Abstract
Chlamydiae replicate in a vacuole within epithelial cells and commonly induce cell damage and a deleterious inflammatory response of unknown molecular pathogenesis. The chlamydial protease-like activity factor (CPAF) translocates from the vacuole to the cytosol, where it cleaves several cellular proteins. CPAF is synthesized as an inactive precursor that is processed and activated during infection. Here, we show that CPAF can be activated in uninfected cells by experimentally induced oligomerization, reminiscent of the activation mode of initiator caspases. CPAF activity induces proteolysis of cellular substrates including two novel targets, cyclin B1 and PARP, and indirectly results in the processing of pro-apoptotic BH3-only proteins. CPAF activation induces striking morphological changes in the cell and, later, cell death. Biochemical and ultrastructural analysis of the cell death pathway identify the mechanism of cell death as nonapoptotic. Active CPAF in uninfected human cells thus mimics many features of chlamydial infection, implicating CPAF as a major factor of chlamydial pathogenicity, Chlamydia-associated cell damage, and inflammation.
Introduction
Chlamydia trachomatis, an obligate intracellular bacterium, is the leading cause of bacterially sexually transmitted disease and a main cause of preventable blindness. C. trachomatis has a biphasic developmental cycle. The infectious elementary body (EB) infects primarily epithelial cells where it develops within a membrane-bound vacuole (called an inclusion) into a replicating noninfectious reticulate body (RB). Within two days, the RB redifferentiates into an EB and is released from the infected cell. Chlamydiae thus develop in a compartment that is separate from the rest of the human or animal host cell. However, the bacteria impact on a number of signaling pathways in the host cell, and cause substantial changes to cellular transcription as well as cell damage (McClarty, 1994; Wyrick, 2000; Fields and Hackstadt, 2002). How Chlamydia achieves this is not known in great detail. Several chlamydial species have been found to possess the components of a functional type III secretion system, which likely enables the bacteria to inject effector proteins into the host cytosol, and a number of such candidate proteins have been identified (for review see Peters et al., 2007).
Infection with Chlamydia causes massive stress to the host cell, and cytolytic activity associated with Chlamydia infection has been described for more than 30 years (Friis, 1972; Todd and Storz, 1975; Chang and Moulder, 1978; Wyrick et al., 1978). By electron microscopy, massive changes to organelles were noticed at later stages of infection, such as dilation and vacuolation of ER, distortion of mitochondria, and nuclear condensation (Todd and Storz, 1975; Todd et al., 1976). Although some of these changes resemble features of apoptosis (Ojcius et al., 1998; Perfettini et al., 2002; Ying et al., 2006), further characterization of signaling pathways indicates that the apoptotic pathway is not activated by Chlamydia and the cytopathic changes observed are nonapoptotic (Ying et al., 2006). Cytopathicity and cell death may be a defense mechanism of the cell to block bacterial replication or may aid bacterial spreading and cause infection-associated inflammation.
It has been entirely unclear how Chlamydia induces cell death. The gene of a potential cytotoxin has been identified in the C. trachomatis genome (Belland et al., 2001), but was later found to be nonfunctional in many serovars (Carlson et al., 2004).
One chlamydial protein has been directly purified from the cytosol of infected cells. Named chlamydial protease-like activity factor (CPAF), this protein was isolated as a factor that can degrade two host transcription factors, RFX5 and USF-1 (Zhong et al., 2001). Because these transcription factors are involved in the expression of major histocompatibility complex (MHC) molecules, it has been speculated that CPAF may contribute to immune evasion of Chlamydia (Zhong et al., 2001). Similarly, it has been suggested that CPAF plays a role in the loss of expression of the MHC-like protein CD1d during infection, which may also enhance escape from host immune surveillance (Kawana et al., 2007). The cytoskeletal protein cytokeratin (CK) 8, a component of intermediate filaments, has also been identified as a substrate of CPAF proteolysis (Dong et al., 2004c), and recently it has been proposed that the pro-apoptotic BH3-only proteins, which are degraded during chlamydial infection (Fischer et al., 2004; Ying et al., 2005), are CPAF substrates (Pirbhai et al., 2006). Because Chlamydia cannot be genetically modified, direct proof of the role of CPAF has been difficult.
CPAF is synthesized in the chlamydial inclusion as one polypeptide, but is rapidly processed into two subunits that assemble into heterodimers and are proteolytically active in the host cytosol (Dong et al., 2004a,b). Expression of the CPAF precursor in human cells did not induce CPAF processing and yielded no proteolytic activity (Dong et al., 2004a).
Although potential functions had thus been assigned to CPAF, we reasoned that an active protease, free in the cytosol of a human cell, may be expected to damage the cell, and that CPAF therefore should be a candidate factor for cytopathic activity. However, until now it has not been possible to express active CPAF in human cells in the absence of infection, and meaningful analysis of CPAF effects in infected cells is very difficult due to the presence of numerous bacterial components in the cell.
We here show that CPAF can be activated by “induced proximity” in the absence of infection. The induced proximity model was proposed to explain the activation of initiator caspases during induction of apoptosis (Salvesen and Dixit, 1999; Pop et al., 2006; Bao and Shi, 2007). According to this model, the dimerization and activation of initiator caspases requires their adaptor-mediated clustering, followed by caspase processing. Similarly, we describe that forced clustering of CPAF leads to its processing and activation in human cells. Activation of CPAF caused massive morphological changes and nonapoptotic death of human host-cells, strongly resembling the changes and the form of cell death induced by chlamydial infection. CPAF should therefore be regarded as a major factor of chlamydial pathogenicity. CPAF activity may help Chlamydia at earlier stages to establish the growing inclusion in the cell and at later stages may facilitate release of newly replicated bacteria.
Results
Activation of CPAF by induced proximity
To study the role of CPAF in cytopathicity, a model had to be developed to express active CPAF. We had noticed certain parallels of caspase-9 and CPAF. Both are synthesized as inactive precursors, both are proteolytically processed into two subunits during their physiological activation, and in both cases the active enzyme is made up of a complex of both subunits. We therefore aimed at achieving experimental oligomerization of CPAF to model the physiological clustering of caspase-9 after its recruitment into the apoptosome during apoptosis (Bao and Shi, 2007). The open reading frame of C. trachomatis CPAF was fused to an N-terminal partner consisting of the FLAG antibody epitope and a triple repeat of a fragment of bacterial gyrase (gyr) B. GyrB binds the cell-permeable synthetic ligand coumermycin (CM). Because CM has two gyrB binding sites, it can bind two gyrB proteins simultaneously, inducing dimerization of the fusion partner of gyrB (Fig. 1 A). A triple repeat rather than an individual gyrB molecule was chosen to enhance complex formation upon CM addition. We and others have used this system in the past for conditional complex formation of intracellular signaling proteins (Farrar et al., 1996; Hacker et al., 2006). Because transient transfections indicated that the protein was toxic to human cells (unpublished data), the fusion construct was placed under the control of a tetracycline (TET)-inducible promoter, where expression was silenced in cells carrying the tet-repressor but induced upon addition of tetracycline (“tet-on”).
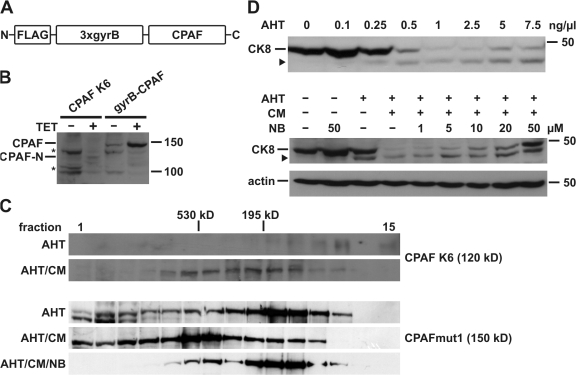
Figure 1.
