux control (13 and 0.1%, resp.) because they have overcapacity over the glycolytic flux [142]. The amebal model allowed evaluating the effect on flux of “inhibiting” the pathway enzymes. The model predicted that in order to diminish by 50% the glycolytic flux (and the ATP concentration; data not shown), HK and PGAM should be inhibited by 24 and 55%, respectively, or both enzymes by 18% (Figure 11). In contrast, to attain the same reduction in flux by inhibiting PPi-PFK and PPDK, they should be decreased >70% (Figure 11). Therefore, the kinetic model results indicate that HK can be an appropriate drug target because its specific inhibition can compromise the energy levels in the parasite. They also indicate that although PPi-PFK and PPDK remain as promising drug targets because of their divergence from the human glycolytic enzymes, highly potent and very specific inhibitors should be designed for these enzymes in order to affect the parasite's energy metabolism.
Figure 11.
Modeled flux behavior when inhibiting pathway enzymes. The predicted flux when varying the enzyme activity was obtained using the kinetic model for Entamoeba histolytica glycolysis [142]. In this case, 100% enzyme activity is the enzyme activity present in amebal extracts, and 100% flux is the ethanol flux displayed by amoebae incubated with glucose. PPi-PFK, PPi-dependent phosphofructokinase; PPDK, pyruvate phosphate dikinase; PGAM, 2,3 bisphosphoglycerate independent 3-phosphoglycerate mutase.
5.5. In vitro reconstitution of metabolic pathways
Another experimental approach for determining the enzyme control coefficients is the in vitro reconstitution of segments of metabolic pathways. It is recalled that for determining the flux control coefficient exerted by a given step on a metabolic pathway the enzyme activity has to be varied, without altering the other components in the system, and the flux variations are to be measured (Figure 4). Such an experiment can be readily made if a pathway is reconstituted with purified enzymes. Some advantages of this approach are that the pathway structure is known, in which the components concentration may be manipulated and analyzed separately, and the enzyme effectors can be assayed. As the system composition is strictly controlled, the results may be highly reproducible. The main disadvantage is that the enzyme concentrations in the assays are diluted and thus the enzyme interactions are not favored. If this interaction is important for activity, the in vitro reconstitution may limit the extrapolation to the metabolic pathway inside the cell.
There are not many studies describing this type of experiments, most probably due to the fact that for applying MCA the pathway must be working under steady-state conditions. In a reconstituted system, only a quasi steady state may be reached because there is net substrate, and cofactors consumption, as well as product accumulation, since it is difficult to attain a constant substrate supply and release of products.
One of the first experimental reports on control coefficient determination in a reconstituted system was carried out for the upper glycolytic segment with the commercially available rabbit muscle HK, HPI, PFK-1, ALDO, and TPI [144]. Each enzyme was separately titrated and the flux variation to glycerol-3-phosphate (by coupling the reconstituted system to an excess of α-GPDH) was measured in the presence of CK to maintain the ATP concentration constant. The flux control coefficients were determined as described in Figure 4. The results showed that PFK-1 and HK exerted the main flux control (65% and 20%, resp.), whereas the remaining 15% resided in the other enzymes. These authors observed that the addition of F1,6BP, a PFK-1 activator slightly diminished the flux control exerted by PFK-1 and increases that of HK. The validation of the summation theorem was also demonstrated in this work [144].
The lower glycolytic segment has also been reconstituted with commercial enzymes for determining the flux control coefficients [145]. The results showed that flux was mainly controlled by PYK (60–100%), although under some conditions control was shared with PGAM; ENO did not contribute to the flux control.
Another important limitation of the reconstitution experiments is that the commercial availability of the purified enzymes from the same organism is restricted or inexistent. However, by using the information from the genome sequence projects and the recombinant DNA technology, it is now possible to access all the enzyme genes from a metabolic pathway in the same organism, thus facilitating their cloning, overexpression, and purification. With this strategy, we cloned, overexpressed, and purified the 10 glycolytic enzymes of Entamoeba histolytica [143] for studying the flux control distribution in this organism by using kinetic modeling [142] and pathway reconstitution.
The reconstitution experiments of the lower amebal glycolytic segment, under near physiological conditions of pH, temperature, and enzyme activity (Figure 12) showed that PGAM and, to a lesser extent, PPDK exert the main flux control (these amebal enzymes are genetically and kinetically different from their human counterparts) with ENO exhibiting negligible control [143]. In turn, reconstitution of the upper amebal glycolytic segment has revealed that HK and, to a much lesser extent HPI, PPi-PFK, and ALD, exerted the main flux control, with TPI having negligible control [146]. These results strongly correlate with the enzyme catalytic efficiencies previously reported [143], in which HK is highly sensitive to AMP inhibition, ALD, and PGAM have the lowest catalytic efficiencies among the glycolytic enzymes, leading to high flux control coefficients and thus becoming suitable candidates for therapeutic intervention. The reconstitution results also agree with the pathway modeling predictions previously analyzed (Section 5.4), in which HK and PGAM are two of the main controlling steps [142].
Figure 12.
Determination of flux control coefficients in an in vitro reconstitution of the final section of Entamoeba histolytica glycolysis. Enzymatic assay with the three recombinant enzymes from the ameba: EhPGAM, EhENO, and EhPPDK. LDH, commercial lactate dehydrogenase. The flux control coefficient was determined at the *marked position. 2PG, 2-phosphoglycerate; 3PG, 3-phosphoglycerate. Modified from [143].
The in vitro reconstitution experiments are also useful for studying the effect on control redistribution of an enzyme modulation that is particularly difficult to manage in vivo; the main controlling steps identified with the reconstitution experiments should be further analyzed with other experimental strategies such as elasticity analysis in the in vivo systems.
5.6. Genetic engineering to manipulate the in vivo protein levels
This experimental approach for determining the control coefficients could be part of the genetic approach analyzed in Section 5.1, but it was separated due to its recent methodological development and because it actually belongs to the molecular genetics rather than to the Mendelian genetics.
5.6.1. Repression of gene expression
This approach is based on the in vivo modulation of the enzyme levels using the RNA antisense technology. There are at least three strategies to inhibit gene expression: (a) the use of single stranded antisense oligonucleotides, which form a double stranded RNA that might be degraded by RNAse H; (b) target RNA degradation with catalytically active oligonucleotides, known as ribozymes that bind to their specific RNA; and (c) RNA degradation using siRNAs (21–23 nucleotides) [147].
The RNA antisense technology was applied for control coefficient determination of the ribulose-bisphosphate-carboxylase (Rubisco) that fixes CO2 in the plant Calvin cycle. This enzyme considered the rate-limiting step of the Calvin cycle and of the whole photosynthetic process, despite its high concentration (4 mM) in the chloroplasts stroma that compensates its low catalytic efficiency.
Attempts to make Rubisco a nonlimiting step, either by modifying its catalytic efficiency or by overexpressing it, have been unsuccessful. Stitt et al. [148] determined the CrubiscoJphotosynthesis of tobacco plants by decreasing its activity with DNA antisense. The plants were transformed with DNA antisense against the mRNA of the enzyme's small subunit, thus promoting its degradation. For Calvin cycle enzymes, the pleiotropic effects were minimal. The results showed that Rubisco may indeed be the photosynthesis limiting step with a CrubiscoJphotosynthesis = 0.69–0.83 when plants are exposed to high illumination (1050 μmol quanta m−2s−1), high humidity (85%), and low CO2 concentrations (25 Pa). However, this flux control decreases to 0.05–0.12 under moderate illumination or high CO2 levels [148]. Unfortunately, the authors did not determine the control coefficients of the other pathway enzymes or the branches fluxes which may be significant.
