Abstract
Developmental and physiological responses are regulated by light throughout the entire life cycle of higher plants. To sense changes in the light environment, plants have developed various photoreceptors, including the red/far-red light-absorbing phytochromes and blue light-absorbing cryptochromes. A wide variety of physiological responses, including most light responses, also are modulated by circadian rhythms that are generated by an endogenous oscillator, the circadian clock. To provide information on local time, circadian clocks are synchronized and entrained by environmental time cues, of which light is among the most important. Light-driven entrainment of the Arabidopsis circadian clock has been shown to be mediated by phytochrome A (phyA), phytochrome B (phyB), and cryptochromes 1 and 2, thus affirming the roles of these photoreceptors as input regulators to the plant circadian clock. Here we show that the expression of PHYB∷LUC reporter genes containing the promoter and 5′ untranslated region of the tobacco NtPHYB1 or Arabidopsis AtPHYB genes fused to the luciferase (LUC) gene exhibit robust circadian oscillations in transgenic plants. We demonstrate that the abundance of PHYB RNA retains this circadian regulation and use a PHYB∷Luc fusion protein to show that the rate of PHYB synthesis is also rhythmic. The abundance of bulk PHYB protein, however, exhibits only weak circadian rhythmicity, if any. These data suggest that photoreceptor gene expression patterns may be significant in the daily regulation of plant physiology and indicate an unexpectedly intimate relationship between the components of the input pathway and the putative circadian clock mechanism in higher plants.
Keywords: Arabidopsis, tobacco, oscillation, luciferase, transgenic plants
Phytochromes are a major photoreceptor family that controls plant development from germination to flowering (1). In the majority of plants phytochrome is encoded by a small multigene family, in Arabidopsis by five genes (2), of which PHYB is thought to be the closest one to the ancestral PHY gene. phyB is the dominant phytochrome in light-grown plants and has been shown to control stem and petiole elongation, chloroplast development, and flowering time (3). Gene-dosage experiments indicate that a 2-fold change in PHYB expression levels causes well-defined, biological responses (4, 5). Light-dependent, physiological processes controlled by phytochrome and cryptochrome (cry) also are regulated by circadian rhythms (6). Circadian clocks are ubiquitous, 24-hr biological timers described in prokaryotic and eukaryotic organisms (6, 7). Circadian-regulated physiological and behavioral responses are thought to ensure the organism’s optimal adaptation to the environment (8). To fulfill their role circadian clocks must be synchronized (entrained) to their local environment. The most important entraining signal is light, and in Arabidopsis several forms of phytochrome, including phyA and phyB, have been shown to mediate light input to the circadian clock (9, 10).
Within the circadian oscillator, light input signals have been shown to alter the level of critical clock components, such as the TIMELESS protein in Drosophila (11). Rhythmic accumulation of the RNA and protein products of “canonical” clock genes, including TIMELESS, is thought to form the biochemical basis of biological timing (7). The nuclear import of the clock proteins allows down-regulation of transcription from the promoters of their cognate genes (7). In the simplest theoretical models, components of the light input pathway were assumed to be dispensable for this oscillator and were thought only to link the oscillator to the environment. More recent findings indicate that photoreceptors and putative photoreceptors function more centrally in the circadian oscillator mechanism; mutations in genes such as CRY in Drosophila (12, 13) and mouse (14, 15) or wc-2 in Neurospora (16) cause arhythmia, not merely insensitivity to light signals.
Parallels between phyB and other components of the circadian system recently have been described. Phytochrome proteins contain a bipartite PAS domain (17), which is present in circadian clock proteins of other species but also in proteins without known circadian functions. The PAS domain is required for in vitro interaction of phytochrome with the potential signaling partner PIF3, a predicted basic helix–loop–helix transcription factor that has been shown to affect light responses in vivo (18). The light-dependent nuclear import of PHYA (19) and PHYB proteins (19–21) corroborates the functional significance of this interaction. The clock proteins in other eukaryotic organisms not only exhibit regulated nuclear translocation, but several protein interactions required for the oscillator mechanism also are known to be mediated by PAS domains (7).
We were interested to test whether the expression of phytochromes, specifically phyB, are under the control of a circadian rhythm, as expected of a component of the oscillator mechanism or an output signaling component. To this end, we determined the expression pattern of the PHYB promoter by using the luciferase (LUC) reporter and the patterns of PHYB RNA and PHYB protein abundance during the circadian cycle. Here we report that reminiscent of canonical clock components, expression of the PHYB gene and synthesis of PHYB protein exhibit a circadian rhythm.