CPAF is activated by induced proximity. (A) Schematic representation of the gyrB-CPAF construct. CPAF was placed under the control of a tetracycline-inducible promoter. FLAG, FLAG tag; 3xgyrB, three consecutive copies of an N-terminal fragment of gyrase B from Escherichia coli; CPAF, CPAF from Chlamydia trachomatis (amino acid residues 18–601). (B) CPAF expression in T-Rex-293 cells. Expression of CPAF was induced by tetracycline (TET) either in CPAF K6 cells stably expressing gyrB-CPAF or T-REx-293 cells transfected with the gyrB-CPAF construct. CPAF-N indicates an N-terminal fragment of gyrB-CPAF after proteolytic activation. Triton X-100 cell extracts were analyzed by Western blotting with an antibody specific for the FLAG tag. Asterisk, unspecific signal. Molecular size markers (in kD) are indicated. (C) Size exclusion chromatography of CPAF. Cell extracts of either CPAF K6 cells (top) or T-REx-293 cells transfected with the CPAFmut1 (carrying the S491A active-site mutation; bottom) were separated on a Superose 200 gel filtration column. Anhydrotetracycline (AHT), coumermycin (CM), or novobiocin (NB) were used as indicated. The elution fractions and eluted molecular size markers are indicated. (D) Activation of CPAF by induced proximity. CPAF expression was induced in CPAF K6 cells with increasing amounts (top) of AHT or using 0.5 ng/ml (bottom). Before addition of CM, cells were preincubated with indicated amounts of NB. Samples were analyzed by Western blotting using anti-CK8 antibodies. The arrowhead indicates a cleavage product of CK8. Detection of actin served as a loading control.
When the gyrB-CPAF construct was transfected into 293T cells stably carrying the tet-repressor (T-REx-293), the addition of tetracycline or its analogue anhydrotetracycline (AHT) induced the appearance of a protein of the expected size of the full-length protein as well as a smaller product, which ran at the expected size of a protein containing the N-terminal tag fused to the N-terminal CPAF-fragment, indicating processing of the protein (Fig. 1 B).
T-REx-293 cells were then transfected and clones were selected that stably carried the TET-inducible gyrB-CPAF construct. Five clones were identified, in which addition of TET/AHT induced expression of a protein of the same size as the smaller product during transient transfection, again very likely corresponding to the fragment of gyrB and the N-terminal fragment of CPAF (Fig. 1 B and unpublished data). No band corresponding to the intact protein was detected in these clones, suggesting more efficient processing. All of the clones showed the same phenotype upon induction of gyrB-CPAF (see following paragraph). Expression of gyrB-CPAF thus leads to spontaneous processing of the protein.
To test for CM-mediated oligomerization, cell extracts from the stable clone K6 expressing gyrB-CPAF were analyzed by size exclusion chromatography. GyrB-N-CPAF was detectable with a peak around the expected molecular weight of the protein (120 kD). Addition of CM caused a shift in the protein to higher molecular weight fractions (Fig. 1 C). Because of the low expression of wild-type gyrB-CPAF, we performed additional experiments with a cleavage-deficient mutant of gyrB-CPAF (CPAFmut1 [see following paragraph], which is expressed at considerably higher levels), expressed by transient transfection and induction in T-REx-293 cells. As shown in Fig. 1 C, most of the protein eluted at about the predicted size of the unprocessed monomer (150 kD), although easily detectable amounts appeared in fractions containing higher molecular weight proteins. Addition of CM caused a shift of the protein peak, which then eluted around 500 kD and might correspond to a trimer or tetramer of the protein. A substantial fraction of the protein appeared to be engaged in formation of even higher molecular weight complexes. Most of these CM-induced complexes could be disrupted by addition of an excess of the monomeric ligand of gyrase B, novobiocin (NB) (Fig. 1 C). These results show the expected oligomerization of gyrB-CPAF by CM, as well as substantial spontaneous complex formation. The spontaneous oligomerization was probably due to the gyrase B domains because novobiocin could reduce the extent of complex formation (compare lanes AHT and AHT/CM/NB in gyrB-CPAFmut1; Fig. 1 C).
We next measured the proteolytic activity of gyrB-CPAF, using cleavage of CK8, one of the reported cellular CPAF substrates, as a read-out. As shown in Fig. 1 D, titration of AHT on the CPAF K6 clone caused increasing cleavage of CK8 even in the absence of CM-induced oligomerization, suggesting that spontaneous aggregation was sufficient for activation of gyrB-CPAF. Addition of CM to cells induced with a lower concentration of AHT caused a higher level of CK8 degradation, which could be partly blocked by increasing concentrations of novobiocin (Fig. 1 D). GyrB-CPAF thus shows some spontaneous aggregation and activity, which can be enhanced by CM; the effect of CM can be prevented by NB.
CPAF activity requires its processing site and an intact protease motif
CPAF activity in lysates from infected cells was inhibited by an inhibitor of the cellular proteasome, lactacystin, but not another one, MG-132 (Zhong et al., 2001). We used CPAF K6 cell lysate as a source of CPAF and lysate from cells transfected with a construct encoding myc-tagged CK8 as a substrate, to test for the effects of standard protease inhibitors on CPAF. Lactacystin but not MG-132, nor any other tested inhibitor, prevented degradation of CK8 (Fig. 2 A and unpublished data). (The following inhibitors were tested: E64, pepstatin A, PMSF, TPCK, and a standard mix of protease inhibitors [Roche].) These results confirm the inhibitory activity of lactacystin previously reported in lysates from infected cells and identify an unusual inhibitor profile for CPAF.
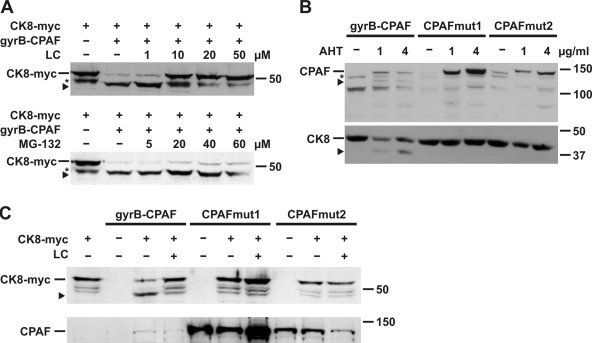
Figure 2.
Analysis of proteolytic activity of CPAF. (A) Inhibition of CPAF activity by proteasome inhibitors. Cell extracts of CPAF K6 cells (gyrB-CPAF) or T-REx-293 cells expressing myc-tagged cytokeratin 8 (CK8-myc) were combined in the presence of various amounts of the proteasome inhibitors lactacystin (LC; top) or MG-132 (bottom) as indicated, and analyzed by Western blotting using an antibody specific for the myc tag. The arrowhead indicates a cleavage product of CK8-myc (asterisk marks an unspecific signal). (B) Analysis of the proteolytic activity of CPAF mutants in vivo. T-REx-293 cells were transfected with either the gyrB-CPAF construct or one of two CPAF mutants. In CPAFmut1, the Tsp-active site was mutated by the replacement S491A. In mutant 2 (CPAFmut2), two amino acid residues (L273G, S275V) were exchanged to prevent autocatalytic cleavage of CPAF. CPAF expression was induced by AHT as indicated. Cell extracts were analyzed by Western blotting using either FLAG tag (top) or CK8 antibodies (bottom). The arrowhead in the top panel shows the cleavage product very likely corresponding to gyrB-CPAF-N; the arrowhead in the bottom panel indicates a cleavage product of CK8. An unspecific signal is marked by an asterisk. (C) Analysis of the proteolytic activity of CPAF mutants in vitro. T-REx-293 cells were transfected with the CPAF constructs as indicated, and cell lysates were incubated with extracts containing myc-tagged CK8, and analyzed as described in A (top). Expression of the CPAF constructs was confirmed by Western blotting using a FLAG tag antibody (bottom).
