Abstract
Although odorants are known to activate olfactory receptor neurons through cAMP, the long-term effects of odorant detection are not known. Our recent findings indicate that there is also a delayed and sustained cAMP response, with kinetics sufficient to mediate long-term cellular responses. This cAMP response is mediated by cGMP through activation of adenylyl cyclase by protein kinase G (PKG). Therefore, we investigated the ability of odorants to regulate gene expression in rat olfactory epithelium. The cAMP-responsive binding protein (CREB) is a well-characterized transcription factor regulated by cAMP. We examined CREB activity in rat olfactory epithelium and olfactory receptor neurons (ORNs) after stimulation with odorants. Odorants increased levels of phosphorylated CREB in olfactory epithelium in vivo, and this increase was localized to ORNs in vitro. Incubation with 8-bromo-cGMP or sodium nitroprusside, a guanylyl cyclase activator, also increased phosphorylated CREB. In vitro, cAMP-dependent protein kinase phosphorylated CREB. In contrast, PKG failed to phosphorylate CREB directly in vitro. Our results demonstrate that the delayed odorant-induced cAMP signal activates CREB, which in turn may modulate gene expression in ORNs. In addition, cGMP indirectly affects CREB activation. This effect of cGMP on CREB activity through cAMP provides another mechanism for the modulation of CREB.
cAMP is a key second messenger in odorant detection in olfactory receptor neurons (ORNs). Many aspects of the mechanism of odor detection in the sensory cilia of ORNs are well established from patch clamp recordings (1) and molecular-cloning studies (2–5). Signal transduction is initiated when odorants interact with specific receptors on cilia (6–9). Receptors subsequently couple to a G protein, which activates adenylyl cyclase (10–12). cAMP levels increase and open a cyclic nucleotide-gated channel, resulting in an influx of Na+ and Ca2+ (1, 13). The immediate response is the generation of a graded receptor potential (14, 15). In addition to signaling involved in immediate stimulus detection and generation of action potential, a delayed response to odorants has also been reported (16–18). This delayed and sustained response cannot mediate odorant detection, but may influence neuronal survival, axon outgrowth, or changes in synaptic strength (19).
In addition to cAMP, recent studies demonstrated an odorant-induced cGMP response in primary cultures of rat ORNs (16, 20) and in cilia (17, 21). Odorants elevated cGMP levels in a slow and sustained manner. These kinetics suggested that cGMP was not involved in initial signaling events, but rather in desensitization, or in other long-term activity-dependent effects (18, 22). We have shown subsequently that the odorant-induced cGMP response mediated the delayed cAMP response by activation of cGMP-dependent protein kinase (PKG) (17). We postulated that the cGMP-mediated delayed cAMP response might regulate long-term cellular responses to odorant detection, including gene expression. Precedent for this pathway exists. PKG was demonstrated to regulate c-fos induction in a baby hamster kidney cell line transfected with a human fos promoter-chloramphenicol acetyltransferase reporter construct (23). However, neither the nuclear localization of PKG nor direct evidence for the phosphorylation of the cAMP-responsive element (CRE) binding protein (CREB) by PKG was demonstrated.
Although not investigated previously in primary sensory neurons, the concept that stimulus detection from extracellular signals, such as hormones, growth factors, and neuronal activity, modulates transcriptional events to produce long-term changes in cellular activity is well-established (24). cAMP is central to this pathway (25, 26). Persistent activation of cAMP-dependent protein kinases is suggested to be critical for establishing long-term synaptic facilitation (27). In many cases, changes in transcription occur through the reversible phosphorylation of transcription factors (28–30). One of the most studied transcription factors is the CREB (26, 31, 32). CREB is a multifunctional transcription factor important for neurotransmitter and growth factor regulation of gene expression and can be phosphorylated by protein kinase A, calmodulin-dependent kinases, or growth factor-sensitive kinases, and may serve to permit signal convergence from different pathways.
In this study, we investigated whether the odorant-induced cAMP signal could regulate CREB phosphorylation in olfactory epithelium in vivo and in ORNs in vitro. We further studied the mechanism by which odorant-induced second messenger responses might be coupled to CREB activation. As our previous results indicated that cGMP was a mediator of the delayed cAMP response, we examined the role of cGMP in the activation of CREB in ORNs.
Materials and Methods
Abs.
Rabbit anti-phosphorylated CREB (anti-pCREB).
Anti-pCREB Ab was a gift from D. D. Ginty at Johns Hopkins University (Baltimore, MD). The anti-pCREB Ab was generated against a synthetic phosphopeptide corresponding to residues 123–136 [KRREILSRRP(pS)YRK] of rat CREB, and affinity-purified. This Ab specifically recognizes the phosphorylated form of CREB at Ser-133.
Mouse anti-β-tubulin (βIII).
βIII Ab was purchased from Babco (Richmond, CA). The Ab (clone TuJ1) was raised against microtubules derived from rat brain and was affinity-purified (33). This Ab is highly reactive to neuron-specific class III β-tubulin (NST), but does not identify β-tubulin found in glial cells. Our previous studies determined that TuJ1 specifically recognized ORNs in olfactory epithelium (34) and in primary cultures of ORNs (35).
CRE Probe.
The CRE probe was obtained from D. D. Ginty. The sequence of the double-stranded DNA CRE probe is: 5′-GTCAGTCGTGAC⋅GTCAATCGGTCA-3′. The DNA-binding sites are indicated in boldface. The probe was labeled at 5′ termini with T4 polynucleotide kinase (United States Biochemical) and [γ-32P]ATP (50 μCi).
