Abstract
A series of chimeral genes, consisting of the yeast GAL10 promoter, yeast ACC1 leader, wheat acetyl-CoA carboxylase (ACCase; EC 6.4.1.2) cDNA, and yeast ACC1 3′-tail, was used to complement a yeast ACC1 mutation. These genes encode a full-length plastid enzyme, with and without the putative chloroplast transit peptide, as well as five chimeric cytosolic/plastid proteins. Four of the genes, all containing at least half of the wheat cytosolic ACCase coding region at the 5′-end, complement the yeast mutation. Aryloxyphenoxypropionate and cyclohexanedione herbicides, at concentrations below 10 μM, inhibit the growth of haploid yeast strains that express two of the chimeric ACCases. This inhibition resembles the inhibition of wheat plastid ACCase observed in vitro and in vivo. The differential response to herbicides localizes the sensitivity determinant to the third quarter of the multidomain plastid ACCase. Sequence comparisons of different multidomain and multisubunit ACCases suggest that this region includes part of the carboxyltransferase domain, and therefore that the carboxyltransferase activity of ACCase (second half-reaction) is the target of the inhibitors. The highly sensitive yeast gene-replacement strains described here provide a convenient system to study herbicide interaction with the enzyme and a powerful screening system for new inhibitors.
Acetyl-CoA carboxylase (ACCase; EC 6.4.1.2), a biotin-dependent carboxylase, catalyzes the first committed step of de novo fatty acid biosynthesis, the carboxylation of acetyl-CoA to produce malonyl-CoA, using bicarbonate as a source of the carboxyl group and ATP as a source of energy. It also provides malonyl-CoA for the synthesis of very long chain fatty acids and for secondary metabolism. Two half-reactions are catalyzed by ACCase:
(i) ATP-dependent activation of CO2 and attachment of the CO2 to biotin on the carboxyl carrier protein by the biotin carboxylase activity;
(ii) Transfer of the CO2 from biotin to acetyl-CoA by the carboxyltransferase activity.
Two forms of ACCase have been identified: a multidomain enzyme in eukaryotes, and a multisubunit enzyme in prokaryotes and plastids of some plants. Plants have two forms of ACCase (reviewed in ref. 1). One, located in plastids, the primary site of plant fatty acid synthesis, can be either a high molecular weight, multidomain enzyme (e.g., wheat and maize), or a multisubunit enzyme (e.g., pea, soybean, tobacco, and Arabidopsis thaliana). The cytosolic plant ACCase is the multidomain type. In Gramineae, genes for both cytosolic and plastid multidomain ACCases are nuclear, e.g., in maize (2) and wheat (3–5). The cytoplasmic and plastid ACCases from wheat are 2260 and 2311 aa long, respectively, and their sequences are 67% identical. A chloroplast targeting signal is present at the N terminus of the multidomain plastid ACCase from wheat (3), maize (2), and Brassica napus (6).
In plants, the contribution of ACCase to the control of flux through the fatty acid pathway is postulated to be very significant (7, 8). This level of control is reflected in the response of sensitive plants to herbicides that target ACCase; fatty acid biosynthesis is inhibited to the point that the plant dies. Aryloxyphenoxypropionates and cyclohexanediones inhibit fatty acid biosynthesis in Gramineae by the strong inhibition of their multidomain plastid ACCase (reviewed in refs. 9–11). We have shown this process to be the case in wheat (12). Natural resistance to these herbicides has become widespread in Lolium and Avena spp. A Lolium rigidum biotype with resistance to sethoxydim has an altered ACCase (see, e.g., refs.13 and 14). Plants other then Gramineae (e.g., dicots) are resistant to these compounds, as are most other eukaryotes and prokaryotes, including yeast.
Two of the present authors, R. H. and P. G., and E. Zuther, J. J. Johnson, and R. McLeod) have shown that the major ACCase in Toxoplasma gondii is sensitive to aryloxyphenoxypropionates (15). T. gondii has two ACCases of the multidomain type, one of which we believe to function in the apicomplexan plastid, a site of fatty acid biosynthesis in these parasites (16). The molecular mechanism of inhibition of the enzyme by these herbicides is not yet known. However, earlier biochemical studies indicated that the carboxyltransferase partial reaction is affected by these inhibitors (17). The structures of representative inhibitors appear in figure 1 of ref. 15.