As described in Section 5.4, the results of the T. brucei glycolysis modeling indicated that GLUT was the main flux control step (CGLUTJ ∼ 50%), [125, 126]. This model predicted a large overcapacity for HK, PFK-1, ALDO, GAPDH, PGAM, ENO, and PYK over the glycolytic flux leading to low flux control coefficients [125, 126]. To validate the modeling results, the concentrations of HK, PFK-1, PGAM, ENO, and PYK were changed with siRNAs in growing parasites [149]. These knockdown expression experiments showed overcapacity of HK and PYK over the flux, although at lower levels than predicted by the model. A good correlation for PGAM and ENO was obtained between model predictions and experimental results. However, a large difference (9 folds) was obtained for PFK-1. This discrepancy is perhaps related to pleiotropic effects of PFK-1 downregulation, as these mutants also displayed diminution in the activities of other enzymes (HK, ENO, and PYK). The combination of these two approaches, in silico modeling and in vivo experimentation, is complementary: on one hand, modeling identifies the enzymes (out of 19 that contain the model) that display the highest flux control coefficients, whereas in vivo experimentation validates the accuracy of the model to establish predictions about the pathway's behavior.
5.6.2. Fine tuning of cellular protein expression
The knockdown experiments described above usually yield only two experimental points of the plot shown in Figure 4: the wild-type and the knockdown strain protein levels or enzyme activities. Thus, with such an approach high levels of inhibition (>80%) are mostly analyzed, whereas intermediate levels of downregulation (if obtained) are generally overlooked. Therefore, knockdown experiments are not very useful to obtain the complete set of experimental data (above and below the wild-type levels of enzyme activity with the corresponding flux) for determining reliable control coefficients.
A strategy to determine flux control coefficients from several protein levels has been developed by using adenovirus-mediated glucose-6-phosphatase (G6Pase) overexpression under the control of the cytomegalovirus promoter in rat hepatocytes. A 2-fold G6Pase overexpression did not alter CG6PaseGlycolysis or CGKGlycolysis (GK, glucokinase). However, if G6Pase is overexpressed by 4 folds, then CGKGlycogen-synthesis diminished from 2.8 to 1.8 and there was a 35% lowering in glycogen synthesis [150]. However, this approach allows titration of flux only above the basal enzyme activities found in the cell, but not below.
These experimental inconveniences have been circumvented by using inducible gene expression systems based in the lac, Lambda, nisin, GAL, tetracycline, and other inducible promoters, in bacteria and yeast [151, 152]. However, a problem frequently encountered with inducible promoters is that a steady-state of protein expression is difficult to attain [151, 152].
Recently, Jensen and Hammer described the design of synthetic promoter libraries (SPL), in particular for L. lactis metabolic optimization [153]. These promoters maintain constant the array of the known consensus sequences for L. lactis gene transcription (−10 and −35 boxes), while the nucleotide sequence between these boxes (a spacer sequence of 17 ± 1 bp) is randomized, thus producing a set of promoters with different transcriptional strength. These promoter libraries allow the transcription and protein expression several folds above and below the wild-type levels of enzyme activity [153], thus enhancing the usefulness of this approach for MCA studies.
The control distribution of glycolysis in E. coli and L. lactis, as discussed in Section 3.2 [17, 24, 27, 151], has been determined by using the SPL technology. SPL for yeast, mammalian and plant cells are also under development [151, 152]. Certainly, the advances in genetic engineering in combination with MCA allow better experimental designs for metabolic optimization of micro-organisms of biotechnological interest.
Concluding remarks —
The frequently recurred idea of manipulating the key enzyme or rate-limiting step (a concept based on a qualitative and rather intuitive background) to change metabolism is incorrect. As MCA has demonstrated, flux control is shared by multiple steps and it is not usually localized in only one step. MCA determines quantitatively the control that a given enzyme exerts on the flux and on intermediary concentration and helps to explain why an enzyme does or does not exert control.
A metabolic pathway is manipulated to change the rate of the end-product formation (i.e., the flux) or the concentration of a relevant intermediary. As it is demonstrated in many unsuccessful experiments, it is not enough to overexpress one enzyme (the rate-limiting step) or many arbitrarily selected sites of the pathway. MCA proposes an initial experimental analysis that determines the structural control of the pathway and identifies the sites (enzymes and transporters) with higher control coefficients values (i.e., targets to be manipulated). For example, if there is a system composed of six enzymes and three of them have flux control coefficients with values of 0.2 or higher and the other three with values of 0.1 or lower, the three enzymes with high control coefficients must be overexpressed (if a flux increase is desired) or repressed (if flux inhibition is the objective) and not only one of them. If one of the selected enzymes is strongly inhibited by its product or has allosteric inhibition, the overexpression of this enzyme might not be enough to increase the flux, as it may also be necessary to moderately vary the product and allosteric modulator consuming enzymes.
If the aim of the researcher is a metabolite concentration increase, which is not the end product of the pathway, MCA suggests the overexpression of those enzymes or transporters in the supply block with the highest control coefficients and/or the repression of those enzymes in the demand block with the highest control coefficients. These manipulations may become complicated if the metabolite of interest has allosteric interactions with enzymes and transporters (inhibition and activation) of both the supply and demand blocks. It is recalled that ethanol production in yeast and lactate and acetate production in lactobacteria do not increase by overexpressing PFK-1, an allosteric enzyme and the presumed rate-limiting step of glycolysis. In fact, the flux was diminished with an excessive PFK-1 overexpression. However, the analysis of these results reveals that the F1,6BP concentration is indeed increased many times over the control level. Another strategy for eliminating the feedback inhibition might be the introduction of mutations on the enzymes that are closer to the metabolite of interest.
ACKNOWLEDGMENT
The present work was partially supported by CONACyT-Mexico Grants no. 60517 to RMS and 46719-Q to ES.
ABBREVIATIONS
- ADH:
alcohol dehydrogenase
- CK:
creatine kinase
- ENO:
enolase
- GAPDH:
glyceraldehyde-3 phosphate dehydrogenase
- HPI:
hexose phosphate isomerase
- LDH:
lactate dehydrogenase
- PDC:
pyruvate decarboxylase
- PGK:
phosphoglucokinase
- PGAM:
phosphoglycerate mutase
- TPI:
triose phosphate isomerase
- PPi-PFK:
pyrophosphate-dependent phosphofructokinase
- α-GPDH:
α-glycerophosphate dehydrogenase
- F6B:
fructose-6-phosphate
- F1,6BP:
fructose-1,6-bisphosphate
- G1P:
glucose-1-phosphate
- G6P:
glucose-6-phosphate
- GSH:
reduced glutathione
- γ-EC:
γ-glutamylcysteine
- MCA:
metabolic control analysis
- siRNA:
small interfering RNA.