Materials and Methods
Growth Conditions and Plant Materials.
Transgenic tobacco and Arabidopsis seedlings were grown in sterile culture as described (22), in temperature-controlled growth rooms with 80 μmol photons/m2 per sec of fluorescent white light (23). The tobacco NtPHYB∷LUC lines have been described (24). The tobacco NtPHYB∷PHYB∷LUC fusion was constructed as follows. A 1,485-bp NtPHYB promoter fragment containing the entire 5′ untranslated region but not the ATG was amplified by PCR to generate unique SalI (5′) and BamHI (3′) sites and cloned into the pPCV812 binary vector (25). The NtPHYB cDNA then was cloned into this construct as a BamHI–SmaI fragment, after modification as described (19) to facilitate the fusion and remove the NtPHYB stop codon. Finally, unique SmaI (5′) and SacI (3′) sites were created in the LUC reporter gene by PCR, and this fragment was added to the construct. The Arabidopsis AtPHYB∷LUC lines (ecotype C24) carried a fusion of 2,292 bp of the Arabidopsis PHYB promoter (26) to the luciferase reporter (22) in the binary vector pPCV812. The tobacco and Arabidopsis chlorophyll a/b-binding protein gene (CAB) 2∷LUC lines have been described (22, 27).
Luminescence and RNA Assays.
Luciferase luminescence was measured by low-light video imaging, using intensified (Hamamatsu VIM, Hamamatsu City, Japan) and liquid-nitrogen cooled (Princeton Instruments, Trenton, NJ, LN/CCD-512-TKB) cameras, essentially as described (27, 28). The luminescence data shown is representative of 3–4 replicate experiments, incorporating at least two independently transformed lines, all of which gave very similar results. Total RNA was extracted as described (29). For PHYB RNA quantification, 100 μg of total RNA per lane was hybridized to an end-labeled oligonucleotide probe and digested with S1 nuclease, as described (30). For CAB RNA quantification, 20 μg of total RNA per lane was analyzed by RNA gel blot hybridization, as described (22), with the Arabidopsis CAB2 coding region probe that will hybridize to most members of the CAB multigene family. The RNA signals from replicate gels were quantified by using a PhosphorImager (Molecular Dynamics).
Protein Assays.
PHYB protein levels were tested in total cell protein extracts, prepared by grinding 1 g of plant tissue in liquid nitrogen for 2 min, adding 1 ml of extraction buffer [100 mM Mops, pH 7.6/50% ethylene glycol/5 mM EDTA/14 mM 2-mercaptoethanol with a Complete mini tablet (Boehringer Mannheim), with 100 μl of 1 M iodoacetamide added per 10 ml of extraction buffer before use], and grinding for an additional 2 min. The mixture was centrifuged at 4°C for 20 min at 20,000 rpm, then 0.5 ml of supernatant was added to 0.1 ml of 6× Laemmli buffer (300 mM Tris⋅HCl, pH 6.8/0.6% bromophenol blue/60% glycerol/12% SDS, with DTT powder added to 600 mM before use) and boiled for 3 min. Ten-microliter aliquots of each extract were analyzed by SDS/PAGE. Equal loading of total protein and even transfer to a Hybond-C membrane were confirmed by Ponceau S staining of the membrane. The membrane was hybridized overnight with either the polyclonal antiserum pRTB (31) or a histone H2b-specific antibody (32). PHYB antigen was quantified by hybridization with a mouse IgG-peroxidase conjugate (according to the manufacturer’s instructions; Sigma), chemiluminescent visualisation (ECL system, Amersham Pharmacia), and densitometric scanning (Molecular Dynamics). The data shown are representative of two independent experiments, each of which gave very similar results. Protein extracts from plants harvested in constant darkness showed rhythmic PHYB abundance in some experiments, but with low amplitude and variable phase; among the replicate experiments, mean PHYB abundance was not rhythmic. Similar results were obtained by using the MAT-1 mAb, which is also specific for PHYB (33).
Results
Luminescent Reporters for PHY Gene Expression.