It has been shown that a small amount of CPAF is cleaved when expressed in Escherichia coli, and the processing site has been mapped; a cleavage site mutant was not processed and was inactive when expressed in E. coli (Dong et al., 2004a). It has further been noted that CPAF contains a domain characteristic of bacterial Tail-specific proteases (Tsp) (Shaw et al., 2002), a class of serine proteases originally described in E. coli (Silber et al., 1992). We therefore generated gyrB-CPAF constructs with point mutations either in the active site of the Tsp domain (S491A, CPAFmut1) or in the processing site (L273G, S275V, CPAFmut2). When expressed transiently in T-REx-293 cells, neither mutant was processed (Fig. 2 B, top) and neither was able to cleave endogenous CK8 (Fig. 2 B, bottom) or myc-tagged CK8 in a cell-free system using lysate from transfected cells (Fig. 2 C). This suggests that CPAF is cleaved autocatalytically and the Tsp domain containing the active-site serine residue is indeed required for its proteolytic activity.
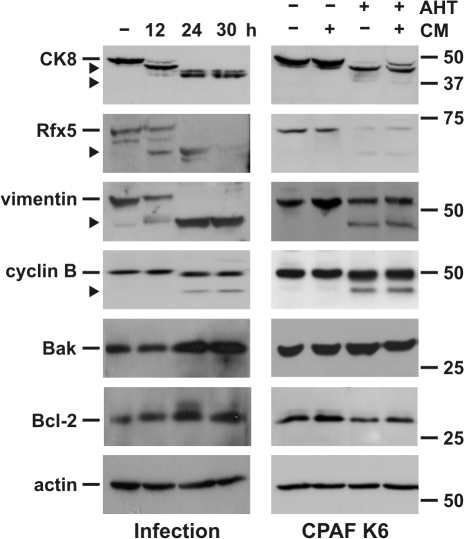
A number of host cell proteins have been reported to be cleaved by CPAF during chlamydial infection, including CK8, the transcription factors RFX5 and USF-1, and the BH3-only proteins Bim, Puma, and Bik (Zhong et al., 2001; Dong et al., 2004c). Another protein, the component of intermediate filaments, vimentin, was also recently shown to be cleaved by CPAF (Valdivia, R., personal communication). We therefore tested the cleavage of these proteins upon CPAF activation in K6 cells. Cleavage of CK8, RFX5, and vimentin occurred upon CPAF activation in K6 cells and yielded fragments of the same sizes as during infection with C. trachomatis (Fig. 3), whereas USF1 was not detectable by Western blotting in these cells. The cell cycle protein cyclin B1 is also degraded during chlamydial infection (Balsara et al., 2006). Degradation products of the same sizes as during infection were generated upon activation of CPAF, suggesting that cleavage of cyclin B1 during infection is mediated by CPAF. A number of control proteins (Bak, Bcl-2, actin) were not degraded (Fig. 3; see following paragraph regarding cleavage of BH3-only proteins). The expression of active CPAF thus recapitulates the known proteolytic activities observed during infection with whole chlamydiae. Based on the levels of substrate cleavage, the amount of active gyrB-CPAF produced corresponds to the levels of CPAF expressed relatively early in the developmental cycle, at least in T-REx-293 cells. The amounts generated during later stages of infection are probably substantially higher and cause more complete degradation of cellular substrates (Fig. 3).
Figure 3.
Infection with C. trachomatis or expression of CPAF causes cleavage of host cell proteins. (Left) Infection with C. trachomatis. T-REx-293 cells were infected with C. trachomatis for indicated periods of time. RIPA buffer extracts of the cells were prepared. (Right) CPAF K6 cells were treated with either 5 ng/μl AHT, CM, or both for 12–14 h as indicated. Triton X-100 cell extracts were prepared and all extracts were analyzed by Western blotting using indicated antibodies. The arrowheads indicate specific cleavage products.
Degradation of anti-apoptotic BH3-only proteins is likely an indirect consequence of CPAF expression
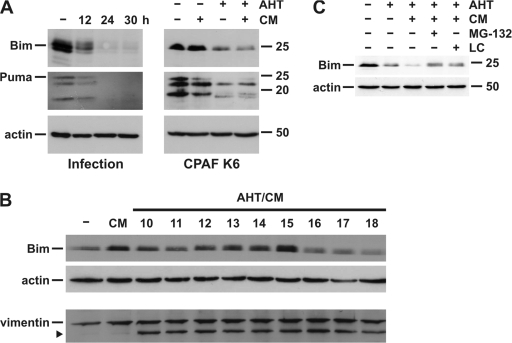
BH3-only proteins are essential mediators of mitochondrial apoptosis (Hacker and Weber, 2007). These proteins are degraded during chlamydial infection (Fischer et al., 2004), which can account for the protection against apoptosis of infected cells. Re-expression of active BH3-only proteins overcomes the Chlamydia-imposed block of apoptosis (Fischer et al., 2004), indicating that this loss is functionally relevant. It has recently been reported that CPAF can degrade BH3-only proteins in cell lysates (Pirbhai et al., 2006). In our initial analyses, we failed to see degradation of the BH3-only proteins Bim and Puma (unpublished data), which were easily detectable in CPAF K6 cells, although both proteins were degraded during infection with C. trachomatis (Fig. 4 A). However, longer periods of CPAF induction in K6 cells did lead to the degradation of Bim and Puma (Fig. 4 A). More extensive time-course studies revealed that Bim degradation occurred later than cleavage of the other substrates (Fig. 4 B). Although vimentin was already cleaved at 10 h after CPAF induction/activation with AHT/CM, the Bim levels were unchanged or even increased up to ∼15 h of CPAF activation, after which point they began to decrease. At 18 h after activation, Bim levels were clearly reduced. No smaller fragments of Bim were detected (Fig. 4 B; vimentin cleavage starts around 4 h under this protocol; unpublished data). This suggested that the degradation of Bim was not mediated directly by CPAF but by subsequent proteolytic events that had been initiated by CPAF. This interpretation is supported by another finding: the degradation of Bim by prolonged activation of CPAF was blocked by lactacystin and MG-132 (Fig. 4 C). The proteolytic activity of CPAF, however, is only inhibited by lactacystin but not MG-132 (Zhong et al., 2001; Fig. 2 A). It is thus unlikely that CPAF directly degrades BH3-only proteins. Nevertheless, the CPAF-induced degradation of BH3-only proteins is the main reason for apoptosis inhibition in infected cells, and CPAF is therefore the main anti-apoptotic factor of Chlamydia.
Figure 4.