Animal Treatment for in Vivo Odorant-Induced CREB Phosphorylation.
Two rats were isolated in separate cages [40 × 24 × 15 cm (width × depth × height)] and exposed to the vapor phase of an odorant mixture (isobutylmethoxypyrazine (IBMP), citralva, and isovaleric acid; 1 mM each dissolved in 2% ethanol) or an equal volume of 2% ethanol solution for 30 min. The nasal turbinates of each rat were dissected immediately and then subjected to SDS/PAGE. pCREB was visualized by Western blot analysis.
Primary Culture of ORNs.
Cultures were prepared as previously described (36) with some modifications. Cells were plated at a density of 3 × 105 cells per cm2 into tissue culture dishes (Falcon) or Labtek tissue culture slides (Nunc) coated with MEM containing d-valine (MDV; GIBCO) and 25 μg/ml laminin (Collaborative Research). Cultures were placed in a humidified 37°C incubator receiving 5% CO2. On day 2 and every day thereafter, cells were fed with MDV containing 15% dialyzed FCS (GIBCO), gentamicin, kanamycin, and 25 ng/ml nerve growth factor (NGF). Two days prior to use, the culture medium was changed to a similar formulation, except that it contained 2.5 ng/ml NGF.
Immunofluorescence.
Immunofluorescence was performed as previously described (37) with modifications. Primary cultures of ORNs in two chamber slides were fixed with an ice-cold mixture of methanol:acetone (1:1) for 20 min at −20°C. After rinsing three times with PBS, cells were blocked with 4% normal donkey serum in 3% BSA for 60 min at room temperature, and incubated with rabbit anti-pCREB (1:2000) and mouse anti-NST (1:1000) Abs overnight at 4°C. On the next day, cells were incubated with donkey anti-rabbit fluorescein-conjugated Ab (Jackson ImmunoResearch) (1:50) for 1 hr at room temperature. After washing three times with PBS, cells were incubated with donkey anti-mouse rhodamine-conjugated Ab (Jackson ImmunoResearch) (1:100) for 1 hr at room temperature. The slides were rinsed with PBS three times and mounted with Aquapolymount (Polysciences).
Gel Electrophoresis and Western Blot Analysis.
Western blot analyses were performed according to Laemmli (38) and Towbin (39) with modifications. Total nasal turbinates of an adult rat (200 μg) or total lysates of primary cultures were solubilized in SDS sample buffer containing 100 mM Tris⋅HCl (pH 6.8), 2% SDS, 10% 2-mercaptoethanol, and 20% glycerol and subjected to SDS/PAGE on a 10% gel. The separated proteins were transferred to nitrocellulose membrane (BA-S 83, 0.2 μm; Schleicher & Schuell), and the membrane was probed with primary Ab at the following Ab concentrations: mouse anti-NST, 1:1000; rabbit anti-pCREB, 1:3000. The secondary Abs were horseradish peroxidase-conjugated goat anti-rabbit Ig (Roche Molecular Biochemicals) used at 1:10,000 dilution or horse anti-mouse Ig (Roche Molecular Biochemicals) used at 1:5,000 dilution. The immunoblots were detected by using chemiluminescence reagent (DuPont). The molecular weight sizes are indicated on the left side of the gels. Bands were visualized by using the enhanced chemiluminescence (Amersham Pharmacia) reagents after exposing the blots to X-Omat film (Kodak). Western blots were scanned and quantitated by densitometer (Personal Densitometer SI; Molecular Dynamics) and the ImageQuant analysis program (ImageQuant v1.2; Molecular Dynamics).
Preparation of Nuclear Extracts.
Nuclear extracts were isolated from primary cultures of ORNs according to the method of Andrews and Faller (40) with 1 mM Na3VO4, 10 μg/ml leupeptin, 5 μg/ml pepstatin A, and 10 μg/ml aprotinin. Samples were divided into aliquots and stored at −70°C until use. Protein concentrations were determined with protein assay kit (Bio-Rad).
Gel Retardation and Supershift Assay.
DNA–protein reaction mixtures (24 μl) contained the following reagents: buffer [10 mM Hepes/10% glycerol/50 mM KCl/0.05 mM EDTA/0.125 mM PMSF (pH 7.9)], 10 μg of nuclear extract, 1 μg of poly[d(I-C)], and radiolabeled probe (106 cpm). Mixtures were incubated at room temperature for 15 min. For supershift assays, nuclear extracts were mixed with appropriate Abs: anti-CREB, anti-pCREB, or preimmune anti-CREB at dilutions of 1:25. The mixtures were incubated at room temperature for an additional 15 min. Samples were loaded onto nondenaturing 4% polyacrylamide gels (3.3% bisacrylamide). After running, the gels were dried and exposed to a PhosphoImaging screen (Molecular Dynamics). The results were saved on computer for further analysis. For permanent records, the gels were also exposed to X-Omat films (Kodak) with one intensifying screen.
In Vitro Phosphorylation.