Wheat cytosolic ACCase can be expressed in yeast and can complement a yeast ACC1 null mutation (18). Furthermore, gene-replacement strains, depending for growth on the wheat cytosolic ACCase, are resistant to Haloxyfop and Sethoxydim, but sensitive to Cethoxydim. In this paper, we report the construction of yeast strains expressing wheat cytosolic/plastid chimeric ACCases whose sensitivity to herbicides reflects the origin of the carboxyltransferase domain of the enzyme.
Materials and Methods
Gene Assembly.
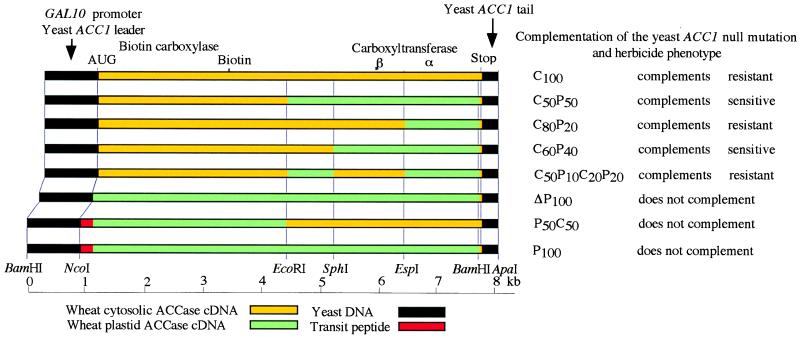
Full-length cDNA, encoding wheat plastid and chimeric ACCases, was assembled with standard molecular biology techniques. PCR-cloned cDNAs described before (3), as well as PCR-generated fragments with new restriction sites, ere digested with useful restriction enzymes and ligated appropriately. Fragments obtained by partial digestion of DNA were used in some cases. The assembly process was monitored by sequencing selected regions of each construct. Previously, we constructed a synthetic gene, gyccwy (here renamed C100), consisting of the yeast GAL10 promoter, yeast ACC1 leader, wheat cytosolic ACCase cDNA, and a yeast ACC1 3′-tail (18). A synthetic gene, encoding wheat plastid ACCase gypcwy (here renamed P100), was constructed by replacing almost the entire coding sequence of gyccwy, between the NcoI site at the start codon and the BamHI site, 53 nucleotides upstream of the stop codon, with the coding sequence of the plastid isozyme (Fig. 1). The NcoI site was engineered by PCR, and the BamHI site is present in both sequences. Genes encoding chimeric ACCases were created by swapping large restriction fragments between conserved EcoRI, SphI, and EspI sites, and a unique ApaI site in the polylinker. A gene encoding plastid ACCase that lacked the N-terminal fragment that contained the putative plastid transit peptide was created from gypcwy by removing the first 67 aa and replacing Q68 with M, and with the concomitant introduction of a new NcoI site. Yeast shuttle vector pRS423 (19) was used for all the constructs. The structures of the chimeral genes in this study are shown in Fig. 1.
Figure 1.
Chimeral genes constructed for expression of wheat cytosolic, plastid, and cytosolic/plastid ACCases in yeast. Construct names reflect composition of the encoded proteins (C, cytosolic ACCase, P, plastid ACCase). Locations of key restriction sites used in the constructions are shown. The major functional domains (biotin carboxylase and carboxyltransferase) and the biotin attachment sites are located as described before (3, 4).
Complementation of a yeast ACC1 null mutation was performed as described previously (18). Saccharomyces cerevisiae strain W303D-ACC1ΔLeu2 (heterozygous strain ACC1/acc1∷ LEU2; see Table 1) was provided by S. D. Kohlwein (Technical Univ. Graz, Austria). Sporulation was induced and 20–60 asci were dissected in a standard tetrad analysis, as described (18). All complementation experiments were carried out at 23°C and 30°C.