References
- 1.Newsholme EA, Start C. Regulation of Metabolism. London, UK: John Wiley & Sons; 1973. [Google Scholar]
- 2.Fell D. Understanding the Control of Metabolism. London, UK: Portland Press; 1997. [Google Scholar]
- 3.David M, Rasched IR, Sund H. Studies of glutamate dehydrogenase. Methionine-169: the preferentially carboxymethylated residue. European Journal of Biochemistry. 1977;74(2):379–385. doi: 10.1111/j.1432-1033.1977.tb11402.x. [DOI] [PubMed] [Google Scholar]
- 4.Teng H, Segura E, Grubmeyer C. Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis. Novel chemistry required for enzymatic aldehyde oxidation. Journal of Biological Chemistry. 1993;268(19):14182–14188. [PubMed] [Google Scholar]
- 5.Wells TNC, Payton MA, Proudfoot AE. Inhibition of phosphomannose isomerase by mercury ions. Biochemistry. 1994;33(24):7641–7646. doi: 10.1021/bi00190a018. [DOI] [PubMed] [Google Scholar]
- 6.Poolman B, Bosman B, Kiers J, Konings WN. Control of glycolysis by glyceraldehyde-3-phosphate dehydrogenase in Streptococcus cremoris and Streptococcus lactis . Journal of Bacteriology. 1987;169(12):5887–5890. doi: 10.1128/jb.169.12.5887-5890.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno-Sánchez R, Torres-Márquez ME. Control of oxidative phosphorylation in mitochondria, cells and tissues. International Journal of Biochemistry. 1991;23(11):1163–1174. doi: 10.1016/0020-711x(91)90212-6. [DOI] [PubMed] [Google Scholar]
- 8.Webster KA. Evolution of the coordinate regulation of glycolytic enzyme genes by hypoxia. Journal of Experimental Biology. 2003;206(17):2911–2922. doi: 10.1242/jeb.00516. [DOI] [PubMed] [Google Scholar]
- 9.Heinisch J. Isolation and characterization of the two structural genes coding for phosphofructokinase in yeast. Molecular and General Genetics. 1986;202(1):75–82. doi: 10.1007/BF00330520. [DOI] [PubMed] [Google Scholar]
- 10.Davies SEC, Brindle KM. Effects of overexpression of phosphofructokinase on glycolysis in the yeast Saccharomyces cerevisiae . Biochemistry. 1992;31(19):4729–4735. doi: 10.1021/bi00134a028. [DOI] [PubMed] [Google Scholar]
- 11.Hauf J, Zimmermann FK, Müller S. Simultaneous genomic overexpression of seven glycolytic enzymes in the yeast Saccharomyces cerevisiae . Enzyme and Microbial Technology. 2000;26(9-10):688–698. doi: 10.1016/s0141-0229(00)00160-5. [DOI] [PubMed] [Google Scholar]
- 12.Schaaff I, Heinisch J, Zimmermann FK. Overproduction of glycolytic enzymes in yeast. Yeast. 1989;5(4):285–290. doi: 10.1002/yea.320050408. [DOI] [PubMed] [Google Scholar]
- 13.Hornberg JJ, Binder B, Bruggeman FJ, Schoeberl B, Heinrich R, Westerhoff HV. Control of MAPK signalling: from complexity to what really matters. Oncogene. 2005;24(36):5533–5542. doi: 10.1038/sj.onc.1208817. [DOI] [PubMed] [Google Scholar]
- 14.Smits HP, Hauf J, Müller S, et al. Simultaneous overexpression of enzymes of the lower part of glycolysis can enhance the fermentative capacity of Saccharomyces cerevisiae . Yeast. 2000;16(14):1325–1334. doi: 10.1002/1097-0061(200010)16:14<1325::AID-YEA627>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15.Emmerling M, Bailey JE, Sauer U. Glucose catabolism of Escherichia coli strains with increased activity and altered regulation of key glycolytic enzymes. Metabolic Engineering. 1999;1(2):117–127. doi: 10.1006/mben.1998.0109. [DOI] [PubMed] [Google Scholar]
- 16.Emmerling M, Bailey JE, Sauer U. Altered regulation of pyruvate kinase or co-overexpression of phosphofructokinase increases glycolytic fluxes in resting Escherichia coli . Biotechnology and Bioengineering. 2000;67(5):623–627. doi: 10.1002/(sici)1097-0290(20000305)67:5<623::aid-bit13>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 17.Solem C, Koebmann BJ, Jensen PR. Glyceraldehyde-3-phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. Journal of Bacteriology. 2003;185(5):1564–1571. doi: 10.1128/JB.185.5.1564-1571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruijter GJG, Panneman H, Visser J. Overexpression of phosphofructokinase and pyruvate kinase in citric acid-producing Aspergillus niger . Biochimica et Biophysica Acta. 1997;1334(2-3):317–326. doi: 10.1016/s0304-4165(96)00110-9. [DOI] [PubMed] [Google Scholar]
- 19.Urbano AM, Gillham H, Groner Y, Brindle KM. Effects of overexpression of the liver subunit of 6-phosphofructo-1-kinase on the metabolism of a cultured mammalian cell line. Biochemical Journal. 2000;352, part 3:921–927. [PMC free article] [PubMed] [Google Scholar]
- 20.Bonini BM, Van Dijck P, Thevelein JM. Uncoupling of the glucose growth defect and the deregulation of glycolysis in Saccharomyces cerevisiae tps1 mutants expressing trehalose-6-phosphate-insensitive hexokinase from Schizosaccharomyces pombe . Biochimica et Biophysica Acta. 2003;1606(1–3):83–93. doi: 10.1016/s0005-2728(03)00086-0. [DOI] [PubMed] [Google Scholar]
- 21.Rivoal J, Hanson AD. Metabolic control of anaerobic glycolysis. Overexpression of lactate dehydrogenase in transgenic tomato roots supports the Davies-Roberts hypothesis and points to a critical role for lactate secretion. Plant Physiology. 1994;106(3):1179–1185. doi: 10.1104/pp.106.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas S, Mooney PJF, Burrell MM, Fell DA. Metabolic control analysis of glycolysis in tuber tissue of potato (Solanum tuberosum): explanation for the low control coefficient of phosphofructokinase over respiratory flux. Biochemical Journal. 1997;322, part 1:119–127. doi: 10.1042/bj3220119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Doherty RM, Lehman DL, Seoane J, Gómez-Foix AM, Guinovart JJ, Newgard CB. Differential metabolic effects of adenovirus-mediated glucokinase and hexokinase I overexpression in rat primary hepatocytes. Journal of Biological Chemistry. 1996;271(34):20524–20530. doi: 10.1074/jbc.271.34.20524. [DOI] [PubMed] [Google Scholar]
- 24.Andersen HW, Pedersen MB, Hammer K, Jensen PR. Lactate dehydrogenase has no control on lactate production but has a strong negative control on formate production in Lactococcus lactis . European Journal of Biochemistry. 2001;268(24):6379–6389. doi: 10.1046/j.0014-2956.2001.02599.x. [DOI] [PubMed] [Google Scholar]
- 25.Koebmann BJ, Solem C, Jensen PR. Control analysis as a tool to understand the formation of the las operon in Lactococcus lactis . FEBS Journal. 2005;272(9):2292–2303. doi: 10.1111/j.1742-4658.2005.04656.x. [DOI] [PubMed] [Google Scholar]