BH3-only proteins are degraded by infection with C. trachomatis or upon prolonged expression of active CPAF. (A) (Left) T-REx-293 cells were infected with C. trachomatis for indicated time points and RIPA buffer extracts were prepared. (Right) CPAF K6 cells were treated with either 5 ng/μl AHT, CM, or both as indicated. Cell extracts were analyzed by Western blotting using antibodies specific for Bim, Puma, or actin as loading control. (B) Time course of cleavage of CPAF substrates. CPAF K6 cells were treated with 5 ng/μl AHT and CM to induce CPAF expression for the indicated time periods. Cell extracts were analyzed by Western blotting. The arrowheads indicate cleavage product of vimentin. (C) Inhibition of Bim degradation by proteasome inhibitors. CPAF K6 cells were treated with 5 ng/ml AHT or 5 ng/ml AHT plus CM. 6 h before cell harvesting, either 40 μM MG-132 or 5 μM clasto-lactacystin β-lactone (LC) were added. Cell extracts were analyzed by Western blotting.
CPAF expression leads to nonapoptotic cell death
These results showed that gyrB-CPAF could reproduce the known proteolytic events of chlamydial infection. We then turned to the question of the cellular consequences of CPAF activity. On the one hand, CPAF induced the degradation of BH3-only proteins and is therefore an anti-apoptotic effector. On the other hand, free proteases in the cytosol have the potential to cause cell death. This has been shown not only for the specialized caspases, but also, for example, for lysosomal peptidases (Lockshin and Zakeri, 2004) and even the promiscuous protease proteinase-K (Wilhelm and Hacker, 1999). Chlamydial infection causes massive morphological changes to the host cell as well as nonapoptotic cell death (Ojcius et al., 1998; Belland et al., 2001; Perfettini et al., 2002; Ying et al., 2006). We therefore asked whether CPAF might contribute to this cytopathicity.
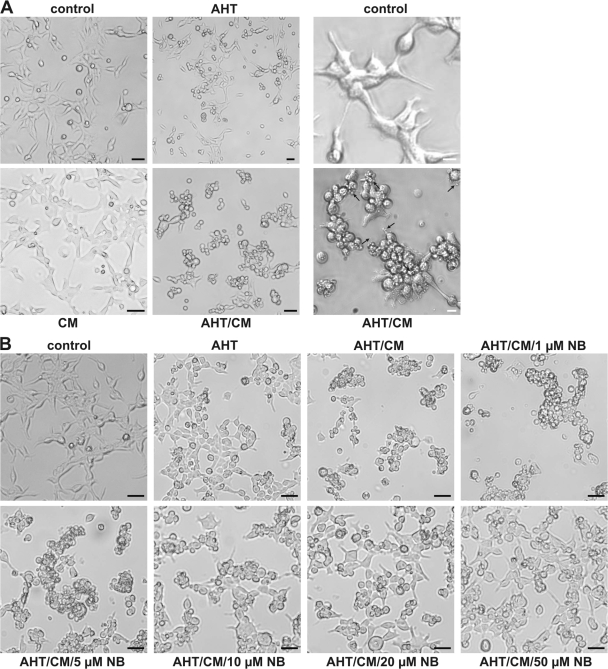
The expression and activation of CPAF in K6 cells (Fig. 5 A) caused striking morphological changes in the cells. The cells rounded up and began to detach from the culture dish. They formed clusters and at later stages, smaller vesicular fragments appeared (Fig. 5 A). Similar albeit less pronounced changes were observed upon transient transfection and activation of gyrB-CPAF in T-REx-293 or T-REx-HeLa cells (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200804023/DC1). Indeed, the morphology resembled the changes observed during chlamydial infection of T-REx-293 cells (Fig. S2). Although not conclusive, this similarity suggests that CPAF is involved in at least some of the morphological changes induced during chlamydial infection. AHT on its own caused less dramatic changes in CPAF K6 cells than when CM was included, and an excess of novobiocin could reduce the phenotypical changes (Fig. 5 B). Cleavage of cellular proteins by CPAF may therefore be one mechanism by which Chlamydia induces the morphological changes observed in the host cell.
Figure 5.
CPAF expression leads to changes in cellular morphology. (A) Changes in cell morphology during CPAF expression. CPAF K6 cells were incubated either with 6 ng/ml AHT, CM, or both for 16 h. Cells were analyzed by light microscopy (left). Arrows indicate smaller vesicular fragments. Black bar, 10 μm. The right panel shows an enlarged section of either a control sample or a sample treated with AHT and CM. White bar, 3 μm. (B) Inhibition of CPAF oligomerization by novobiocin. CPAF expression was induced in CPAF K6 cells by adding AHT, or cells were left untreated. 30 min before addition of CM, the cells were incubated with indicated amounts of NB. Cells were analyzed by light microscopy. Bar, 10 μm.
The dramatic changes as observed by microscopy suggested that K6 cells expressing active CPAF were dying. Cell death induction in cell culture by chlamydial infection has been previously documented, and our recent analysis suggests that this cell death occurs by a nonapoptotic process (Ying et al., 2006). When cell death was measured as loss of cellular metabolic activity by MTT assay, it became apparent that most cells were dead after 20 h of expression of active CPAF. No decrease in viability was measured at 7 h, although very clear morphological changes were already apparent (Fig. 6 A and unpublished data). Cell death in this assay was not inhibited by the caspase inhibitor zVAD-fmk (Fig. 6 A), consistent with the lack of effect of zVAD-fmk in host cell death induced by chlamydial infection (Ojcius et al., 1998; Perfettini et al., 2002; Ying et al., 2006). Because caspase activity is required for apoptotic cell death, this is suggestive of a nonapoptotic form of cell death. Plasma membrane integrity was relatively well maintained, with only ∼20% of cells taking up the vital dye propidium iodide, despite a reduction of metabolic activity of ∼80% at 20 h of treatment with AHT/CM (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200804023/DC1).
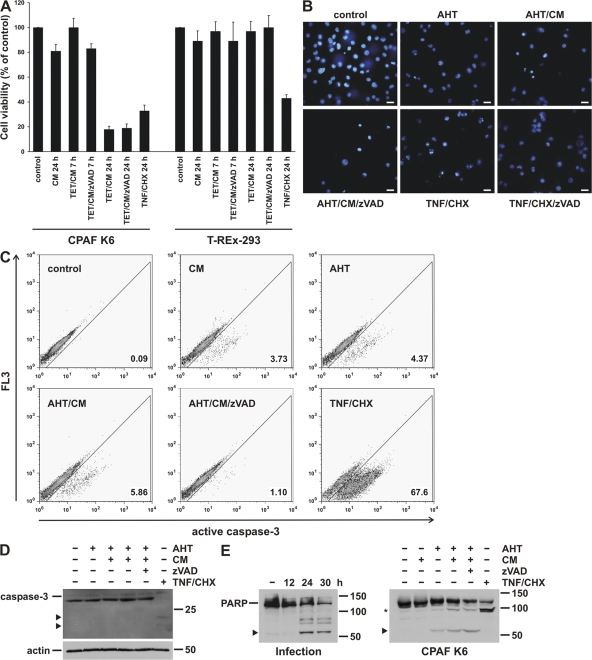
Figure 6.