Phosphorylation reactions were done according to the method of Steiner et al. (41) with modifications. Total primary cultures were scraped from culture dishes and collected in lysis buffer [50 mM Tris/0.5% SDS/200 μg/ml trazalol/2% 2-mercaptoethanol (pH 8.0)]. The cells were lysed in lysis buffer for 1 hr on ice. The lysed cells were diluted in RIPA1 [50 mM Tris/150 mM NaCl/10 mM EDTA/2 mM NaF/1 mM ammonium molybdate/20 μg/ml PMSF/40 μg/ml aprotinin/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS (pH 7.5)] and centrifuged at 100,000 × g for 15 min. The supernatants were collected and incubated with one of the protein kinases for 30 min at room temperature in 200 μl of a reaction buffer containing 50 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 0.4 mM EGTA, 1 mM ATP, and 0.5 μCi (18.5 kBq) of [γ-32P]ATP. Protein kinases were purchased from Promega. cAMP-dependent protein kinase (PKA) catalytic subunits were used at 300 units per reaction. One unit is defined as the amount of enzyme required to incorporate 1 pmol of phosphate into the substrate in 1 min. To inhibit PKA activity, 0.5 μM KT5720 (PKA inhibitor) was mixed with PKA catalytic subunit, and then applied. PKG from Promega is an α isozyme of PKG purified from bovine lungs. PKG (brain) was purified from rat brain tissues in our laboratory. In contrast to the catalytic subunit of PKA, PKG requires an appropriate stimulator to be converted to active PKG. Therefore, 2 μM 8-bromo-cGMP (8-br-cGMP) was added to 300 units of PKG per reaction. In all cases, the activities of the kinases were verified by incubation of the kinase with a known substrate for that kinase. The samples were then subjected to immunoprecipitation.
Immunoprecipitation of CREB.
Immunoprecipitation of CREB from primary culture of rat ORNs was done according to the method of Ginty et al. (42) with modifications. The phosphorylated samples were incubated with rabbit anti-CREB Ab (1:200) for 1 hr on ice. Protein A-Sepharose was added and incubated on ice for an additional hour. The immune complexes were centrifuged in a microcentrifuge, and the pellets were recovered. The pellets were rinsed twice with RIPA2 [50 mM Tris/0.5 M LiCl/0.5% Nonidet P-40 (pH 7.4)] and once with RIPA3 [10 mM Tris/0.5 M LiCl/0.5% Nonidet P-40 (pH 7.4)]. The immune complex samples were added to the same volume of SDS sample buffer containing 100 mM Tris⋅HCl (pH 6.8), 2% SDS, 10% 2-mercaptoethanol, and 20% glycerol. The samples were boiled and subjected to SDS/PAGE with a 10% acrylamide gel. The gels were vacuum dried and exposed to X-Omat film (Kodak).
Purification and Assay of PKG.
For PKG phosphorylation assays, PKG was purified from rat brains according to Francis et al. (43) and Corbin and Doskeland (44) with modifications. Total rat brain tissues (50 g) were combined with 200 ml of buffer A [10 mM KH2PO4/1 mM EDTA/25 mM 2-mercaptoethanol (pH 6.8)] and homogenized. The homogenized tissues were centrifuged at 12,000 × g for 30 min and filtered through a glass wool-filled column. The filtered supernatant was mixed with 20 ml of DE-52 cellulose (Whatman) and incubated for 2 hr in an end-to-end shaker. The DE-52 cellulose was isolated by using a glass Buchner funnel with a fritted disk (coarse) under a vacuum pump. The resin was washed with 1 liter of buffer A and packed in a glass column. After a wash with 4 liters of buffer A, proteins were eluted with 300 ml of buffer A containing 300 mM NaCl. The eluates were applied in preequilibrated column packed with cAMP-agarose (Sigma) in buffer A. The cAMP-agarose resin was washed sequentially with 200 ml of buffer A containing 2 M NaCl and 10 ml of buffer A containing 10 mM AMP. PKG was eluted by applying buffer A containing 10 mM cAMP.
The eluted PKG was assayed for activity. Ten-microliter aliquots of the eluted PKG were mixed with 50 μl of assay solution [20 mM Tris/200 μM ATP/136 μg/ml PKG substrate/20 mM MgCl2/100 μM 3-isobutyl-1-methylxanthine/1 μM PKA inhibitor/30,000 cpm/μl [γ-32P]ATP (pH 7.4)] and allowed to incubate for 10 min at 30°C. The phosphorylation reaction was terminated by spotting onto nitrocellulose filter papers (HAWP-0.45 μm; Millipore). The nitrocellulose filter papers were washed four times with 75 mM phosphoric acid. The filter papers were rinsed with 100% ethanol and allowed to dry. The dried filters were placed into vials filled with 8 ml of scintillation fluid and their radioactivity was measured in a scintillation counter (Beckman LS6000SC).
Results
Odorants Increase pCREB in Olfactory Tissue in Vivo.
Our recent findings showed that odorants induced a delayed cAMP signal that may mediate activity-driven cellular responses. Because CREB is a transcription factor regulated by several messengers, including cAMP, we determined whether odorants could influence the phosphorylation of CREB in vivo in the olfactory epithelium (Fig. 1).
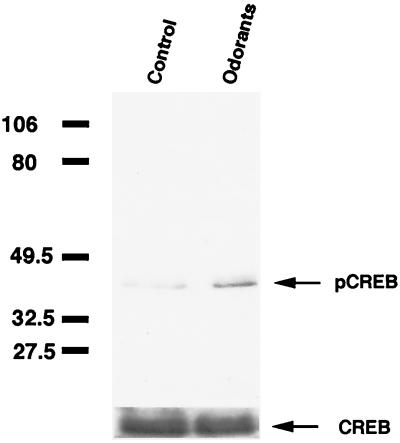
Figure 1.