Table 1.
Yeast strains
| Strain | Construct* | Parent vector | ACC1 | URA3 | LEU2 | HIS3 | TRP1 | Galactose dependence | Doubling time, hr†
|
Herbicide sensitivity | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 23° C | 30° C | |||||||||||
| MJ 1.12 | C100 | pRS424 | − | − | + | − | + | + | 4.8 ± 0.2 | 3.1 ± 0.1 | No | 18 |
| MJ 1.13 | C100 | pRS424 | − | − | + | − | + | + | 4.6 ± 0.2 | 3.2 ± 0.1 | No | 18 |
| MJ 9.11 | Yeast ACC1§ | pRS426 | + | + | + | − | − | − | 4.5 ± 0.2 | 2.2 ± 0.2 | No | 18 |
| MJ 9.14 | Yeast ACC1§ | pRS426 | + | + | + | − | − | − | 5.3 ± 0.4 | 2.4 ± 0.1 | No | 18 |
| MJ 9.13 | Yeast ACC1¶ | None | + | − | − | − | − | − | 3.9 ± 0.2 | 2.4 ± 0.1 | No | 18 |
| TN 1/2-1 | C50P50 | pRS423 | − | − | + | + | − | + | 6.0 ± 0.3 | 4.3 ± 0.2 | Yes | This work |
| TN 1/2-3 | C50P50 | pRS423 | − | − | + | + | − | + | 4.7 ± 0.2 | 4.4 ± 0.1 | Yes | This work |
| PG 3.34 | C80P20 | pRS423 | − | − | + | + | − | + | 8.9 ± 0.3 | 13.3 ± 1.7 | No | This work |
| PG 3.43 | C80P20 | pRS423 | − | − | + | + | − | + | 8.8 ± 0.2 | 13.5 ± 1.8 | No | This work |
| PG 4.41 | C60P40 | pRS423 | − | − | + | + | − | + | 14.6 ± 1.2 | ND | Yes | This work |
| PG 4.43 | C60P40 | pRS423 | − | − | + | + | − | + | 12.4 ± 0.9 | ND | Yes | This work |
| PG 5.21 | C50P10C20P20 | pRS423 | − | − | + | + | − | + | 9.2 ± 0.3 | ND | No | This work |
| PG 5.22 | C50P10C20P20 | pRS423 | − | − | + | + | − | + | 14.3 ± 1.0 | ND | No | This work |
All strains were obtained by sporulation of W303D-ACC1ΔLeu2 transformed with an appropriate plasmid. Haploid strains: 1.12 and 1.13, 9.11 and 9.14, 4.41 and 4.43, 5.21 and 5.22 were obtained from single tetrads. W303D-ACC1ΔLeu2 relevant genotype: MATa/MATα, leu2-3, 112/leu2-3, 112 his3-11,15/his311, 15 ade2-1/ade2-1 ura3-1/ura3-1 trp-1/trp1-1 can1-100/can1-100 ACC1/acc1∷LEU2.
Structures of the constructs are shown in Fig. 1, except ACC1, which are annoted below.
† In YPRG medium; ND, no growth detected.
‡ Strong sensitivity to aryloxyphenoxypropionate and cyclohexanedione herbicides.
§ Plasmid-borne.
¶ Genomic.
Growth inhibition of the gene-replacement yeast strains by herbicides was assessed as described before (18), by measuring culture optical density at 600 nm. One OD unit corresponds to culture density of approximately 1.3 × 107 cells per ml. Yeast growth and inhibition by the herbicides was measured at 23°C and 30°C. Clodinafop, Quizalafop, Propaquizafop, Sethoxydim, and Cethoxydim (our name for CGA215684), were provided by Novartis (Research Triangle Park, NC), and Haloxyfop was provided by DowElanco (Indianapolis, IN). Herbicides were added as 100-fold concentrated stock solutions in DMSO.