CPAF expression causes nonapoptotic cell death. (A) CPAF reduces cell viability. CPAF K6 or T-REx-293 cells were treated with the indicated combinations of TET, CM, and the caspase inhibitor zVAD-fmk (zVAD). As a positive control, cells were treated with TNF-α (TNF) and cycloheximide (CHX). After indicated time points, cell viabilities were measured by MTT assay. Relative cell viability was calculated (untreated cells were set to 100%). Data are normalized means/SEM of three independent experiments. (B) Analysis of nuclear morphology after CPAF expression by Hoechst staining. CPAF K6 cells were treated with 10 ng/ml AHT, CM, or zVAD-fmk as indicated. As a positive control of apoptosis, cells were treated with TNF/CHX (as described in A). After 16 h, cells were stained with the Hoechst 33342 dye and analyzed by fluorescence microscopy. Bar, 15 μm. (C) Caspase-3 activation during CPAF-expression. CPAF K6 cells were treated as described in B and analyzed by flow cytometry using an antibody specific for active caspase-3. (D) Analysis of caspase-3 activation by Western blotting. CPAF K6 cells were treated as described in B, and cell extracts were analyzed by Western blotting using an antibody specific for active caspase-3. Arrowheads indicate specific cleavage products of activated caspase-3. Detection of actin served as loading control. (E) Cleavage of PARP by infection with C. trachomatis or expression of active CPAF. (Left) CPAF K6 cells were infected with C. trachomatis for the indicated periods of time. (Right) CPAF K6 cells were treated as described in B. Cell extracts were analyzed by Western blotting using a PARP antibody. The arrowheads indicate cleavage products resulting from chlamydial infection or CPAF expression. The asterisk indicates a specific PARP cleavage product due to caspase activation by TNF/CHX.
During apoptosis, the chromatin condenses and the nuclei are fragmented. As these changes in nuclear morphology are a good marker of apoptosis, we next analyzed the dying cells for changes in nuclear morphology. CPAF activation caused nuclear condensation in some cells; however, the morphology was not quite typical for apoptosis and distinct from the appearance of nuclei in the same cells undergoing apoptosis upon treatment with TNF-α/cycloheximide (TNF/CHX) (Fig. 6 B). Higher magnification photographs of the nuclear morphology of cells dying due to treatment with TNF/CHX, upon activation of CPAF or upon chlamydial infection, are shown in Fig. S4 (available at http://www.jcb.org/cgi/content/full/jcb.200804023/DC1). Furthermore, unlike the changes induced by the apoptosis-inducing protocol with TNF/CHX, the CPAF-induced nuclear morphological changes were not prevented by the caspase inhibitor zVAD-fmk (Fig. 6 B). These results reproduced the features of Chlamydia-induced cell death and suggested that CPAF induced cell death through a nonapoptotic mechanism.
Apoptosis is the result of the activation of the apoptotic signal transduction pathway, and we therefore tested directly whether this pathway was activated. Caspase-3 is a central protease in the apoptotic pathway, and caspase-3 is regularly activated proteolytically during apoptosis. An antibody directed against an active caspase-3 fragment showed only few positive cells upon activation of CPAF (treatment with AHT/CM) (Fig. 6 C). Western blotting further failed to detect the cleaved form of caspase-3 seen during apoptosis in cells expressing active CPAF (Fig. 6 D), in agreement with the absence of caspase-3 activation in cells infected with C. caviae or C. trachomatis (Ojcius et al., 1998; Ying et al., 2006). During apoptosis, caspase-3 cleaves the nuclear enzyme poly (ADP-ribose) polymerase (PARP) (Nicholson et al., 1995), and PARP cleavage can thus be used as a marker of apoptosis. Surprisingly, PARP was degraded both during chlamydial infection and upon CPAF activation (Fig. 6 E). However, the prominent cleavage band was different from the one generated during apoptosis; a very light band corresponding to apoptotic cleavage appears to be visible on the blot, although the appearance of this band was not sensitive to caspase inhibition (Fig. 6 E). Induction of apoptosis by treatment of the cells with TNF/CHX yielded the typical caspase-dependent PARP fragment (Fig. 6 E). Although PARP cleavage is thus seen upon CPAF activation, it is not mediated by caspase-3 but probably by CPAF itself. The relevance of the cleavage of this new CPAF substrate for the infection remains to be seen.
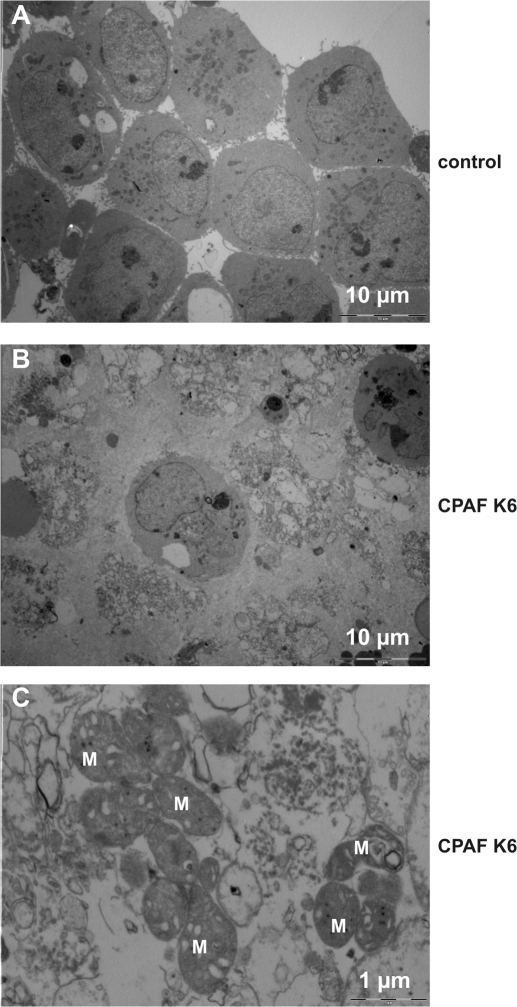
We finally used electron microscopy for the analysis of structural changes in cells containing active CPAF. At earlier stages (after 7 h of CPAF expression/activity), although the cells had already rounded up and detached from the plate, no ultrastructural changes were observed (unpublished data). This is perhaps somewhat surprising given the strong morphological changes, but is in accordance with the unaffected metabolic activity at this time. At 20 h, however, only relatively few cells were still morphologically intact while the majority were in a process of disintegration (Fig. 7 A, B). Only few apoptotic cells were detected in this analysis but many necrotic cells, confirming that CPAF induced nonapoptotic (necrotic) cell death in CPAF K6 cells. At higher magnification, dying cells showed numerous lamellar structures of unknown composition as well as many mitochondria that still seemed only slightly affected (Fig. 7 C). Ectopic expression and activation of CPAF thus causes a nonapoptotic form of cell death that appears indistinguishable from cell death induced during infection with C. trachomatis.
Figure 7.
CPAF expression leads to changes in cellular ultrastructure. CPAF K6 cells were treated with 5 ng/ml AHT and CM for 20 h (middle and bottom), or left untreated (control; top). Cells were harvested, pelleted, and analyzed by electron microscopy. The bottom panel shows a 10-fold magnification. Scale bars are indicated. M, mitochondria.
Discussion
This study shows that C. trachomatis CPAF can be activated by oligomerization to induce degradation of host cell proteins. On the one hand, CPAF caused the degradation of pro-apoptotic BH3-only proteins and is therefore a mediator of chlamydial anti-apoptotic activity. On the other hand, CPAF induced cell death in the absence of hallmarks of apoptosis. Activation of the apoptotic pathway was observed only in a small minority of dying cells, identifying this cell death as nonapoptotic or necrotic. All the observed features recapitulated the changes seen in cells infected by C. trachomatis. Infections with Chlamydia commonly induce strong inflammatory responses. These reactions may thus be the result of the CPAF-mediated release of not only bacterial but also pro-inflammatory cellular molecules.