- 26.Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. Journal of Bacteriology. 2002;184(14):3909–3916. doi: 10.1128/JB.184.14.3909-3916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koebmann BJ, Andersen HW, Solem C, Jensen PR. Experimental determination of control of glycolysis in Lactococcus lactis . Antonie van Leeuwenhoek. 2002;82(1–4):237–248. [PubMed] [Google Scholar]
- 28.Koebmann BJ, Solem C, Pedersen MB, Nilsson D, Jensen PR. Expression of genes encoding F1-ATPase results in uncoupling of glycolysis from biomass production in Lactococcus lactis . Applied and Environmental Microbiology. 2002;68(9):4274–4282. doi: 10.1128/AEM.68.9.4274-4282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas S, Fell DA. A control analysis exploration of the role of ATP utilisation in glycolytic-flux control and glycolytic-metabolite-concentration regulation. European Journal of Biochemistry. 1998;258(3):956–967. doi: 10.1046/j.1432-1327.1998.2580956.x. [DOI] [PubMed] [Google Scholar]
- 30.Hofmeyr J-HS, Cornish-Bowden A. Regulating the cellular economy of supply and demand. FEBS Letters. 2000;476(1-2):47–51. doi: 10.1016/s0014-5793(00)01668-9. [DOI] [PubMed] [Google Scholar]
- 31.Mendoza-Cózatl D, Loza-Tavera H, Hernández-Navarro A, Moreno-Sánchez R. Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants. FEMS Microbiology Reviews. 2005;29(4):653–671. doi: 10.1016/j.femsre.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Cornish-Bowden A, Cárdenas ML. Information transfer in metabolic pathways. Effects of irreversible steps in computer models. European Journal of Biochemistry. 2001;268(24):6616–6624. doi: 10.1046/j.0014-2956.2001.02616.x. [DOI] [PubMed] [Google Scholar]
- 33.Meister A. Glutathione metabolism. Methods in Enzymology. 1995;251:3–7. doi: 10.1016/0076-6879(95)51106-7. [DOI] [PubMed] [Google Scholar]
- 34.Noctor G, Arisi A-CM, Jouanin L, Foyer CH. Manipulation of glutathione and amino acid biosynthesis in the chloroplast. Plant Physiology. 1998;118(2):471–482. doi: 10.1104/pp.118.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu YL, Pilon-Smits EAH, Tarun AS, Weber SU, Jouanin L, Terry N. Cadmium tolerance and accumulation in Indian mustard is enhanced by overexpressing γ-glutamylcysteine synthetase. Plant Physiology. 1999;121(4):1169–1177. doi: 10.1104/pp.121.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Moon JS, Ko T-S, Petros D, Goldsbrough PB, Korban SS. Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiology. 2003;131(2):656–663. doi: 10.1104/pp.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilon-Smits EAH, Hwang S, Mel Lytle C, et al. Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction, and tolerance. Plant Physiology. 1999;119(1):123–132. doi: 10.1104/pp.119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatzfeld Y, Cathala N, Grignon C, Davidian J-C. Effect of ATP sulfurylase overexpression in bright yellow 2 tobacco cells: regulation of ATP sulfurylase and So42− transport activities. Plant Physiology. 1998;116(4):1307–1313. doi: 10.1104/pp.116.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito K, Kurosawa M, Tatsuguchi K, Takagi Y, Murakoshi I. Modulation of cysteine biosynthesis in chloroplasts of transgenic tobacco overexpressing cysteine synthase [O-acetylserine(thiol)-lyase] Plant Physiology. 1994;106(3):887–895. doi: 10.1104/pp.106.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harms K, von Ballmoos P, Brunold C, Höfgen R, Hesse H. Expression of a bacterial serine acetyltransferase in transgenic potato plants leads to increased levels of cysteine and glutathione. The Plant Journal. 2000;22(4):335–343. doi: 10.1046/j.1365-313x.2000.00743.x. [DOI] [PubMed] [Google Scholar]
- 41.Inoue Y, Sugiyama K-I, Izawa S, Kimura A. Molecular identification of glutathione synthetase (GSH2) gene from Saccharomyces cerevisiae . Biochimica et Biophysica Acta. 1998;1395(3):315–320. doi: 10.1016/s0167-4781(97)00199-1. [DOI] [PubMed] [Google Scholar]
- 42.Kim S-J, Shin YH, Kim K, Park E-H, Sa J-H, Lim C-J. Regulation of the gene encoding glutathione synthetase from the fission yeast. Journal of Biochemistry and Molecular Biology. 2003;36(3):326–331. doi: 10.5483/bmbrep.2003.36.3.326. [DOI] [PubMed] [Google Scholar]
- 43.Zhu YL, Pilon-Smits EAH, Jouanin L, Terry N. Overexpression of glutathione synthetase in Indian mustard enhances cadmium accumulation and tolerance. Plant Physiology. 1999;119(1):73–79. doi: 10.1104/pp.119.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Creissen G, Firmin J, Fryer M, et al. Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. The Plant Cell. 1999;11(7):1277–1291. doi: 10.1105/tpc.11.7.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant CM, MacIver FH, Dawes IW. Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide γ-glutamylcysteine. Molecular Biology of the Cell. 1997;8(9):1699–1707. doi: 10.1091/mbc.8.9.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortiz DF, Ruscitti T, McCue KF, Ow DW. Transport of metal-binding peptides by HMT1, a fission yeast ABC-type vacuolar membrane protein. Journal of Biological Chemistry. 1995;270(9):4721–4728. doi: 10.1074/jbc.270.9.4721. [DOI] [PubMed] [Google Scholar]
- 47.Niederberger P, Prasad R, Miozzari G, Kacser H. A strategy for increasing an in vivo flux by genetic manipulations: the tryptophan system of yeast. Biochemical Journal. 1992;287, part 2:473–479. doi: 10.1042/bj2870473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katsumata R, Ikeda M. Hyperproduction of tryptophan in Corynebacterium glutamicum by pathway engineering. Nature Biotechnology. 1993;11(8):921–925. [Google Scholar]
- 49.Morbach S, Sahm H, Eggeling L. Use of feedback-resistant threonine dehydratases of Corynebacterium glutamicum to increase carbon flux towards L-isoleucine. Applied and Environmental Microbiology. 1995;61(12):4315–4320. doi: 10.1128/aem.61.12.4315-4320.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koffas MAG, Jung GY, Aon JC, Stephanopoulos G. Effect of pyruvate carboxylase overexpression on the physiology of Corynebacterium glutamicum . Applied and Environmental Microbiology. 2002;68(11):5422–5428. doi: 10.1128/AEM.68.11.5422-5428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Padilla L, Krämer R, Stephanopoulos G, Agosin E. Overproduction of trehalose: heterologous expression of Escherichia coli trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in Corynebacterium glutamicum . Applied and Environmental Microbiology. 2004;70(1):370–376. doi: 10.1128/AEM.70.1.370-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radmacher E, Vaitsikova A, Burger U, Krumbach K, Sahm H, Eggeling L. Linking central metabolism with increased pathway flux: L-valine accumulation by Corynebacterium glutamicum . Applied and Environmental Microbiology. 