The High Fidelity PCR System (Boehringer Mannheim) was used for DNA amplification. Protein extract preparation and Western analysis with [35S]streptavidin and with antibodies specific for wheat ACCase were described previously (20). Yeast media and growth conditions were as described (18). The YPRG yeast complex medium for vegetative growth contained 1% Bacto-Yeast Extract, 2% Bacto-Peptone, 0.1% adenine sulfate, 2% galactose, and 2% raffinose.
Results
We assembled a series of chimeral genes, consisting of the yeast GAL10 promoter, yeast ACC1 leader, wheat ACCase cDNA, and yeast ACC1 3′-tail in high-copy-number, yeast expression vectors of the pRS series (18). The ACCase coding sequence was assembled from fragments of different cDNA clones, isolated from a cDNA library or cloned by PCR, based on the genomic sequence of wheat cytosolic and plastid ACCase genes, as described before (3, 18). To assemble the genes described in this paper, we took advantage of the modular design and restriction sites engineered for this purpose in the original construct (18). In addition to the synthetic gene encoding the cytosolic isozyme (construct C100), we constructed new genes, encoding the full-length plastid enzyme, with and without the putative chloroplast transit peptide (construct P100 and ΔP100, respectively), as well as five chimeric cytosolic/plastid enzymes (Fig. 1). These chimeric genes were introduced into heterozygous S. cerevisiae strain W303D-ACC1ΔLeu2 (in which one copy of the ACC1 gene was replaced with a LEU2 cassette), and their ability to complement the yeast mutation was analyzed by standard tetrad analysis. We have shown previously that the construct encoding wheat cytosolic ACCase complements a yeast ACC1 null mutation (18).
At least several sets of three or four viable spores, derived from a single tetrad of about 20 tested, were obtained for strains carrying some of the chimeric genes. This result indicates that these genes can complement the ACC1 mutation, because haploid ACC1 disruptants do not grow beyond a few cell divisions (21). Some synthetic genes did not yield more than two viable spores from a single tetrad of about 60 tested at either 23°C or 30°C. It was concluded that these synthetic genes did not complement the yeast mutation. A synthetic gene expressing wheat cytosolic ACCase (C100; Fig. 1), known to complement the mutation (18), was used as a positive control. Marker analysis of haploid strains obtained from tetrad dissection confirmed their genotypes.
Four of the new artificial genes, all containing at least half of the wheat cytosolic ACCase coding region at the 5′-end, complement the yeast mutation when grown at 23°C. Two of these genes fail to complement the mutation at 30°C (constructs C60P40 and C50P10C20P20; Fig. 1). The other two (constructs C50P50 and C80P20; Fig. 1) complement at both 23°C and 30°C. Three of the new artificial genes (ΔP100, P50C50, and P100; Fig. 1), all containing at least half of the wheat plastid ACCase coding region at the 5′-end, fail to complement the ACC1 mutation at either temperature.
The absence of detectable complementation for some chimeric constructs could be caused by a failure of expression of these synthetic genes at the transcription or translation level, or by defects in protein function, stability, or modification, or by mistargeting to a wrong cellular compartment. Western blots, probed with [35S]streptavidin and with antibodies specific for wheat ACCase (data not shown), confirmed the ability of all the new artificial genes to drive expression of full-size ACCase in yeast, even those for which no complementation was observed. These wheat peptides are biotinylated, as determined by their ability to bind streptavidin. Lack of complementation by the gene encoding plastid ACCase is unlikely to be caused by misdirection of the protein to the wrong cellular compartment, because a chimeric gene encoding full-length plastid ACCase lacking the transit peptide (ΔP100) fails to complement the yeast mutation as well. Lack of complementation may be caused by low enzymatic activity of plastid ACCase in yeast, e.g., because there is a much lower intracellular pH in yeast (below 6.0) than in plant chloroplasts (above 8.0).