Phosphorylation of CREB by application of odorants. Western blot analysis of pCREB and total CREB levels in rat nasal turbinates after a 15-min stimulation with odorant mixtures (IBMP, citralva, isovaleric acid; 1 mM each) in a vapor phase. Arrows point to the 43-kDa immunoreactive pCREB and CREB proteins. The molecular mass (in kDa) is indicated at left.
When rats were exposed to an odorant mixture (IBMP, citralva, and isovaleric acid), the amount of pCREB was increased in odorant-stimulated rat nasal tissue, although a basal level of pCREB was detectable (Fig. 1). To standardize gel loading and the relative increase in immunoreactivity by odorant application, samples were immunoblotted with anti-pCREB Ab, stripped, and reprobed with anti-CREB Ab. The increase in pCREB immunoreactivity in turbinates isolated from individual rats that were exposed to these three odorants ranged from 16% to 63% of control.
Odorant-Induced Increases in pCREB Localized to ORNs.
To determine the localization of the odorant-induced increases in pCREB, primary cultures of ORNs were exposed for 15 min to odorants (IBMP, citralva, and isovaleric acid) and then double-labeled with polyclonal anti-pCREB and monoclonal anti-NST Abs (Fig. 2). Immunoreactivities were visualized by using rhodamine-conjugated anti-mouse IgG and fluorescein-conjugated anti-rabbit IgG. Images were overlaid so that regions expressing both pCREB and NST appeared yellow. The numbers of pCREB-positive cells among NST-positive cells were determined.
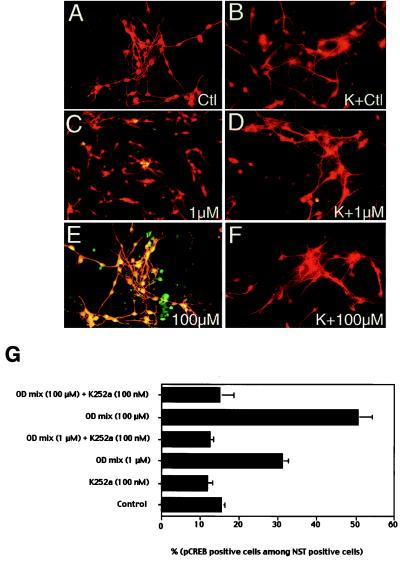
Figure 2.
Double immunofluorescence with anti-pCREB and anti-NST in primary cultures of ORNs treated with odorants. (A–F) Primary cultures were incubated with MEM/d-Val for 1.5 hr and then switched to medium (A, C, and E) or medium containing 100 nM K252a (B, D, and F) for 30 min before odorant stimulation. Concentrations of odorant mixtures that were applied for 15 min are indicated in each panel. K, K252a preincubation. pCREB and NST were visualized by immunofluorescence. pCREB was localized by fluorescein-conjugated anti-rabbit Ab (green) and NST was localized by rhodamine-conjugated anti-mouse Ab (red). Colocalization is visualized as yellow. (G) Quantitative analyses of odorant (OD)-induced pCREB increase in ORNs. Relative pCREB immunoreactivity indicates pCREB-positive cells among NST-positive cells. Cells were counted from at least six randomly selected areas of each slide from three independent experiments. SEM is represented as line bars.
Stimulation of cells with odorant mixtures resulted in a significant increase in the number of pCREB-positive cells/NST-positive cells (Fig. 2 A, C, and E). At baseline, pCREB immunoreactivity was detected in a percentage of cells, although this immunostaining for pCREB was not as intense as in odorant-treated cells. These neurons that had low levels of pCREB-immunoreactivity were still scored as positive. The number of pCREB-positive cells was increased almost 2-fold by exposure of cells to a 1 μM total concentration mixture of odorants (199%, P < 0.001) (Fig. 2G). A higher concentration of odorant mixture (100 μM) further increased the number of pCREB-positive cells more than 3-fold (324%, P < 0.001) (Fig. 2G). About 50% of the NST-positive cells were positive for pCREB when cells were exposed to 100 μM odorant mixtures (Fig. 2G). To determine whether this staining depended on protein kinase activity, cells were incubated with K252a, a cell-permeant protein kinase inhibitor (Fig. 2 B, D, and F) (45, 46). The increase in pCREB by odorants could be completely blocked by preincubation with K252a (P < 0.001, in both conditions). The basal levels of CREB phosphorylation observed (16% ± 0.83%) were also blocked by incubation with K252a by 24% (P < 0.05). Nuclei containing phosphorylated CREB in NST-negative cells were also observed. These cells may represent mature ORNs that have a decreased amount of NST, consistent with our previous in vivo observation that NST immunoreactivity decreases with ORN maturation (34).
Odorants Phosphorylate CREB in Primary Culture of ORNs in Time-Dependent and Dose-Dependent Manners.