2002;68(5):2246–2250. doi: 10.1128/AEM.68.5.2246-2250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simic P, Willuhn J, Sahm H, Eggeling L. Identification of glyA (encoding serine hydroxymethyltransferase) and its use together with the exporter ThrE to increase L-threonine accumulation by Corynebacterium glutamicum . Applied and Environmental Microbiology. 2002;68(7):3321–3327. doi: 10.1128/AEM.68.7.3321-3327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wisselink HW, Moers APHA, Mars AE, Hoefnagel MHN, de Vos WM, Hugenholtz J. Overproduction of heterologous mannitol 1-phosphatase: a key factor for engineering mannitol production by Lactococcus lactis . Applied and Environmental Microbiology. 2005;71(3):1507–1514. doi: 10.1128/AEM.71.3.1507-1514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ladero V, Ramos A, Wiersma A, et al. High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering. Applied and Environmental Microbiology. 2007;73(6):1864–1872. doi: 10.1128/AEM.02304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solem C, Koebmann BJ, Yang F, Jensen PR. The las enzymes control pyruvate metabolism in Lactococcus lactis during growth on maltose. Journal of Bacteriology. 2007;189(18):6727–6730. doi: 10.1128/JB.00902-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, Saavedra E. Energy metabolism in tumor cells. FEBS Journal. 2007;274(6):1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 58.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84(6):1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 59.Discher DJ, Bishopric NH, Wu X, Peterson CA, Webster KA. Hypoxia regulates β-enolase and pyruvate kinase-M promoters by modulating Sp1/Sp3 binding to a conserved GC element. Journal of Biological Chemistry. 1998;273(40):26087–26093. doi: 10.1074/jbc.273.40.26087. [DOI] [PubMed] [Google Scholar]
- 60.Egea M, Metón I, Baanante IV. Sp1 and Sp3 regulate glucokinase gene transcription in the liver of gilthead sea bream (Sparus aurata) Journal of Molecular Endocrinology. 2007;38(3-4):481–492. doi: 10.1677/jme.1.02176. [DOI] [PubMed] [Google Scholar]
- 61.Murphy BJ, Andrews GK, Bittel D, et al. Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Research. 1999;59(6):1315–1322. [PubMed] [Google Scholar]
- 62.Riddle SR, Ahmad A, Ahmad S, et al. Hypoxia induces hexokinase II gene expression in human lung cell line A549. American Journal of Physiology. 2000;278(2):L407–L416. doi: 10.1152/ajplung.2000.278.2.L407. [DOI] [PubMed] [Google Scholar]
- 63.Parolin ML, Spriet LL, Hultman E, Hollidge-Horvat MG, Jones NL, Heigenhauser GJF. Regulation of glycogen phosphorylase and PDH during exercise in human skeletal muscle during hypoxia. American Journal of Physiology. 2000;278(3):E522–E534. doi: 10.1152/ajpendo.2000.278.3.E522. [DOI] [PubMed] [Google Scholar]
- 64.Michels PAM, Bringaud F, Herman M, Hannaert V. Metabolic functions of glycosomes in trypanosomatids. Biochimica et Biophysica Acta. 2006;1763(12):1463–1477. doi: 10.1016/j.bbamcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 65.Opperdoes FR, Michels PAM. Enzymes of carbohydrate metabolism as potential drug targets. International Journal for Parasitology. 2001;31(5-6):481–489. doi: 10.1016/s0020-7519(01)00155-2. [DOI] [PubMed] [Google Scholar]
- 66.Azema L, Claustre S, Alric I, et al. Interaction of substituted hexose analogues with the Trypanosoma brucei hexose transporter. Biochemical Pharmacology. 2004;67(3):459–467. doi: 10.1016/j.bcp.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 67.Lakhdar-Ghazal F, Blonski C, Willson M, Michels P, Perie J. Glycolysis and proteases as targets for the design of new anti-trypanosome drugs. Current Topics in Medicinal Chemistry. 2002;2(5):439–456. doi: 10.2174/1568026024607472. [DOI] [PubMed] [Google Scholar]
- 68.Bakker BM, Westerhoff HV, Opperdoes FR, Michels PAM. Metabolic control analysis of glycolysis in trypanosomes as an approach to improve selectivity and effectiveness of drugs. Molecular and Biochemical Parasitology. 2000;106(1):1–10. doi: 10.1016/s0166-6851(99)00197-8. [DOI] [PubMed] [Google Scholar]
- 69.Fairlamb AH, Cerami A. Metabolism and functions of trypanothione in the Kinetoplastida. Annual Review of Microbiology. 1992;46:695–729. doi: 10.1146/annurev.mi.46.100192.003403. [DOI] [PubMed] [Google Scholar]
- 70.Müller S, Liebau E, Walter RD, Krauth-Siegel RL. Thiol-based redox metabolism of protozoan parasites. Trends in Parasitology. 2003;19(7):320–328. doi: 10.1016/s1471-4922(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 71.Le Quesne SA, Fairlamb AH. Regulation of a high-affinity diamine transport system in Trypanosoma cruzi epimastigotes. Biochemical Journal. 1996;316, part 2:481–486. doi: 10.1042/bj3160481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dumas C, Ouellette M, Tovar J, et al. Disruption of the trypanothione reductase gene of Leishmania decreases its ability to survive oxidative stress in macrophages. The EMBO Journal. 1997;16(10):2590–2598. doi: 10.1093/emboj/16.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tovar J, Cunningham ML, Smith AC, Croft SL, Fairlamb AH. Down-regulation of Leishmania donovani trypanothione reductase by heterologous expression of a trans-dominant mutant homologue: effect on parasite intracellular survival. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(9):5311–5316. doi: 10.1073/pnas.95.9.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tovar J, Wilkinson S, Mottram JC, Fairlamb AH. Evidence that trypanothione reductase is an essential enzyme in Leishmania by targeted replacement of the tryA gene locus. Molecular Microbiology. 1998;29(2):653–660. doi: 10.1046/j.1365-2958.1998.00968.x. [DOI] [PubMed] [Google Scholar]
- 75.Krieger S, Schwarz W, Arlyanayagam MR, Fairlamb AH, Krauth-Siegel RL, Clayton C. Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Molecular Microbiology. 2000;35(3):542–552. doi: 10.1046/j.1365-2958.2000.01721.x. [DOI] [PubMed] [Google Scholar]
- 76.Kelly JM, Taylor MC, Smith K, Hunter KJ, Fairlamb AH. Phenotype of recombinant Leishmania donovani and Trypanosoma cruzi which over-express trypanothione reductase. Sensitivity towards agents that are thought to induce oxidative stress. European Journal of Biochemistry. 1993;218(1):29–37. doi: 10.1111/j.1432-1033.1993.tb18348.x. [DOI] [PubMed] [Google Scholar]