Other more trivial explanations may account for these observations. A possible mutation in the N-terminal half of the plastid ACCase, introduced by PCR during the construction of the chimeric gene, cannot be excluded. Sequence comparisons of the plastid ACCase, encoded by P100, with the plastid ACCases encoded by genes Acc-1,1 and Acc-1,2 (3), revealed 5 and 3 changes, respectively, within the first 619 aa. cDNAs used for the construction were cloned by using mRNA from hexaploid wheat, for which only one complete and one partial gene sequence are available. Therefore, it is impossible to establish which, if any, of these amino acid differences resulted from PCR errors. Obviously, such possible mutations are not present in the C-terminal half, because the cytosolic/plastid chimeric gene (C50P50; Fig. 1) produces active ACCase and complements the yeast mutation.
Two haploid gene-replacement strains were selected for each of the constructs for further analysis. Their growth rates vary significantly, depending on the structure of the chimeric ACCase (Table 1). The strains that did not grow in liquid culture at 30°C are the same ones that did not grow on plates under comparable conditions in the tetrad analysis. In addition, strains 3.34 and 3.43 (C80P20 construct; Fig. 1) grew faster at 23°C than at 30°C, a reversal of the growth rates at the two temperatures of strains harboring the wild-type yeast ACC1 gene (9.11 and 9.14). No differences in cell morphology were observed among the strains at the level of the light microscope, except for a broader cell size variation when compared with the wild type, especially evident for the slowest growing strains (PG 4.41 and PG 4.43, and PG 5.21 and PG 5.22).
Growth inhibition of gene-replacement yeast strains by herbicides was measured after 48 hr of growth under full galactose induction in yeast extract/peptone/raffinose/galactose (YPRG) liquid medium (Fig. 2). Under these conditions, the fastest-growing strains reached the end of the logarithmic phase at approximately 16 × 107 cells per ml. All other strains grew more slowly. There was significant variation from experiment to experiment of the culture densities measured in this way. However, the growth of strains carrying the same construct was similar, and the growth and inhibition patterns of different strains were reproducible.
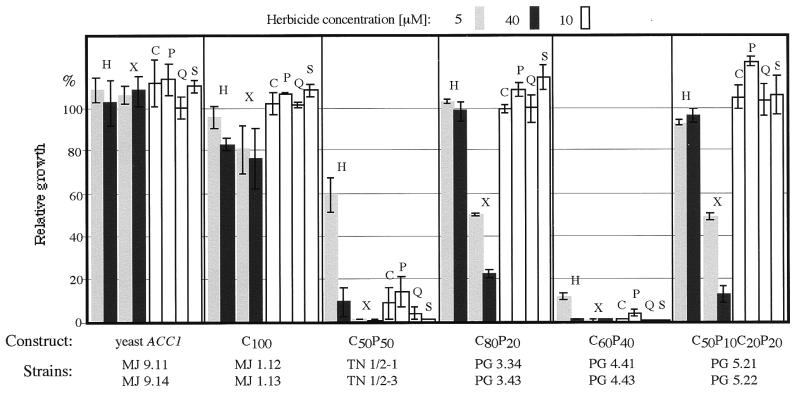
Figure 2.
Growth inhibition by herbicides of yeast gene-replacement haploid strains (Table 1) expressing chimeric wheat ACCases. Growth without herbicide in medium containing 1% DMSO, 100%. Average values for two strains containing the same construct obtained in two or three experiments are shown. Error bars indicate variation observed for each pair of strains. H, Haloxyfop; X, Cethoxydim, C, Clodinafop, P, Propaquizafop; Q, Quizalafop; S, Sethoxydim.
Consistent with the results reported previously, strains 1.12 and 1.13 (with a synthetic gene expressing wheat cytosolic ACCase) grew almost as well as strains 9.11, 9.13, and 9.14 (with the wild-type yeast ACC1 gene) at 30°C (Table 1). At this temperature, growth of strains 1.12 and 1.13 is inhibited by Cethoxydim (by 35% and 75%, at 40 μM for cultures grown under full and limited galactose induction, respectively) but not by Haloxyfop, and strains 9.11 and 9.14 are not affected by either of these herbicides (ref. 18) and data not shown). Strains containing constructs C50P50 and C80P20 tested at 30°C gave a Haloxyfop and Cethoxydim sensitivity pattern similar to that described below for 23°C (data not shown).