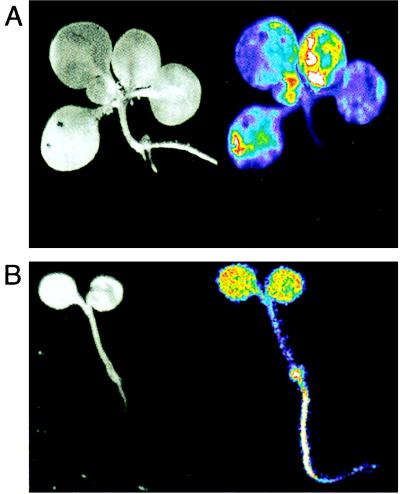
The bioluminescence patterns of transgenic tobacco seedlings carrying a tobacco PHYB1 fusion to luciferase (NtPHYB∷LUC) were characterized by in vivo imaging, after growth in darkness or in light-dark cycles (LDs). The NtPHYB∷LUC construct was principally expressed in the aerial tissues (Fig. 1). Arabidopsis plants carrying an Arabidopsis PHYB∷LUC fusion (AtPHYB∷LUC) were luminescent in all tissues (Fig. 1). These data are consistent with earlier observations of RNA abundance and β-glucuronidase reporter fusions in both species (34–36).
Figure 1.
Organ specificity of luminescence in transgenic PHYB∷LUC seedlings. (A) NtPHYB∷LUC expression was imaged in 3-week-old, LD-grown tobacco seedlings. (B) AtPHYB∷LUC expression was imaged in 1-week-old, LD-grown Arabidopsis seedlings. (Left) Reflected-light image. (Right) Luminescence image presented in false-color (blue, low intensity; red, high intensity).
PHY Gene Expression Is Controlled by the Circadian Clock.
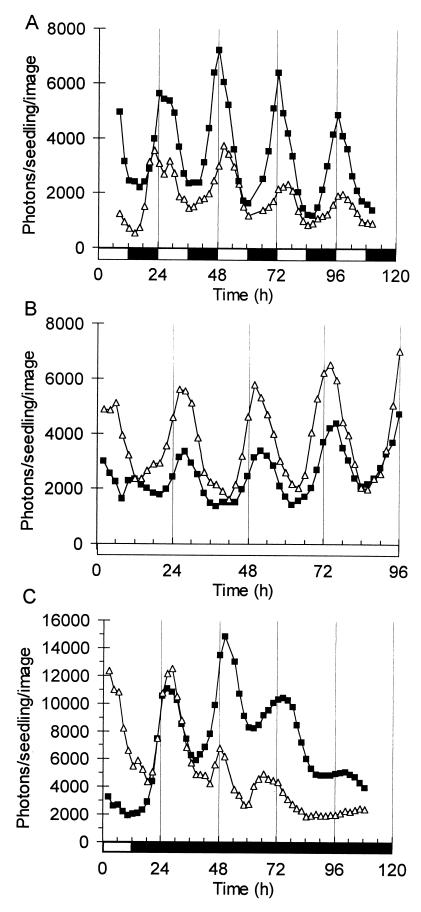
NtPHYB∷LUC tobacco seedlings were grown for 2 weeks and imaged under LDs, together with CAB2∷LUC controls (Fig. 2A). The peak of CAB2∷LUC activity occurred 2–4 hr after lights on, showing the anticipation of lights on and lights off that is typical of circadian-regulated genes. The PHYB promoter directed diurnal cycling of luciferase activity, with maximal expression at lights on and minimal activity at lights off. The rhythm of NtPHYB∷LUC expression in LD unexpectedly had a higher amplitude than CAB2∷LUC. The pattern is consistent with circadian regulation at an earlier phase than CAB or with a negative regulation of PHYB expression by light, which previously has been described for PHYA rather than PHYB (29).
Figure 2.
Circadian regulation of PHY gene expression in tobacco seedlings. Luminescence of NtPHYB∷LUC (■) and CAB2∷LUC (▵) was imaged in seedlings (A) grown under LDs, or after transfer to (B) constant light or (C) constant darkness. Open box on time axis, light interval; filled box, dark interval.