- 77.Comini MA, Guerrero SA, Haile S, Menge U, Lünsdorf H, Flohé L. Validation of Trypanosoma brucei trypanothione synthetase as drug target. Free Radical Biology and Medicine. 2004;36(10):1289–1302. doi: 10.1016/j.freeradbiomed.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 78.Ariyanayagam MR, Oza SL, Guther ML, Fairlamb AH. Phenotypic analysis of trypanothione synthetase knockdown in the African trypanosome. Biochemical Journal. 2005;391, part 2:425–432. doi: 10.1042/BJ20050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huynh TT, Huynh VT, Harmon MA, Phillips MA. Gene knockdown of γ-glutamylcysteine synthetase by RNAi in the parasitic protozoa Trypanosoma brucei demonstrates that it is an essential enzyme. Journal of Biological Chemistry. 2003;278(41):39794–39800. doi: 10.1074/jbc.M306306200. [DOI] [PubMed] [Google Scholar]
- 80.Guimond C, Trudel N, Brochu C, et al. Modulation of gene expression in Leishmania drug resistant mutants as determined by targeted DNA microarrays. Nucleic Acids Research. 2003;31(20):5886–5896. doi: 10.1093/nar/gkg806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shahi SK, Krauth-Siegel RL, Clayton CE. Overexpression of the putative thiol conjugate transporter TbMRPA causes melarsoprol resistance in Trypanosoma brucei . Molecular Microbiology. 2002;43(5):1129–1138. doi: 10.1046/j.1365-2958.2002.02831.x. [DOI] [PubMed] [Google Scholar]
- 82.Kacser H, Burns JA. The control of flux. Symposia of the Society for Experimental Biology. 1973;27:65–104. [PubMed] [Google Scholar]
- 83.Kacser H, Burns JA. Molecular democracy: who shares the controls? Biochemical Society Transactions. 1979;7(5):1149–1160. doi: 10.1042/bst0071149. [DOI] [PubMed] [Google Scholar]
- 84.Heinrich R, Rapoport SM, Rapoport TA. Metabolic regulation and mathematical models. Progress in Biophysics and Molecular Biology. 1978;32:1–82. [PubMed] [Google Scholar]
- 85.Rapoport TA, Heinrich R, Jacobasch G, Rapoport S. A linear steady state treatment of enzymatic chains. A mathematical model of glycolysis of human erythrocytes. European Journal of Biochemistry. 1974;42(1):107–120. doi: 10.1111/j.1432-1033.1974.tb03320.x. [DOI] [PubMed] [Google Scholar]
- 86.Flint HJ, Tateson RW, Barthelmess IB, Porteous DJ, Donachie WD, Kacser H. Control of the flux in the arginine pathway of Neurospora crassa. Modulations of enzyme activity and concentration. Biochemical Journal. 1981;200(2):231–246. doi: 10.1042/bj2000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cohen NS, Cheung C-W, Sijuwade E, Raijman L. Kinetic properties of carbamoyl-phosphate synthase (ammonia) and ornithine carbamoyltransferase in permeabilized mitochondria. Biochemical Journal. 1992;282, part 1:173–180. doi: 10.1042/bj2820173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Powers-Lee SG, Mastico RA, Bendayan M. The interaction of rat liver carbamoyl phosphate synthetase and ornithine transcarbamoylase with inner mitochondrial membranes. Journal of Biological Chemistry. 1987;262(32):15683–15688. [PubMed] [Google Scholar]
- 89.Middleton RJ, Kacser H. Enzyme variation, metabolic flux and fitness: alcohol dehydrogenase in Drosophila melanogaster . Genetics. 1983;105(3):633–650. doi: 10.1093/genetics/105.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rodríguez-Enríquez S, Torres-Márquez ME, Moreno-Sánchez R. Substrate oxidation and ATP sypply in AS-30D hepatoma cells. Archives of Biochemistry and Biophysics. 2000;375(1):21–30. doi: 10.1006/abbi.1999.1582. [DOI] [PubMed] [Google Scholar]
- 91.Garcia C, Pardo JP, Moreno-Sánchez R. Control of oxidative phosphorylation supported by NAD-linked substrates in rat brain mitochondria. Biochemical Archives. 1996;12(3):157–176. [Google Scholar]
- 92.Gellerich FN, Kunz WS, Bohnensack R. Estimation of flux control coefficients from inhibitor titrations by non-linear regression. FEBS Letters. 1990;274(1-2):167–170. doi: 10.1016/0014-5793(90)81355-r. [DOI] [PubMed] [Google Scholar]
- 93.Moreno-Sánchez R, Devars S, López-Gómez F, Uribe A, Corona N. Distribution of control of oxidative phosphorylation in mitochondria oxidizing NAD-linked substrates. Biochimica et Biophysica Acta. 1991;1060(3):284–292. doi: 10.1016/s0005-2728(05)80318-4. [DOI] [PubMed] [Google Scholar]
- 94.Moreno-Sánchez R. Contribution of the translocator of adenine nucleotides and the ATP synthase to the control of oxidative phosphorylation and arsenylation in liver mitochondria. Journal of Biological Chemistry. 1985;260(23):12554–12560. [PubMed] [Google Scholar]
- 95.Rossignol R, Letellier T, Malgat M, Rocher C, Mazat J-P. Tissue variation in the control of oxidative phosphorylation: implication for mitochondrial diseases. Biochemical Journal. 2000;347, part 1:45–53. [PMC free article] [PubMed] [Google Scholar]
- 96.López-Gómez FJ, Torres-Márquez ME, Moreno-Sánchez R. Control of oxidative phosphorylation in AS-30D hepatoma mitochondria. International Journal of Biochemistry. 1993;25(3):373–377. doi: 10.1016/0020-711x(93)90627-q. [DOI] [PubMed] [Google Scholar]
- 97.Wisniewski E, Kunz WS, Gellerich FN. Phosphate affects the distribution of flux control among the enzymes of oxidative phosphorylation in rat skeletal muscle mitochondria. Journal of Biological Chemistry. 1993;268(13):9343–9346. [PubMed] [Google Scholar]
- 98.Latsis T, Andersen B, Agius L. Diverse effects of two allosteric inhibitors on the phosphorylation state of glycogen phosphorylase in hepatocytes. Biochemical Journal. 2002;368, part 1:309–316. doi: 10.1042/BJ20021070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gupte SA, Arshad M, Viola S, et al. Pentose phosphate pathway coordinates multiple redox-controlled relaxing mechanisms in bovine coronary arteries. American Journal of Physiology. 2003;285(6):H2316–H2326. doi: 10.1152/ajpheart.00229.2003. [DOI] [PubMed] [Google Scholar]
- 100.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 101.Zu XL, Guppy M. Cancer metabolismml: facts, fantasy, and fiction. Biochemical and Biophysical Research Communications. 2004;313(3):459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 102.Erecinska M, Dagani F. Relationships between the neuronal sodium/potassium pump and energy metabolism. Effects of K+, Na+, and adenosine triphosphate in isolated brain synaptosomes. Journal of General Physiology. 1990;95(4):591–616. doi: 10.1085/jgp.95.4.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brown GC, Lakin-Thomas PL, Brand MD. Control of respiration and oxidative phosphorylation in isolated rat liver cells. European Journal of Biochemistry. 1990;192(2):355–362. doi: 10.1111/j.1432-1033.1990.tb19234.x. [DOI] [PubMed] [Google Scholar]