We were prompted to test for the possibility of activation by induced proximity by similarities between the known characteristics of CPAF and the well-established process of activation of initiator caspases, especially caspase-9. Both CPAF and caspase-9 are synthesized as zymogens that have low proteolytic activity. Both are activated by what has been shown (caspase-9) or suspected (CPAF) to be autocatalysis, and both form, during physiological activation, complexes of the two subunits derived from intramolecular cleavage. Our analysis shows that they differ in that a cleavage-defective mutant of CPAF is inactive, whereas auto-cleavage defective caspase-9 still can cleave its substrates (Stennicke et al., 1999). A mutant carrying a point mutation in the CPAF processing site was defective in both processing and activity. These results strongly suggest that CPAF is activated by an autocatalytic process. Although we cannot exclude the possibility that a cellular protease is involved in gyrB-CPAF processing, this seems unlikely.
The induced proximity model has been initially proposed to explain the activation of initiator caspases by adaptor-induced clustering during apoptosis. This mode of protease activation is conserved between the nematode Caenorhabditis elegans and mammals, and has been worked out in great detail (Bao and Shi, 2007). Our results suggest that this principle of protease activation is even much older in evolutionary terms, as it is already found in bacteria. CPAF is efficiently translocated from the bacteria-harboring vacuole into the cytosol, although the mechanism of this translocation is unclear. Our data indicate that oligomerization of CPAF is an essential step in its maturation. This is supported by the observation that CPAF secreted from the vacuole into the host cytosol during infection with C. trachomatis was also found in a complex of ∼200 kD, which probably corresponds to 3–4 subunits (unpublished data). When and how CPAF oligomerizes is unclear. One possibility is that a bacterial chaperone binds and translocates CPAF into the host cytosol, during which process the spatial requirements for CPAF activation might be met. It is also conceivable that CPAF may transiently associate with the inclusion membrane at a sufficiently high local concentration to cause its own activation. Initiator caspases are activated by clustering induced by specific adaptors. In the best characterized example, caspase-9 is clustered after heptamerization of its adaptor, Apaf-1 (Bao and Shi, 2007). The existence of a specific bacterial adaptor for CPAF is therefore also possible.
Limited homology between CPAF and Tsp has been observed. Our results show that the Tsp-like active site in C. trachomatis CPAF is required for its activity. Chlamydia has a protein, CT441 in C. trachomatis, that shows higher similarity to known bacterial Tsp and that has recently been characterized (Lad et al., 2007). The role of Tsp in bacteria is not well understood, but these enzymes appear to be involved with proteolytic modifications and degradation of various bacterial proteins (Paetzel and Dalbey, 1997). The conservation of the active site in CPAF might be an indication of evolutionary origin, but the low overall homology suggests that it has assumed other functions. The translocation of CPAF into the host cytosol also indicates that its physiological targets may not be bacterial but host cell proteins.
It is interesting to note that CPAF both has anti-apoptotic activity and induces nonapoptotic cell death, reproducing two salient features of chlamydial infection. Although the relevance of cell death modulation for chlamydial pathogenesis is not known, it is likely that apoptosis inhibition may contribute to the ability of chlamydiae to complete their developmental cycle and perhaps to maintain persistent human infections. During viral infection, the host cell defense often includes the induction of apoptosis through the mitochondrial, BH3-only protein controlled pathway (Everett and McFadden, 2002). Although Chlamydia differs from viruses in important aspects, it shares their dependency on cellular integrity for its replication. The cell's response to chlamydial infection may include the attempt to undergo apoptosis through activation of BH3-only proteins. These proteins are general sensors of cell stress and can be activated to induce apoptosis in many different situations (Strasser, 2005). It is therefore even conceivable that they are activated in response to CPAF activity. CPAF might thus induce apoptosis but at the same time counter it by causing the degradation of BH3-only proteins. In this scenario, as BH3-only proteins are mostly degraded, the net result of the activity of CPAF would be the observed nonapoptotic cell death. CPAF-induced cell death could therefore be a masked form of apoptosis that serves as a cellular defense reaction.
The molecular mechanism for the cell death–inducing activity of CPAF is still unknown. The appearance of lamellar structures in the cell might suggest changes to organelles and intracellular membranes, such as lysosomes and the ER, and their membranes. The ultrastructural changes to these organelles that have been described during chlamydial infection (Todd and Storz, 1975; Todd et al., 1976) are thus likely connected to CPAF activity, and it could be the release of, for instance, lysosomal peptidases, that causes the damage and eventual death of the cell.
All sequenced chlamydial strains have a gene coding for CPAF. The genome of an endosymbiont of free-living amoeba, candidatus protochlamydia amoebophila (UWE25), has recently been sequenced, and even this distantly related bacterium carries a recognizable CPAF homologue (Horn et al., 2004; Collingro et al., 2005). This suggests that CPAF serves a function that is similar for the different species and perhaps even the very different requirements of infection of human cells and amoeba. When first discovered, CPAF's activity to degrade transcription factors required for MHC-expression was noted. Although such a mechanism might contribute to immune evasion by Chlamydia, it appears more likely that this is not the evolutionarily selected function of CPAF, especially not in amoeba lacking MHC. Cytoskeletal structures like the intermediate filament components CK8 and vimentin are good candidates as essential targets of CPAF activity. The host cell cytosol has to accommodate the rapidly growing inclusion, and the disruption of intermediate filaments might facilitate expansion of the inclusion and perhaps eventually the release of the inclusion by lysis or extrusion (Hybiske and Stephens, 2007). Whether CPAF-induced cell death is beneficial for the host or for the bacteria is therefore uncertain at this stage. Either way, our results strongly suggest that CPAF is an important factor in chlamydial pathogenicity, and cellular alterations and responses induced by CPAF might be involved in causing protracted infections.
Materials and methods
Cloning of expression vectors
A fragment encoding for amino acid residues 2–221 of gyrB of E. coli was amplified by PCR and cloned into a pcDNA4/TO/myc-His vector (Invitrogen). Consecutively, two additional gyrB fragments, separated by linker sequences of 16 amino acid residues, were inserted 5′ of the first copy. A sequence coding for a FLAG tag was added 3′ of the gyrB fragments (gyrB construct). The coding sequence of CPAF of C. trachomatis (amino acid residues 18–601) was inserted behind the FLAG-gyrB construct (gyrB-CPAF). CPAF mutants were generated by point mutations using a Stratagene XL Mutagenesis kit. The open reading frame of human cytokeratin 8 (CK8; ATCC #61515) was amplified by PCR, thereby adding a myc-tag coding sequence to the 3′ end and cloned into a pENTR/SD/D-TOPO vector (Invitrogen). Subsequently, the CK8-myc fragment was shuffled into a pcDNA6.2/V5-DEST (CK8-myc construct) by Gateway LR reaction, according to the manufacturer's instructions (Invitrogen).
Cell lines and cell culture
The human embryonic kidney cell line, T-REx-293, and T-REx-HeLa cells, which stably express the tetracycline repressor (Invitrogen), were grown and maintained in humidified air containing 5% CO2 at 37°C in DMEM supplemented with 10% fetal calf serum (tetracycline negative; PAA Laboratories), 50 μg/μl penicillin/streptomycin, and 5 μg/μl blasticidin. T-REx-293 clones stably expressing gyrB-CPAF were generated by electroporation with the construct and antibiotic selection.
Chlamydial infections of T-REx-293 cells
The C. trachomatis strain L2 was obtained from ATCC. Before infection, the culture medium was replaced with DMEM without FCS and antibiotics, and cells were infected at a MOI = 3. After 2 h, 10% FCS were added. At indicated time points, cells were harvested and lysed by incubation with RIPA buffer (1% Triton X-100, 0.5% SDS, 0.5% deoxycholate,1 mM EDTA, 150 mM NaCl, and 50 mM Tris, pH 8.0), supplemented with a protease inhibitor cocktail (Roche).