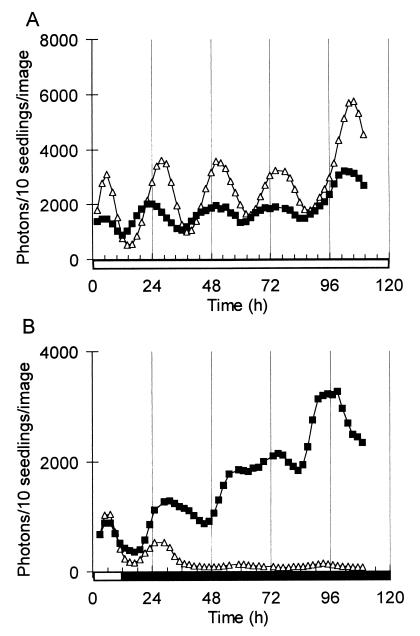
To distinguish between these modes of regulation, seedlings of the same transgenic lines were assayed after transfer to constant light or darkness. NtPHYB∷LUC luminescence was rhythmic under both conditions with a 3- to 4-fold amplitude, indicating that PHYB expression is regulated by a circadian rhythm similar to that of CAB2∷LUC (Fig. 2 B and C). In darkness, the amplitude of the NtPHYB∷LUC rhythm initially rose, and a high amplitude was maintained for at least one cycle longer than the rhythm of CAB2∷LUC luminescence (Fig. 2C), indicating that the peak level of PHYB expression is not down-regulated in darkness in the same manner as CAB expression. Seven-day-old Arabidopsis transformants carrying the native AtPHYB∷LUC fusion were assayed in the same protocol (Fig. 3). AtPHYB∷LUC also was regulated by the circadian clock, with subtle differences compared with the expression pattern in tobacco. In constant light, AtPHYB∷LUC had a slightly lower amplitude and earlier phase than CAB2∷LUC (Fig. 3A). The level of AtPHYB∷LUC activity increased throughout the experiment in darkness, in sharp contrast to the rapid dampening of CAB2∷LUC luminescence levels (Fig. 3B). These data indicate that PHYB expression may be down-regulated by light in green seedlings, in addition to its circadian regulation.
Figure 3.
Circadian regulation of PHYB gene expression in Arabidopsis. Luminescence of AtPHYB∷LUC (■) and CAB2∷LUC (▵) was imaged in seedlings grown under LDs, after transfer to (A) constant light or (B) constant darkness. Open box on time axis, light interval; filled box, dark interval.
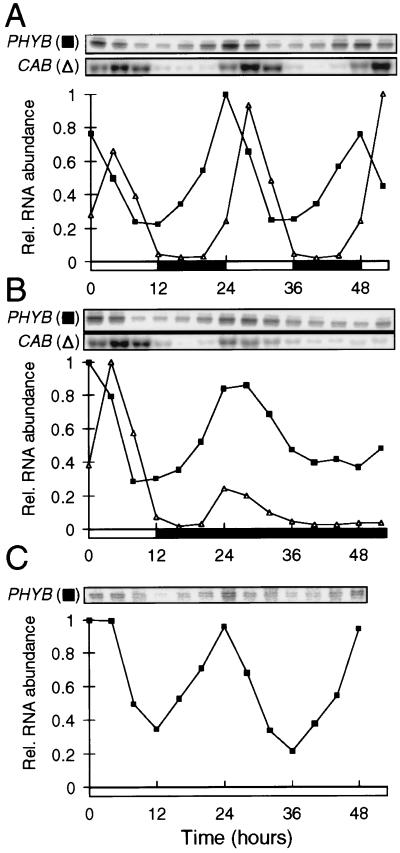
To determine whether the rhythmic promoter activity was maintained at the level of RNA accumulation, PHYB RNA levels were measured by S1 nuclease protection assays, in extracts of 3-week-old tobacco plantlets grown in LD and transferred to constant light or to darkness. RNA from the CAB multigene family also was tested by RNA gel blot hybridization (Fig. 4). The accumulation of native PHYB RNA was rhythmically regulated under all conditions, in a pattern that closely followed the activity of the NtPHYB∷LUC fusion. The peak phase of the PHYB RNA rhythm was similar to that of the CAB family RNA, or about 4 hr earlier. The amplitude of the rhythm in RNA level was 4- to 5-fold, as measured by PhosphorImager analysis. This finding is similar to the amplitude of rhythmic NtPHYB∷LUC activity, indicating that the pattern of PHYB RNA accumulation largely reflects the circadian regulation of the PHYB promoter.
Figure 4.