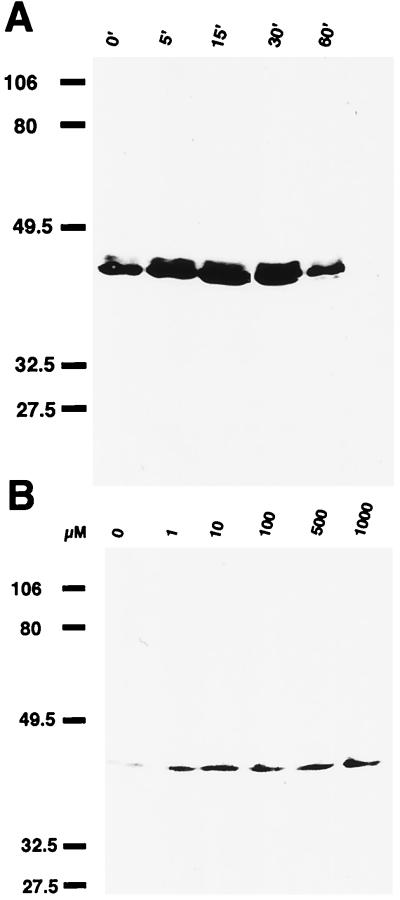
To determine whether the changes in pCREB seen in ORNs in response to the odorants occurred in time- and dose-dependent manners, primary cultures of ORNs were incubated for 0–60 min in the presence of 1 μM IBMP (Fig. 3A). Incubations were terminated by addition of boiling SDS sample buffer. Samples at each incubation period were subjected to immunoblot analysis with anti-pCREB Ab. pCREB levels increased significantly over 5–30 min of incubation with IBMP, although discernible basal levels of pCREB were seen, as discussed previously. The pCREB levels decreased by 60 min (Fig. 3A). A dose–response curve was also determined (Fig. 3B). Primary cultures were incubated with a range of IBMP concentrations, from 1 μM to 1 mM, for 30 min. Incubations were quenched by addition of boiling SDS buffer. Levels of pCREB increased with IBMP concentration (Fig. 3B), demonstrating that odorants could induce phosphorylation of CREB in a dose-dependent manner. Similar experiments were done with PC-12 cells exposed to odorants. No increases in CREB phosphorylation were detected with odorant treatment of PC-12 cells (data not shown).
Figure 3.
Time course and dose-dependent curve of odorant-induced pCREB increase in primary cultures of ORNs. (A) Time course of pCREB induction by odorants. Primary cultures of ORN were incubated with 1 μM IBMP for various time periods. Incubation periods (min) are indicated above. The molecular mass (in kDa) is indicated at left. (B) Dose response of pCREB induction by odorants. Primary cultures were incubated for 30 min with IBMP in range of 1 μM to 1 mM. Odorant concentrations were indicated above. The molecular mass (in kDa) is indicated at left.
Anti-pCREB Shifts the CRE–CREB Binding Complex in Nuclear Extracts of Odorant-Stimulated ORNs.
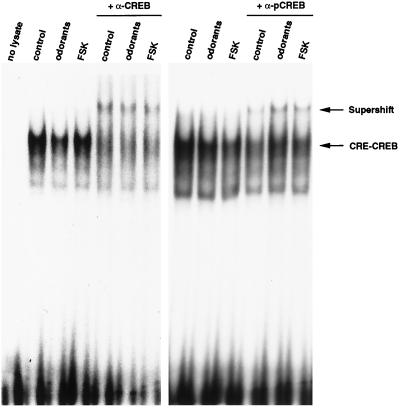
CREB-mediated changes in gene expression are dependent on the Ser-133 phosphorylation status of CREB, which is constitutively bound to CRE sites (30). We determined whether the odorant-induced increase in pCREB in ORNs represented an increase in CREB bound to DNA. Nuclear extracts from control, forskolin, and odorant-treated ORN primary cultures were prepared and subjected to the gel shift assay with radiolabeled CRE as a probe (Fig. 4). Forskolin, a strong adenylyl cyclase activator that in turn activates PKA, was used as a positive control. The same amount of CREB, as visualized by supershift with anti-CREB Ab, was bound to CRE in every treatment condition (Fig. 4 Left). To determine whether odorant can activate CREB by phosphorylation of CREB at Ser-133 and in turn convert CREB into a transcriptionally active form, Ab specific to pCREBSer133 was incubated with the prebound CRE–CREB complexes. Consistent with our previous data, 13% of CREB was phosphorylated at Ser-133 residues and bound to DNA in the basal state (Fig. 4 Right). About 45% of CREB was phosphorylated on Ser-133 residues in the presence of odorants, as determined by density comparison between CRE–CREB complex and supershifted complex, based on densitometer readings. Forskolin induced a 46% increase in CREB phosphorylation, as detected in the CRE–CREB complex.
Figure 4.
Binding to the CRE by nuclear extract of odorant- or forskolin (FSK)-treated primary ORN cultures. Nuclear extracts from control, odorant, or forskolin-treated cells were prepared as described in Materials and Methods. The same amounts (10 μg) of total lysates were incubated with radiolabeled CRE probe for 15 min. For supershift assay, CRE–CREB complexes were further incubated with specific to anti-CREB or anti-pCREB Abs (α) or with preimmune serum (not shown). The CRE–CREB complex and supershifted complex are indicated by arrows. Results are representative of three independent experiments.
PKA, but Not PKG, Directly Mediates the Phosphorylation of CREB in ORNs.
Our previous study suggested that the delayed cAMP response is mediated by cGMP by means of the activation of PKG (17). We hypothesized that the cGMP-mediated cAMP response may be involved with long-term cellular responses to stimulation, such as gene expression. Therefore, we examined the role of cGMP in the odorant-induced activation of CREB. Primary cultures were incubated with 10 μM 8-br-cGMP for 30 min, and the incubation was terminated by addition of boiling SDS buffer. The samples were immunoblotted with anti-pCREB Ab. Incubation with 8-br-cGMP increased pCREB levels, whereas CREB levels remained constant (Fig. 5A). Because the existence of soluble guanylyl cyclase and a functional cGMP pathway in ORNs had been reported earlier (16, 21, 37, 47), we examined whether an NO donor could mimic the effects of 8-br-cGMP. We incubated primary cultures with 100 μM sodium nitroprusside, a potent NO donor. NO, in turn, stimulates soluble guanylyl cyclases to generate cGMP. Incubation with sodium nitroprusside for 30 min increased the pCREB level (Fig. 5A). Taken together, these results indicated that cGMP can induce the activation of CREB.