Transient transfection of T-REx-293 cells and induction of protein expression
Transient transfections of T-REx-293 cells were performed using FuGene HD (Roche), following the manufacturer's instructions. CPAF expression was induced by addition of 0.5 ng/μl anhydrotetracycline (AHT; IBA) for 13–14 h unless otherwise indicated. In some experiments 4 μg/ml tetracycline (TET) was used. For oligomerization experiments 1 μM coumermycin (CM; Sigma-Aldrich) was added 7 h before harvesting to the cells. When indicated, novobiocin (NB; Sigma-Aldrich) was added 30 min before CM addition.
Immunoblotting
Cells were harvested and lysed in Triton X-100 buffer (1% Triton X-100, 1 mM EDTA, 150 mM NaCl, 50 mM Tris, pH 8.0, and protease inhibitor cocktail). Cell extracts were separated using SDS-PAGE and proteins were transferred onto nitrocellulose membranes. Equivalent amounts of protein were loaded and equal loading was confirmed by detection of β-actin or tubulin using specific antibodies (Sigma-Aldrich). Membranes were probed with anti-Bim, anti-cyclin B1, anti-FLAG, anti-myc, anti-PARP, anti-Puma (all from Cell Signaling Technology), anti-Bak, anti-Bcl-2 (both from BD Biosciences), anti-RFX 5, anti-vimentin, anti-CK8 (all three from Acris), or anti-caspase 3 (Abcam) antibodies. Proteins were visualized using peroxidase-conjugated secondary antibodies and a chemoluminescence detection system (GE Healthcare).
Size exclusion chromatography
Cells were lysed in 1% Triton X-100 and 1 mM EDTA in PBS, and lysates were cleared by centrifugation. NB was used at a concentration of 20 μM and was added 30 min before chromatography. All extracts were separated on a Superose 200 gel filtration column (GE Healthcare). Molecular sizes were calculated by plotting the log of the molecular weight of standard marker proteins (Sigma-Aldrich) against their elution volume.
Cell-free cleavage assay
CPAF K6 cells expressing oligomerized CPAF were lysed in NP-40 buffer (1% NP-40, 150 mM NaCl, 1 mM EDTA, and 20 mM MOPS, pH 7.4), and equivalent amounts of the extract were mixed either with PBS as a control or with NP-40 extracts of T-REx-293 cells, transiently transfected with the CK8-myc construct. Extracts were incubated at 37°C for 1 h. CPAF K6 cell extracts were preincubated with the proteasome inhibitors clasto-lactacystin β-lactone (LC; Sigma-Aldrich) or MG-132 (EMD) at 37°C for 20 min before substrate addition.
MTT assay
Cell viability was tested by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma-Aldrich) assay. MTT was added to cells at a concentration of 0.5 mg/μl and incubated at 37°C for 1 h. Generated formazane crystals were dissolved in DMSO, and the OD at a wavelength of 570 nm was measured. As a positive control, cells treated with TNFα (R&D Systems)/cycloheximide (0.5 ng/ml each) for 16 h were used. The caspase inhibitor zVAD-fmk was used at a concentration of 25 μM.
Analysis of cell morphology and detection of apoptosis
Cell morphology was analyzed in culture media by light microscopy at RT (CKX41 inverted microscope, 40x/0.55 lens, U-CMAD3 video adaptor, F-View II Camera, Cell-F Soft Imaging Solution; all from Olympus).
For detection of apoptosis, cells were stained with 1 μg/μl Hoechst 33342 dye (Roche) and incubated for 30 min at 37°C. Cells were harvested, washed with PBS, resuspended in PBS, and embedded in mounting fluid (Labsystems Oy). Nuclei were examined using an Epifluorescence microscope at RT (DMRBE microscope, 40x/0.70 lens, both from Leica; AxioCam MRc camera with AxioVision software; Carl Zeiss, Inc.). Adobe Photoshop and Microsoft PhotoEditor were used to adjust image size and resolution, and to enhance contrast of the whole image for better visibility in some pictures.
Flow cytometry
Caspase-3 activation was detected by flow cytometry analysis. CPAF K6 cells were harvested, fixed in 2% neutral-buffered paraformaldehyde, and permeabilized with 0.5% saponin (Sigma-Aldrich). Active caspase-3 was detected with an anti-active caspase-3 antibody (Abcam) and FITC-conjugated goat anti–rabbit secondary antibody (Dianova). Flow cytometric analysis was performed with a FACSCalibur (Becton Dickinson).
Electron microscopy
Cells were harvested and fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4; Electron Microscopy Sciences) and embedded in epoxy resin (epon 812; Electron Microscopy Sciences). Ultrathin sections were examined with an EM 10 CR transmission electron microscope (Carl Zeiss, Inc.). For image acquisition, a MegaView III camera system (Olympus) was used.
Online supplemental material
Fig. S1 shows the morphological changes in T-REx-293 and T-REx-HeLa cells, respectively, due to transient transfection of CPAF. Fig. S2 compares the changes in cellular morphology between infection with C. trachomatis and the expression of CPAF in CPAF K6 cells. Fig. S3 describes the cell viability of CPAF K6 cells after CPAF induction measured by propidium iodide uptake. Fig. S4 shows nuclear morphology after CPAF expression or infection with C. trachomatis at higher magnification. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200804023/DC1.
Supplementary Material
Acknowledgments
We thank Florian Schmidt for constructing gyrB-CPAF, Dr. Silke Fischer for help with the initial experiments, and Luise Jennen for help with electron microscopy.
This work was supported by the Else Kröner-Fresenius-Stiftung. J.G. Christian is supported by a consortium of the ERA-NET of the European Union (ECIBUG).
Abbreviations used in this paper: AHT, anhydrotetracycline; CHX, cycloheximide; CK8, cytokeratin 8; CM, coumermycin; CPAF, chlamydial protease-like activity factor; gyrB-CPAF, N-FLAG-3xgyrB-CPAF construct; NB, novobiocin; PARP, poly (ADP-ribose) polymerase; TET, tetracycline; TNF, tumor necrosis factor; Tsp, tail-specific protease; zVAD, zVAD-fmk.