Rhythmic accumulation of PHYB RNA. RNA from PHYB (■) and the CAB gene family (▵) were detected by S1 nuclease protection and RNA gel blot hybridization, respectively, in extracts of tobacco seedlings (A) grown under LDs, or after transfer to (B) constant darkness or (C) constant light. Hybridization signals were quantified by PhosphorImager analysis and are normalized to the highest signal. Open box on time axis, light interval; filled box, dark interval.
PHYB Protein Is Rhythmically Synthesized, But Accumulates with a Reduced Amplitude.
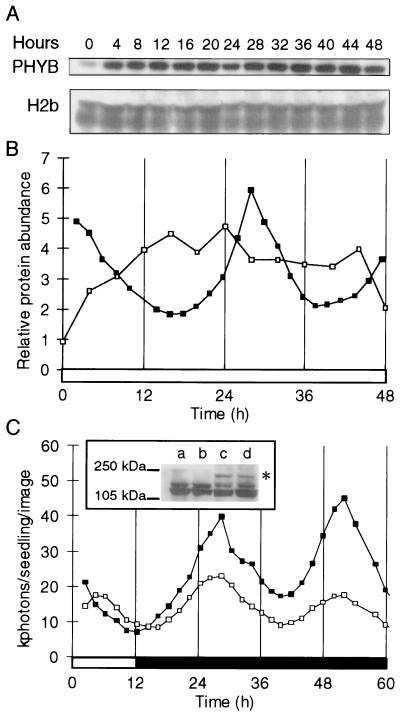
Total protein extracts were isolated from 3-week-old tobacco plantlets under LD and after transfer to constant light or darkness. The abundance of bulk PHYB protein and a nuclear marker protein, histone H2b, was assayed by Western blot analysis. The level of PHYB protein showed very weak rhythmicity, if any, whereas histone H2b levels were not rhythmically regulated (Fig. 5A). The PHYB signal was quantified by densitometry and normalized to the histone as a control, to avoid artefacts of gel loading or nuclear extraction (Fig. 5B). The severe reduction in the amplitude of the PHYB protein rhythm, compared with the rhythm of PHYB RNA, indicated that the rhythm in RNA abundance was masked by either translational or posttranslational regulation.
Figure 5.
Circadian regulation of PHYB protein synthesis and accumulation. (A) The abundance of PHYB and histone H2/2b proteins was determined by Western blot analysis, in extracts of tobacco plants grown under LD and transferred to constant light. (B) Luminescence of NtPHYB∷LUC (■) and PHYB protein quantification (□). Western blots were analyzed by scanning densitometry. PHYB protein levels were normalized to histone levels after local background subtraction. (C) Luminescence of NtPHYB∷LUC (■) and NtPHYB∷PHYB∷LUC (□) tobacco plants was measured by imaging plants grown under LDs and transferred to constant darkness at time 12 h. NtPHYB∷LUC levels are divided by three to facilitate phase comparison. (Inset) The detection of the PHYB∷LUC fusion protein (*) specifically in protein extracts of the NtPHYB∷PHYB∷LUC plants, using the PHYB-specific antiserum. The major band at 120 kDa is PHYB. Lanes a and b, NtPHYB∷LUC; lanes c and d, NtPHYB∷PHYB∷LUC; lanes a and c, harvested after 12 h dark (time 24 h); lanes b and d, harvested after 24 h dark (time 36 h). Open box on time axis, light interval; filled box, dark interval.
To distinguish between these mechanisms, a translational fusion between the coding regions of luciferase and NtPHYB was constructed, under the control of the NtPHYB promoter. The luminescence of transgenic tobacco plants carrying this construct (NtPHYB∷PHYB∷LUC) thus would reflect the rate of synthesis of the PHYB∷LUC fusion protein. NtPHYB∷PHYB∷ LUC plantlets were grown under LD and imaged under constant conditions, as described above. The rhythmic luminescence pattern of these seedlings was very similar to the luminescence driven by the NtPHYB∷LUC fusion, both in the light (data not shown) and in darkness (Fig. 5C). Correct synthesis of the fusion protein was confirmed by Western blotting with PHYB-specific antisera, in extracts of plants harvested after 12 h and 24 h in darkness (Fig. 5C). Even if a portion of the fusion protein is cleaved to release monomeric luciferase, the rhythmic luminescence driven by the NtPHYB∷PHYB∷LUC fusion construct indicates that the synthesis of new PHYB protein is rhythmic. This rhythmicity subsequently is most likely to be masked by posttranslational mechanisms operating at the level of the PHYB protein.