At 23°C, strains harboring the yeast ACC1 gene on a plasmid (9.11 and 9.14) are not affected by any of the six herbicides at concentrations below 40 μM. Strains expressing full-length wheat cytosolic ACCase show some sensitivity to Haloxyfop and Cethoxydim, but no sensitivity to the other three aryloxyphenoxypropionates and one cyclohexanedione at 10 μM (Fig. 2). This is an expected result, because both cytosolic enzymes (yeast and wheat) are known to be resistant to aryloxyphenoxypropionates and cyclohexanediones at low concentrations. These results agree well with earlier observations concerning the sensitivity/resistance of wheat and maize cytosolic and plastid ACCase isozymes (12, 22).
Growth of the strains containing the cytosolic/plastid C50P50 chimeric gene (TN 1/2–1, TN 1/2–3) is strongly inhibited by all four aryloxyphenoxypropionates and two cyclohexanediones tested (Fig. 2). The same sensitive phenotype is observed for strains harboring chimeric gene C60P40 (4.41 and 4.43), and is largely reversed to the resistant phenotype for chimeric genes C80P20 (strains 3.34 and 3.43) and C50P10C20P20 (strains 5.21 and 5.22). The last two strains remain more sensitive to Cethoxydim.
Although none of the genes that include the N-terminal half of plastid ACCase complement the yeast ACC1 null mutation, the analysis of the chimeras that do complement allows us to locate the sensitivity domain and the herbicide-binding site to a 400-aa fragment (amino acid residues 1462 to 1873 of the unprocessed wheat plastid ACCase). This strong inhibition of growth of the sensitive strains reflects the sensitivity of wheat plastid ACCase observed in vitro and in vivo (12, 18).
Discussion
Recently we established an efficient yeast expression system to produce plant ACCase polypeptides in their active form. By testing complementation of a yeast ACC1 mutation, several important questions can be addressed conveniently: Can our cDNA produce full-length polypeptide in yeast? Is it biotinylated? Is a particular chimeric protein an active ACCase? Does a wheat chimeric ACCase render yeast sensitive to herbicides?
In yeast, a single gene (ACC1) encodes ACCase, which provides malonyl-CoA for both fatty acid biosynthesis and subsequent fatty acid elongation. The product of the same gene is needed to provide malonyl-CoA for the biosynthesis of very-long-chain fatty acids for the nuclear envelope (23). Complementation of the ACC1 null mutation by wheat cytosolic ACCase is consistent with the conclusion that the only essential role of yeast ACCase is to provide malonyl-CoA (23).
The differential responses among our chimeric constructs localizes the herbicide sensitivity determinant to a 400-amino-acid fragment of the C-terminal half of the multidomain plastid ACCase. Sequence comparison of different multidomain and multisubunit ACCases suggests that this region contains an essential part of the carboxyltransferase β-subdomain. This domain contains a cluster of several amino acids conserved among bacteria and eukaryotes. These amino acid residues most likely constitute a functional site of the carboxyltransferase domain. One possible explanation of inhibition by the aryloxyphenoxypropionates and cyclohexanediones is their binding and interference with that function.
Inhibitors are valuable tools for studies of enzyme mechanism. Understanding the molecular basis of inhibitor action is an important goal. The two classes of inhibitors used here specifically target plastid ACCase from Graminae (9–11). Inhibitor-sensitive, yeast gene-replacement strains, such as the strains described, can be used to study the mechanisms of these inhibitors. Mutants that confer resistance can be selected and the mutation site(s) identified. These mutations will define inhibitor binding sites studied by other methods.
Aryloxyphenoxypropionates and cyclohexanediones are specific. They have no effect on the growth of wild-type yeast or mammalian cells. Our results on herbicide action on wheat (12), Toxoplasma gondii (15), and yeast gene-replacement strains clearly indicate that in different eukaryotic systems inhibition of ACCase activity leads to a complete inhibition of growth. The yeast gene-replacement strains described here should be helpful in determining the enzyme mechanism and in screening ACCase inhibitors with new specificities for use in agriculture and medicine.