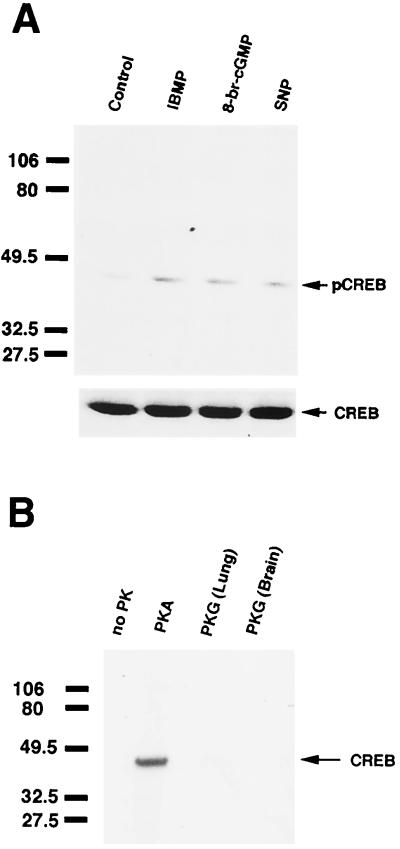
Figure 5.
cGMP-induced pCREB production in primary cultures of ORNs. (A) Western blot analysis of pCREB and CREB in primary cultures of ORNs. Cells were stimulated with 10 μM 8-br-cGMP or 100 μM sodium nitroprusside (SNP). IBMP was applied at 100 μM. Arrows indicate the locations of pCREB and CREB. (B) In vitro phosphorylation of CREB in primary cultures of ORNs. The protein kinases utilized are indicated above the lanes. PKA denotes addition of 300 units of PKA catalytic subunits. “PKG (Lung)” indicates incubation with the α isozyme of PKG purified from bovine lungs. “PKG (Brain)” indicates the addition of PKG purified from rat brain. A total of 2 μM 8-br-cGMP was added to activate PKG. Arrows point to the 43-kDa CREB proteins labeled by 32P. The molecular mass (in kDa) is indicated at left.
To determine whether the effect of cGMP on CREB was direct or indirect, CREB from primary cultures of ORNs was phosphorylated in vitro by PKA or PKG and then immunoprecipitated with anti-CREB Ab. Addition of the catalytic subunit of PKA (300 units) phosphorylated CREB (Fig. 5B), and this phosphorylation was inhibited by incubation with 0.5 μM KT5720, a specific PKA inhibitor (data not shown). For PKG-mediated phosphorylation of CREB, we tested two different PKGs. One was purified from bovine lung, and the other was purified from rat brain. Neither of the PKGs phosphorylated CREB obtained from primary cultures of ORNs (Fig. 5B), suggesting that the effect of cGMP on CREB phosphorylation was indirect. Thus, cGMP may modulate other intermediates in the pathway (possibly the cAMP pathway in olfaction) to lead ultimately to CREB phosphorylation.
Discussion
The main finding of these experiments is that odorants induce phosphorylation of CREB in olfactory turbinates in vivo and in ORNs in vitro. Furthermore, our data implicate a role for cGMP in this pathway. Studies in many systems have demonstrated that signaling pathways that elevate cAMP levels can modulate gene expression (see reviews, refs. 26, 48, and 49). The activation of the cAMP cascade by odorants is well established (see review, ref. 50), but information concerning any long-term effects of odorant detection or odorant-induced cAMP elevations in ORNs is lacking. Here, we demonstrate that the detection of odorants, in a vapor phase, induced CREB phosphorylation in nasal turbinates. This effect was localized to ORNs in culture and suggests that odorant detection can ultimately modulate the regulation of gene expression in these primary sensory neurons. Despite its widespread localization in neurons, the function of cGMP is largely unknown. Our results suggest that, in response to stimulus detection, cGMP levels increase and affect long-term cellular responses in neurons by in turn modulating cAMP levels.
In our previous studies, we have observed that odorant exposure produced a delayed cAMP response in ORNs, in addition to the immediate cAMP response involved in stimulus detection (17). We demonstrated that this second cAMP signal was mediated by cGMP by means of activation of PKG. We speculated that the delayed cAMP signal could regulate long-term cellular events in the ORNs, although this was not demonstrated previously. It was, however, unclear whether the odorant-induced delayed cAMP signal could affect pathways known to be involved in long-term cellular responses (51, 52). A likely target of the delayed odorant-induced cAMP response would be CREB.
CREB is a nuclear DNA binding protein and binds to the CRE site (32). CREB is activated by phosphorylation at Ser-133, and this CREB activation event is accompanied by the binding of related DNA-binding proteins, such as CREB-binding protein (53–55). These components form a functional complex with pCREB bound to the CRE site. Here, we demonstrate that the stimulation of ORNs by odorants is sufficient to induce pCREB and that the odorant-induced pCREB is bound to CRE. Forskolin mimics the effects of odorant stimulation. Interestingly, the amount of pCREB bound to CRE in the presence of relatively low concentrations of odorants is similar to that achieved by forskolin. This is in contrast to the cAMP increases attained during the immediate response to odorants, at which time the forskolin-induced cAMP increase is significantly larger than that induced by a single odorant (11, 12). This suggests that the delayed cAMP response is quite robust in responding cells, or alternatively, that odorants may affect other messengers, such as calcium, that synergize with cAMP to alter pCREB levels. These results indicate that, in ORNs, CREB is activated by odorant stimulation and the activated CREB binds to CRE. The activated CREB bound to CRE may, in turn, form functional complexes with other DNA-binding proteins. The recruitment of additional DNA-binding proteins remains to be established.