References
- Balsara, Z.R., S. Misaghi, J.N. Lafave, and M.N. Starnbach. 2006. Chlamydia trachomatis infection induces cleavage of the mitotic cyclin B1. Infect. Immun. 74:5602–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, Q., and Y. Shi. 2007. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 14:56–65. [DOI] [PubMed] [Google Scholar]
- Belland, R.J., M.A. Scidmore, D.D. Crane, D.M. Hogan, W. Whitmire, G. McClarty, and H.D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. USA. 98:13984–13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, J.H., S. Hughes, D. Hogan, G. Cieplak, D.E. Sturdevant, G. McClarty, H.D. Caldwell, and R.J. Belland. 2004. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 72:7063–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, G.T., and J.W. Moulder. 1978. Loss of inorganic ions from host cells infected with Chlamydia psittaci. Infect. Immun. 19:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro, A., E.R. Toenshoff, M.W. Taylor, T.R. Fritsche, M. Wagner, and M. Horn. 2005. ‘Candidatus Protochlamydia amoebophila’, an endosymbiont of Acanthamoeba spp. Int. J. Syst. Evol. Microbiol. 55:1863–1866. [DOI] [PubMed] [Google Scholar]
- Dong, F., M. Pirbhai, Y. Zhong, and G. Zhong. 2004. a. Cleavage-dependent activation of a chlamydia-secreted protease. Mol. Microbiol. 52:1487–1494. [DOI] [PubMed] [Google Scholar]
- Dong, F., J. Sharma, Y. Xiao, Y. Zhong, and G. Zhong. 2004. b. Intramolecular dimerization is required for the chlamydia-secreted protease CPAF to degrade host transcriptional factors. Infect. Immun. 72:3869–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., H. Su, Y. Huang, Y. Zhong, and G. Zhong. 2004. c. Cleavage of host keratin 8 by a Chlamydia-secreted protease. Infect. Immun. 72:3863–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett, H., and G. McFadden. 2002. Poxviruses and apoptosis: a time to die. Curr. Opin. Microbiol. 5:395–402. [DOI] [PubMed] [Google Scholar]
- Farrar, M.A., I. Alberol, and R.M. Perlmutter. 1996. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 383:178–181. [DOI] [PubMed] [Google Scholar]
- Fields, K.A., and T. Hackstadt. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18:221–245. [DOI] [PubMed] [Google Scholar]
- Fischer, S.F., J. Vier, S. Kirschnek, A. Klos, S. Hess, S. Ying, and G. Hacker. 2004. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J. Exp. Med. 200:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis, R.R. 1972. Interaction of L cells and Chlamydia psittaci: entry of the parasite and host responses to its development. J. Bacteriol. 110:706–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker, G., and A. Weber. 2007. BH3-only proteins trigger cytochrome c release, but how? Arch. Biochem. Biophys. 462:150–155. [DOI] [PubMed] [Google Scholar]
- Hacker, H., V. Redecke, B. Blagoev, I. Kratchmarova, L.C. Hsu, G.G. Wang, M.P. Kamps, E. Raz, H. Wagner, G. Hacker, et al. 2006. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 439:204–207. [DOI] [PubMed] [Google Scholar]
- Horn, M., A. Collingro, S. Schmitz-Esser, C.L. Beier, U. Purkhold, B. Fartmann, P. Brandt, G.J. Nyakatura, M. Droege, D. Frishman, et al. 2004. Illuminating the evolutionary history of chlamydiae. Science. 304:728–730. [DOI] [PubMed] [Google Scholar]
- Hybiske, K., and R.S. Stephens. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA. 104:11430–11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawana, K., A.J. Quayle, M. Ficarra, J.A. Ibana, L. Shen, Y. Kawana, H. Yang, L. Marrero, S. Yavagal, S.J. Greene, et al. 2007. CD1d degradation in Chlamydia trachomatis-infected epithelial cells is the result of both cellular and chlamydial proteasomal activity. J. Biol. Chem. 282:7368–7375. [DOI] [PubMed] [Google Scholar]
- Lad, S.P., G. Yang, D.A. Scott, G. Wang, P. Nair, J. Mathison, V.S. Reddy, and E. Li. 2007. Chlamydial CT441 is a PDZ domain-containing tail-specific protease that interferes with the NF-kappaB pathway of immune response. J. Bacteriol. 189:6619–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin, R.A., and Z. Zakeri. 2004. Caspase-independent cell death? Oncogene. 23:2766–2773. [DOI] [PubMed] [Google Scholar]
- McClarty, G. 1994. Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol. 2:157–164. [DOI] [PubMed] [Google Scholar]
- Nicholson, D.W., A. Ali, N.A. Thornberry, J.P. Vaillancourt, C.K. Ding, M. Gallant, Y. Gareau, P.R. Griffin, M. Labelle, Y.A. Lazebnik, et al. 1995. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 376:37–43. [DOI] [PubMed] [Google Scholar]
- Ojcius, D.M., P. Souque, J.L. Perfettini, and A. Dautry-Varsat. 1998. Apoptosis of epithelial cells and macrophages due to infection with the obligate intracellular pathogen Chlamydia psittaci. J. Immunol. 161:4220–4226. [PubMed] [Google Scholar]
- Paetzel, M., and R.E. Dalbey. 1997. Catalytic hydroxyl/amine dyads within serine proteases. Trends Biochem. Sci. 22:28–31. [DOI] [PubMed] [Google Scholar]
- Perfettini, J.L., J.C. Reed, N. Israel, J.C. Martinou, A. Dautry-Varsat, and D.M. Ojcius. 2002. Role of Bcl-2 family members in caspase-independent apoptosis during Chlamydia infection. Infect. Immun. 70:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, J., D.P. Wilson, G. Myers, P. Timms, and P.M. Bavoil. 2007. Type III secretion a la Chlamydia. Trends Microbiol. 15:241–251. [DOI] [PubMed] [Google Scholar]
- Pirbhai, M., F. Dong, Y. Zhong, K.Z. Pan, and G. Zhong. 2006. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J. Biol. Chem. 281:31495–31501. [DOI] [PubMed] [Google Scholar]
- Pop, C., J. Timmer, S. Sperandio, and G.S. Salvesen. 2006. The apoptosome activates caspase-9 by dimerization. Mol. Cell. 22:269–275. [DOI] [PubMed] [Google Scholar]
- Salvesen, G.S., and V.M. Dixit. 1999. Caspase activation: the induced-proximity model. Proc. Natl. Acad. Sci. USA. 96:10964–10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, A.C., B.B. Vandahl, M.R. Larsen, P. Roepstorff, K. Gevaert, J. Vandekerckhove, G. Christiansen, and S. Birkelund. 2002. Characterization of a secreted Chlamydia protease. Cell. Microbiol. 4:411–424. [DOI] [PubMed] [Google Scholar]
- Silber, K.R., K.C. Keiler, and R.T. Sauer. 1992. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc. Natl. Acad. Sci. USA. 89:295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke, H.R., Q.L. Deveraux, E.W. Humke, J.C. Reed, V.M. Dixit, and G.S. Salvesen. 1999. Caspase-9 can be activated without proteolytic processing. J. Biol. Chem. 274:8359–8362. [DOI] [PubMed] [Google Scholar]
- Strasser, A. 2005. The role of BH3-only proteins in the immune system. Nat. Rev. Immunol. 5:189–200. [DOI] [PubMed] [Google Scholar]
- Todd, W.J., and J. Storz. 1975. Ultrastructural cytochemical evidence for the activation of lysosomes in the cytocidal effect of Chlamydia psittaci. Infect. Immun. 12:638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, W.J., A.M. Doughri, and J. Storz. 1976. Ultrastructural changes in host cellular organelles in the course of the chlamydial developmental cycle. Zentralbl. Bakteriol. (Orig A) 236:359–373. [PubMed] [Google Scholar]
- Wilhelm, S., and G. Hacker. 1999. Proteolytic specificity of caspases is required to signal the appearance of apoptotic morphology. Eur. J. Cell Biol. 78:127–133. [DOI] [PubMed] [Google Scholar]
- Wyrick, P.B. 2000. Intracellular survival by Chlamydia. Cell. Microbiol. 2:275–282. [DOI] [PubMed] [Google Scholar]
- Wyrick, P.B., E.A. Brownridge, and B.E. Ivins. 1978. Interaction of Chlamydia psittaci with mouse peritoneal macrophages. Infect. Immun. 19:1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, S., B.M. Seiffert, G. Hacker, and S.F. Fischer. 2005. Broad degradation of proapoptotic proteins with the conserved Bcl-2 homology domain 3 during infection with Chlamydia trachomatis. Infect. Immun. 73:1399–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, S., S.F. Fischer, M. Pettengill, D. Conte, S.A. Paschen, D.M. Ojcius, and G. Hacker. 2006. Characterization of host cell death induced by Chlamydia trachomatis. Infect. Immun. 74:6057–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.