- 104.Hafner RP, Brown GC, Brand MD. Analysis of the control of respiration rate, phosphorylation rate, proton leak rate and protonmotive force in isolated mitochondria using the ‘top-down’ approach of metabolic control theory. European Journal of Biochemistry. 1990;188(2):313–319. doi: 10.1111/j.1432-1033.1990.tb15405.x. [DOI] [PubMed] [Google Scholar]
- 105.Wanders RJ, Groen AK, van Roermund CW, Tager JM. Factors determining the relative contribution of the adenine-nucleotide translocator and the ADP-regenerating system to the control of oxidative phosphorylation in isolated rat-liver mitochondria. European Journal of Biochemistry. 1984;142(2):417–424. doi: 10.1111/j.1432-1033.1984.tb08303.x. [DOI] [PubMed] [Google Scholar]
- 106.Groen AK, van Roermund CWT, Vervoorn RC, Tager JM. Control of gluconeogenesis in rat liver cells. Flux control coefficients of the enzymes in the gluconeogenic pathway in the absence and presence of glucagon. Biochemical Journal. 1986;237, part 2:379–389. doi: 10.1042/bj2370379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marín-Hernández A, Rodríguez-Enríquez S, Vital-González PA, et al. Determining and understanding the control of glycolysis in fast-growth tumor cells: flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS Journal. 2006;273(9):1975–1988. doi: 10.1111/j.1742-4658.2006.05214.x. [DOI] [PubMed] [Google Scholar]
- 108.Kruckeberg AL, Neuhaus HE, Feil R, Gottlieb LD, Stitt M. Decreased-activity mutants of phosphoglucose isomerase in the cytosol and chloroplast of Clarkia xantiana. Impact on mass-action ratios and fluxes to sucrose and starch, and estimation of flux control coefficients and elasticity coefficients. Biochemical Journal. 1989;261, part 2:457–467. doi: 10.1042/bj2610457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Quant PA, Robin D, Robin P, Girard J, Brand MD. A top-down control analysis in isolated rat liver mitochondria: can the 3-hydroxy-3-methylglutaryl-CoA pathway be rate-controlling for ketogenesis? Biochimica et Biophysica Acta. 1993;1156(2):135–143. doi: 10.1016/0304-4165(93)90128-u. [DOI] [PubMed] [Google Scholar]
- 110.Fell DA, Snell K. Control analysis of mammalian serine biosynthesis. Feedback inhibition of the final step. Biochemical Journal. 1988;256, part 1:97–101. doi: 10.1042/bj2560097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chassagnole C, Fell DA, Rais B, Kudla B, Mazat J-P. Control of the threonine-synthesis pathway in Escherichia coli: a theoretical and experimental approach. Biochemical Journal. 2001;356, part 2:433–444. doi: 10.1042/0264-6021:3560433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Galazzo JL, Bailey JE. Fermentation pathway kinetics and metabolic flux control in suspended and immobilized Saccharomyces cerevisiae . Enzyme and Microbial Technology. 1990;12(3):162–172. [Google Scholar]
- 113.Diderich JA, Teusink B, Valkier J, et al. Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast Saccharomyces bayanus . Microbiology. 1999;145(12):3447–3454. doi: 10.1099/00221287-145-12-3447. [DOI] [PubMed] [Google Scholar]
- 114.Westerhoff HV, Kahn D. Control involving metabolism and gene expression: the square-matrix method for modular decomposition. Acta Biotheoretica. 1993;41(1-2):75–83. doi: 10.1007/BF00712776. [DOI] [PubMed] [Google Scholar]
- 115.Chase JR, Rothman DL, Shulman RG. Flux control in the rat gastrocnemius glycogen synthesis pathway by in vivo 13C/31P NMR spectroscopy. American Journal of Physiology. 2001;280(4):E598–E607. doi: 10.1152/ajpendo.2001.280.4.E598. [DOI] [PubMed] [Google Scholar]
- 116.De Atauri P, Orrell D, Ramsey S, Bolouri H. Is the regulation of galactose 1-phosphate tuned against gene expression noise? Biochemical Journal. 2005;387, part 1:77–84. doi: 10.1042/BJ20041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Poolman MG, Assmus HE, Fell DA. Applications of metabolic modelling to plant metabolism. Journal of Experimental Botany. 2004;55(400):1177–1186. doi: 10.1093/jxb/erh090. [DOI] [PubMed] [Google Scholar]
- 118.Mendes P. GEPASI: a software package for modelling the dynamics, steady states and control of biochemical and other systems. Computer Applications in the Biosciences. 1993;9(5):563–571. doi: 10.1093/bioinformatics/9.5.563. [DOI] [PubMed] [Google Scholar]
- 119.Cornish-Bowden A, Hofmeyr J-HS. MetaModel: a program for modelling and control analysis of metabolic pathways on the IBM PC and compatibles. Computer Applications in the Biosciences. 1991;7(1):89–93. doi: 10.1093/bioinformatics/7.1.89. [DOI] [PubMed] [Google Scholar]
- 120.Sauro HM. SCAMP: a general-purpose simulator and metabolic control analysis program. Computer Applications in the Biosciences. 1993;9(4):441–450. doi: 10.1093/bioinformatics/9.4.441. [DOI] [PubMed] [Google Scholar]
- 121.Sauro HM. JARNAC: a system for interactive metabolic analysis. In: Hofmeyr J-HS, Rohwer JM, Snoep JL, editors. Animating the Cellular Map. Stellenbosch, South Africa: Stellenbosch University Press; 2000. pp. 221–228. [Google Scholar]
- 122.Olivier BG, Rohwer JM, Hofmeyr J-HS. Modelling cellular systems with PySCeS. Bioinformatics. 2005;21(4):560–561. doi: 10.1093/bioinformatics/bti046. [DOI] [PubMed] [Google Scholar]
- 123.Teusink B, Passarge J, Reijenga CA, et al. Can yeast glycolysis be understood terms of vitro kinetics of the constituent enzymes? Testing biochemistry. European Journal of Biochemistry. 2000;267(17):5313–5329. doi: 10.1046/j.1432-1327.2000.01527.x. [DOI] [PubMed] [Google Scholar]
- 124.Pritchard L, Kell DB. Schemes of flux control in a model of Saccharomyces cerevisiae glycolysis. European Journal of Biochemistry. 2002;269(16):3894–3904. doi: 10.1046/j.1432-1033.2002.03055.x. [DOI] [PubMed] [Google Scholar]
- 125.Bakker BM, Michels PAM, Opperdoes FR, Westerhoff HV. Glycolysis in bloodstream form Trypanosoma brucei can be understood in terms of the kinetics of the glycolytic enzymes. Journal of Biological Chemistry. 1997;272(6):3207–3215. doi: 10.1074/jbc.272.6.3207. [DOI] [PubMed] [Google Scholar]
- 126.Bakker BM, Michels PAM, Opperdoes FR, Westerhoff HV. What controls glycolysis in bloodstream form Trypanosoma brucei? Journal of Biological Chemistry. 1999;274(21):14551–14559. doi: 10.1074/jbc.274.21.14551. [DOI] [PubMed] [Google Scholar]
- 127.Albe KR, Wright BE. Carbohydrate metabolism in dictyostelium discoideumml: II. Systems' analysis. Journal of Theoretical Biology. 1994;169(3):243–251. doi: 10.1006/jtbi.1994.1145. [DOI] [PubMed] [Google Scholar]