Preincubation with a protein kinase inhibitor, K252a (45, 46), not only abolished the odorant-induced pCREB increase but also lowered the basal level of pCREB in ORNs. This observation is in agreement with our in vivo data, which showed that a high basal level of pCREB exists in intact rat nasal turbinate tissue, even without experimental application of exogenous odorants. This should not be surprising because CREB can be activated by many different pathways, such as the neurotransmitter-mediated PKA pathway, calcium–calmodulin-dependent kinases, or growth factor-sensitive kinase pathways (56–58). This may reflect the endogenous activation of the CREB pathway by ambient odorants. We observed that some cells are positive for pCREB in the response to high concentration of odorant mixture but negative for NST. These cells may represent mature ORNs that are no longer NST-positive that are responding to odorants. This observation suggests that odor stimulation is not restricted to mature ORNs, but can also occur in “immature” ORNs. Therefore, odor stimulation to ORNs can be crucial for odor detection, as well as ORN development or survival.
Our results also demonstrate that cGMP is involved in CREB activation, although not by a direct mechanism. Incubation with an NO donor mimicked the effect of cGMP on CREB activation. We previously demonstrated that cGMP augmented the cAMP signal in a PKG-dependent manner by direct activation of adenylyl cyclase in ORNs. We suggested that this cGMP/PKG-mediated cAMP signal might regulate gene expression in ORNs for long-term cellular events. The observation that PKG does not directly phosphorylate CREB in ORNs supports this hypothesis. Therefore, the effect of cGMP on CREB phosphorylation may be indirect: cGMP modulates cAMP levels to affect CREB phosphorylation. A similar scheme may exist in other systems and may provide another mechanism for the modulation of CREB activity in response to stimulation.
Acknowledgments
We thank Dr. J. Yoo for her technical assistance and critical reading of this manuscript; Dr. D. Ginty for the kind gifts of CREB Abs and CRE probe; S. Ahn for preparation of PC-12 cells; and Lana Kramer for manuscript preparation. This work was supported by a grant from the W. M. Keck Foundation and National Institute on Deafness and Other Communication Disorders Grants RO1DC2979 (to G.V.R.) and F32DC00243 (to C.M.).
Abbreviations
- ORN
olfactory receptor neuron
- CRE
cAMP-responsive element
- CREB
CRE-binding protein
- PKA
cAMP-dependent protein kinase
- PKG
cGMP-dependent protein kinase
- NST
neuron-specific class III β-tubulin
- IBMP
isobutylmethoxypyrazine
- βIII
anti β-tubulin
- pCREB
phosphorylated CREB
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nakamura T, Gold G H. Nature (London) 1987;325:442–444. doi: 10.1038/325442a0. [DOI] [PubMed] [Google Scholar]
- 2.Jones D T, Reed R R. Science. 1989;244:790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- 3.Dhallan R S, Yau K W, Schrader K A, Reed R A. Nature (London) 1990;347:184–187. doi: 10.1038/347184a0. [DOI] [PubMed] [Google Scholar]
- 4.Kaupp U B. Trends Neurosci. 1991;14:150–157. doi: 10.1016/0166-2236(91)90087-b. [DOI] [PubMed] [Google Scholar]
- 5.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 6.Rhein L D, Cagan R H. Proc Natl Acad SciUSA. 1980;77:4412–4416. doi: 10.1073/pnas.77.8.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck L B. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- 8.Malnic B, Hirono J, Sato T, Buck L. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 9.Dwyer N D, Troemel E R, Sengupta P, Bargmann C I. Cell. 1998;93:455–466. doi: 10.1016/s0092-8674(00)81173-3. [DOI] [PubMed] [Google Scholar]
- 10.Pace U, Hanski E, Salomon Y, Lancet D. Nature (London) 1985;316:255–258. doi: 10.1038/316255a0. [DOI] [PubMed] [Google Scholar]
- 11.Sklar P B, Anholt R R H, Snyder S H. J Biol Chem. 1986;261:15538–15543. [PubMed] [Google Scholar]
- 12.Ronnett G V, Cho H, Hester L D, Wood S F, Snyder S H. J Neurosci. 1993;13:1751–1758. doi: 10.1523/JNEUROSCI.13-04-01751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firestein S, Werblin F S. Science. 1989;244:79–82. doi: 10.1126/science.2704991. [DOI] [PubMed] [Google Scholar]
- 14.Getchell T V, Shepherd G M. J Physiol (London) 1978;282:521–540. doi: 10.1113/jphysiol.1978.sp012479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ottoson D. Acta Physiol Scand. 1956;122:1–83. [PubMed] [Google Scholar]
- 16.Verma A, Hirsch D J, Glatt C E, Ronnett G V, Snyder S H. Science. 1993;259:381–384. doi: 10.1126/science.7678352. [DOI] [PubMed] [Google Scholar]
- 17.Moon C, Jaberi P, Otto-Bruc A, Baehr W, Palczewski K, Ronnett G. J Neurosci. 1998;18:3195–3205. doi: 10.1523/JNEUROSCI.18-09-03195.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroner C, Boekhoff I, Lohmann S M, Genieser H G, Breer H. Eur J Biochem. 1996;236:632–637. doi: 10.1111/j.1432-1033.1996.00632.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Tonegawa S. Annu Rev Neurosci. 1997;20:157–184. doi: 10.1146/annurev.neuro.20.1.157. [DOI] [PubMed] [Google Scholar]