Acknowledgments
The authors are grateful to S. D. Kohlwein for providing valuable materials and to Ian King for his help in assembling full-length cDNA encoding wheat plastid ACCase. We thank Thierry Niderman and Daphne Preuss for valuable discussions. This work was supported by grants from Novartis, by the H. and F. Block Research Fund of the University of Chicago, and by the University of Chicago/Argonne National Laboratory Collaborative Grant Program.
Abbreviation
- ACCase
acetyl-CoA carboxylase
References
- 1.Konishi T, Shinohara K, Yamada K, Sasaki Y. Plant Cell Physiol. 1996;37:117–122. doi: 10.1093/oxfordjournals.pcp.a028920. [DOI] [PubMed] [Google Scholar]
- 2.Egli M, Lutz S, Somers D, Gengenbach B. Plant Physiol. 1995;108:1299–1300. doi: 10.1104/pp.108.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R. Proc Natl Acad Sci USA. 1997;94:14179–14185. doi: 10.1073/pnas.94.25.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gornicki P, Podkowinski J, Scappino L A, DiMaio J, Ward E, Haselkorn R. Proc Natl Acad Sci USA. 1994;91:6860–6864. doi: 10.1073/pnas.91.15.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podkowinski J, Sroga G E, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1996;93:1870–1874. doi: 10.1073/pnas.93.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulte W, Topfer R, Stracke R, Schell J, Martini N. Proc Natl Acad Sci USA. 1997;94:3465–3470. doi: 10.1073/pnas.94.7.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohlrogge J B, Jaworski J G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- 8.Page R A, Okada S, Harwood J L. Biochim Biophys Acta. 1994;1210:369–372. doi: 10.1016/0005-2760(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 9.Golz A, Focke M, Lichtenthaler H K. J Plant Physiol. 1994;143:426–433. [Google Scholar]
- 10.Holt J S, Powles S B, Holtum J A M. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:203–229. [Google Scholar]
- 11.Konishi T, Sasaki Y. Proc Natl Acad Sci USA. 1994;91:3598–3601. doi: 10.1073/pnas.91.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gornicki P, Haselkorn R. Plant Mol Biol. 1993;22:547–552. doi: 10.1007/BF00015984. [DOI] [PubMed] [Google Scholar]
- 13.Burnet M W M, Hart Q, Holtum J A M, Powles S B. Weed Sci. 1994;42:369–377. [Google Scholar]
- 14.Powles S B, Matthews J M. In: Achievements and Developments in Combating Resistance. Denholm I, Devonshire A, Hollomon D W, editors. London: Elsevier; 1992. pp. 75–87. [Google Scholar]
- 15.Zuther E, Johnson J J, Haselkorn R, McLeod R, Gornicki P. Proc Natl Acad Sci USA. 1999;96:13387–13392. doi: 10.1073/pnas.96.23.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waller R F, Keeling P J, Donald R G K, Striepen B, Handman E, Lang-Unnasch N, Cowman A F, Besra G S, Roos D S, McFadden G I. Proc Natl Acad Sci USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rendina A R, Felts J M, Beaudoin J D, Craig-Kennard A C, Look L L, Paraksos S L, Hagenah J A. Arch Biochem Biophys. 1988;265:219–225. doi: 10.1016/0003-9861(88)90387-6. [DOI] [PubMed] [Google Scholar]
- 18.Joachimiak M, Tevzadze G, Podkowinski J, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1997;94:9990–9995. doi: 10.1073/pnas.94.18.9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell D A, Marshall T K, Deschenes R J. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 21.Haslacher M, Ivessa A, Paltauf F, Kohlwein S. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 22.Howard J L, Ridley S M. FEBS Lett. 1990;261:261–264. [Google Scholar]
- 23.Schneiter R, Hitomi M, Ivessa A S, Fasch E-V, Kohlwein S D, Tartakoff A M. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]