- 128.Rohwer JM, Botha FC. Analysis of sucrose accumulation in the sugar cane culm on the basis of in vitro kinetic data. Biochemical Journal. 2001;358, part 2:437–445. doi: 10.1042/0264-6021:3580437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cronwright GR, Rohwer JM, Prior BA. Metabolic control analysis of glycerol synthesis in Saccharomyces cerevisiae . Applied and Environmental Microbiology. 2002;68(9):4448–4456. doi: 10.1128/AEM.68.9.4448-4456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nielsen J, Jorgensen HS. Metabolic control analysis of the penicillin biosynthetic pathway in a high-yielding strain of Penicillium chrysogenum . Biotechnology Progress. 1995;11(3):299–305. doi: 10.1021/bp00033a010. [DOI] [PubMed] [Google Scholar]
- 131.Poolman MG, Fell DA, Thomas S. Modelling photosynthesis and its control. Journal of Experimental Botany. 2000;51:319–328. doi: 10.1093/jexbot/51.suppl_1.319. [DOI] [PubMed] [Google Scholar]
- 132.Hua Q, Yang C, Shimizu K. Metabolic control analysis for lysine synthesis using Corynebacterium glutamicum and experimental verification. Journal of Bioscience and Bioengineering. 2000;90(2):184–192. [PubMed] [Google Scholar]
- 133.Berthon HA, Kuchel PW, Nixon PF. High control coefficient of transketolase in the nonoxidative pentose phosphate pathway of human erythrocytes: NMR, antibody, and computer simulation studies. Biochemistry. 1992;31(51):12792–12798. doi: 10.1021/bi00166a012. [DOI] [PubMed] [Google Scholar]
- 134.de Groot MJL, Prathumpai W, Visser J, Ruijter GJG. Metabolic control analysis of Aspergillus niger L-arabinose catabolism. Biotechnology Progress. 2005;21(6):1610–1616. doi: 10.1021/bp050189o. [DOI] [PubMed] [Google Scholar]
- 135.Hoefnagel MHN, Starrenburg MJC, Martens DE, et al. Metabolic engineering of lactic acid bacteria, the combined approach: kinetic modelling, metabolic control and experimental analysis. Microbiology. 2002;148(4):1003–1013. doi: 10.1099/00221287-148-4-1003. [DOI] [PubMed] [Google Scholar]
- 136.Mendoza-Cózatl DG, Moreno-Sánchez R. Control of glutathione and phytochelatin synthesis under cadmium stress. Pathway modeling for plants. Journal of Theoretical Biology. 2006;238(4):919–936. doi: 10.1016/j.jtbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 137.Meister A. Glutathione synthesis. In: Boyer PD, editor. The Enzymes. Vol. 10. New York, NY, USA: Academic Press; 1994. pp. 671–697. [Google Scholar]
- 138.Speiser DM, Abrahamson SL, Banuelos G, Ow DW. Brassica juncea produces a phytochelatin-cadmium-sulfide complex. Plant Physiology. 1992;99(3):817–821. doi: 10.1104/pp.99.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vande WJG, Ow DW. Accumulation of metal-binding peptides in fission yeast requires h m t2+ . Molecular Microbiology. 2001;42(1):29–36. doi: 10.1046/j.1365-2958.2001.02624.x. [DOI] [PubMed] [Google Scholar]
- 140.Small JR, Kacser H. Responses of metabolic systems to large changes in enzyme activities and effectors. 1. The linear treatment of unbranched chains. European Journal of Biochemistry. 1993;213(1):613–624. doi: 10.1111/j.1432-1033.1993.tb17801.x. [DOI] [PubMed] [Google Scholar]
- 141.Reeves RE. Metabolism of Entamoeba histolytica Schaudinn, 1903. Advances in Parasitology. 1984;23:105–142. doi: 10.1016/s0065-308x(08)60286-9. [DOI] [PubMed] [Google Scholar]
- 142.Saavedra E, Marín-Hernández A, Encalada R, Olivos A, Mendoza-Hernández G, Moreno-Sánchez R. Kinetic modeling can describe in vivo glycolysis in Entamoeba histolytica . FEBS Journal. 2007;274(18):4922–4940. doi: 10.1111/j.1742-4658.2007.06012.x. [DOI] [PubMed] [Google Scholar]
- 143.Saavedra E, Encalada R, Pineda E, Jasso-Chávez R, Moreno-Sánchez R. Glycolysis in Entamoeba histolytica: biochemical characterization of recombinant glycolytic enzymes and flux control analysis. FEBS Journal. 2005;272(7):1767–1783. doi: 10.1111/j.1742-4658.2005.04610.x. [DOI] [PubMed] [Google Scholar]
- 144.Torres NV, Souto R, Meléndez-Hevia E. Study of the flux and transition time control coefficient profiles in a metabolic system in vitro and the effect of an external stimulator. Biochemical Journal. 1989;260, part 3:763–769. doi: 10.1042/bj2600763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Giersch C. Determining elasticities from multiple measurements of flux rates and metabolite concentrations. Application of the multiple modulation mehod to a reconstituted pathway. European Journal of Biochemistry. 1995;227(1-2):194–201. doi: 10.1111/j.1432-1033.1995.tb20376.x. [DOI] [PubMed] [Google Scholar]
- 146.Moreno-Sánchez R, Encalada R, Marín-Hernández A, Saavedra E. Experimental validation of metabolic pathway modeling. An illustration with glycolytic segments from Entamoeba histolytica . FEBS Journal. 2008;275(13):3454–3469. doi: 10.1111/j.1742-4658.2008.06492.x. [DOI] [PubMed] [Google Scholar]
- 147.Kurreck J. Antisense technologies: improvement through novel chemical modifications. European Journal of Biochemistry. 2003;270(8):1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 148.Stitt M, Quick WP, Schurr U, Schulze E-D, Rodermel SR, Bogorad L. Decreased ribulose-1,5-bisphosphate carboxylase-oxygenase in transgenic tobacco transformed with ‘antisense’ rbcS II. Flux-control coefficients for photosynthesis in varying light, Co2, and air humidity. Planta. 1991;183(4):555–566. doi: 10.1007/BF00194277. [DOI] [PubMed] [Google Scholar]
- 149.Albert M-A, Haanstra JR, Hannaert V, et al. Experimental and in silico analyses of glycolytic flux control in bloodstream form Trypanosoma brucei . Journal of Biological Chemistry. 2005;280(31):28306–28315. doi: 10.1074/jbc.M502403200. [DOI] [PubMed] [Google Scholar]
- 150.Aiston S, Trinh KY, Lange AJ, Newgard CB, Agius L. Glucose-6-phosphatase overexpression lowers glucose 6-phosphate and inhibits glycogen synthesis and glycolysis in hepatocytes without affecting glucokinase translocation. Journal of Biological Chemistry. 1999;274(35):24559–24566. doi: 10.1074/jbc.274.35.24559. [DOI] [PubMed] [Google Scholar]
- 151.Koebmann BJ, Tornøe J, Johansson B, Jensen PR. Experimental modulation of gene expression. In: Kholodenko BN, Westerhoff HV, editors. Metabolic Engineering in the Post Genomic Era. Norfolk, UK: Horizon Bioscience; 2004. pp. 155–179. [Google Scholar]
- 152.Hammer K, Mijakovic I, Jensen PR. Synthetic promoter libraries—tuning of gene expression. Trends in Biotechnology. 2006;24(2):53–55. doi: 10.1016/j.tibtech.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 153.Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Applied and Environmental Microbiology. 1998;64(1):82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]