- 20.Ingi T, Ronnett G V. J Neurosci. 1995;15:8214–8222. doi: 10.1523/JNEUROSCI.15-12-08214.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breer H, Klemm T, Boekhoff I. NeuroReport. 1992;3:1030–1031. doi: 10.1097/00001756-199211000-00022. [DOI] [PubMed] [Google Scholar]
- 22.Zufall F, Leinders-Zufall T. J Neurosci. 1997;17:2703–2712. doi: 10.1523/JNEUROSCI.17-08-02703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudi T, Huvar I, Meinecke M, Lohmann S M, Boss G R, Pilz R B. J Biol Chem. 1996;271:4597–4600. doi: 10.1074/jbc.271.9.4597. [DOI] [PubMed] [Google Scholar]
- 24.Hill C S, Treisman R. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 25.Brindle P, Nakjima T, Montminy M. Proc Natl Acad SciUSA. 1995;92:10521–10525. doi: 10.1073/pnas.92.23.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sassone-Corsi P. Annu Rev Cell Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- 27.Chain D G, Casadio A, Schacher S, Hegde A N, Valbrun M, Yamamoto N, Goldberg A L, Bartsch D, Kandel E R, Schwartz J H. Neuron. 1999;22:147–156. doi: 10.1016/s0896-6273(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 28.Hunter T, Karin M. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 29.Karin M, Smeal T. Trends Biochem Sci. 1992;17:418–422. doi: 10.1016/0968-0004(92)90012-x. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto K K, Gonzalez G A, Biggs W H, III, Montminy M R. Nature (London) 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 31.Montminy M R, Bilezikjian L M. Nature (London) 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez G A, Yamamoto K K, Fischer W H, Karr D, Menzel P, Biggs W, Vale W W, Montminy M R. Nature (London) 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- 33.Lee M K, Rebhun L I, Frankfurter A. Proc Natl Acad Sci USA. 1990;87:7195–7199. doi: 10.1073/pnas.87.18.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roskams A J I, Cai X, Ronnett G V. Neuroscience. 1998;83:191–200. doi: 10.1016/s0306-4522(97)00344-8. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham A, Manis P, Reed R, Ronnett G. Neuroscience. 1999;93:1301–1312. doi: 10.1016/s0306-4522(99)00193-1. [DOI] [PubMed] [Google Scholar]
- 36.Ronnett G V, Hester L D, Snyder S H. J Neurosci. 1991;11:1243–1255. doi: 10.1523/JNEUROSCI.11-05-01243.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roskams A J, Bredt D S, Dawson T M, Ronnett G V. Neuron. 1994;13:289–299. doi: 10.1016/0896-6273(94)90347-6. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Towbin H, Staehelin T, Gorden J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steiner J P, Dawson T M, Fotuhi M, Glatt C E, Snowman A M, Cohen N, Snyder S H. Nature (London) 1992;358:584–587. doi: 10.1038/358584a0. [DOI] [PubMed] [Google Scholar]
- 42.Ginty D D, Kornhauser J M, Thompson M A, Bading H, Mayo K E, Takahashi J S, Greenberg M E. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 43.Francis S H, Wolfe L, Corbin J D. Methods Enzymol. 1991;200:332–341. doi: 10.1016/0076-6879(91)00150-u. [DOI] [PubMed] [Google Scholar]
- 44.Corbin J D, Doskeland S O. J Biol Chem. 1983;258:11391–11397. [PubMed] [Google Scholar]
- 45.Kase H, Iwahashi K, Nakanishi S, Matsuda Y, Yamada K, Takahashi M, Murakata C, Sato A, Kaneko M. Biochem Biophys Res Commun. 1987;142:436–440. doi: 10.1016/0006-291x(87)90293-2. [DOI] [PubMed] [Google Scholar]
- 46.Tischler A S, Ruzicka L A, Perlman R L. J Neurochem. 1990;55:1159–1165. doi: 10.1111/j.1471-4159.1990.tb03120.x. [DOI] [PubMed] [Google Scholar]
- 47.Breer H, Shepherd G M. Trends Neurosci. 1993;16:5–9. doi: 10.1016/0166-2236(93)90040-s. [DOI] [PubMed] [Google Scholar]
- 48.Nagamine Y, Reich E. Proc Natl Acad Sci USA. 1985;82:4606–4610. doi: 10.1073/pnas.82.14.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sassone-Corsi P. Int J Biochem Cell Biol. 1998;30:27–38. doi: 10.1016/s1357-2725(97)00093-9. [DOI] [PubMed] [Google Scholar]
- 50.Ronnett G V, Snyder S H. Trends Neurosci. 1992;15:508–512. doi: 10.1016/0166-2236(92)90104-g. [DOI] [PubMed] [Google Scholar]
- 51.Kang H, Schuman E M. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 52.Bailey C H, Bartsch D, Kandel E R. Proc Natl Acad Sci USA. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatina Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 54.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Nature (London) 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 55.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nature (London) 1994;16:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 56.Sheng M E, Thompson M A, Greenberg M E. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalez G A, Montminy M R. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 58.Ginty D D, Bonni A, Greenberg M E. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]