Discussion
Rhythmic Regulation of Phytochrome Genes.
Phytochrome photoreceptors have been closely linked to the circadian system in higher plants, but there is very little information available about the control of phytochromes themselves by the circadian clock. Spectrophotometric assays that reflect the most abundant phytochrome species, phyA, did not reveal any circadian cycling of the total phytochrome in dark-adapted seedlings (37). Analysis of PHYA mRNA levels (38) as well as the luminescence of both NtPHYA∷LUC and NtPHYB∷LUC transgenes (24) resulted in uniform, nonrhythmic patterns in completely dark-grown tobacco seedlings. Other circadian markers such as the accumulation of tobacco CAB21 and CAB40 RNA (38) and the luminescence of Arabidopsis CAB2∷LUC (22, 39) and wheat CAB1∷LUC (24) reporters are clearly rhythmic at this developmental stage. These light-independent rhythms, which are likely to be induced by imbibition (38, 40), evidently do not control expression of the PHY genes.
Published results on PHY expression patterns in plants grown in LDs have been more variable. No oscillation in RNA abundance was reported for Arabidopsis PHY genes (26) and tobacco PHYA (29). In contrast, Hauser et al. (41) detected diurnal cycling in the RNA abundance of specific tomato PHY genes in greenhouse-grown plants but no rhythms under temperature-controlled conditions of constant light or darkness (including in the PHYB1 gene that is most similar to the tobacco PHYB1 used in this work). We have tested the activity of the tobacco NtPHYB∷LUC lines created by Kolar et al. (24) and additional Arabidopsis lines, in which expression of the LUC reporter was driven by the Arabidopsis PHYB promoter (Fig. 1). The activity of the PHYB∷LUC reporters is regulated by a circadian clock when the plants are transferred to constant light or to constant darkness (Figs. 2 and 3). The observed PHYB∷LUC rhythms are very similar to the CAB2∷LUC rhythm in LD and constant light, having identical periods and high amplitudes. The CAB2∷LUC rhythm damps rapidly in darkness, especially in Arabidopsis, because of the overall decrease in the level of CAB transcription. The persistence of the PHYB∷LUC rhythms in constant light and darkness probably reflect the relatively light-insensitive transcription of PHYB genes, which is consistent with previously published results (34, 35). The rhythmic expression of PHYB∷LUC in seedlings grown in LD and transferred to constant darkness (Fig. 2) contrasts with the data for completely dark-grown seedlings, in which expression of the PHYB∷LUC reporter is arhythmic (24).
Functions in Phototransduction.
The significance of the observed rhythms in PHYB∷LUC expression is underlined by the circadian rhythms in the accumulation of PHYB RNA (Fig. 4) and in the synthesis of PHYB protein (Fig. 5C) under LD, constant light, and constant darkness conditions. The synthesis of new PHYB protein was measured by the luminescence driven by the NtPHYB∷PHYB∷LUC transgene, which encodes a PHYB∷LUC fusion protein (Fig. 5C). The rhythm of PHYB synthesis retains a similar amplitude to the rhythm of promoter activity and closely follows the accumulation of PHYB RNA, which indicates that translational control has little effect on PHYB accumulation. When the PHYB∷LUC fusion protein was expressed from the cauliflower mosaic virus 35S promoter, there was no evidence of rhythmic, posttranslational regulation that was antagonistic to the rhythm in synthesis (L.K.B. and A.H., unpublished results). A long half-life in the PHYB protein therefore is most likely to account for the observed suppression of circadian rhythmicity in the accumulation of bulk PHYB (Fig. 5B). Synthesis of PHYB in the morning thus would contribute a daily peak of new PHYB, but the contribution was apparently too small for reliable detection by Western blotting, in the presence of the larger, existing PHYB pool.
The rhythmic synthesis nonetheless might be significant. The phase of the maximal PHYB protein synthesis (ZT 0–2) is similar to the phase of maximal light responsiveness for CAB expression (at ZT4; ref. 42) and to the maximal inhibition of hypocotyl elongation (at ZT0; ref. 23) in Arabidopsis. A 2-fold change in PHYB gene dosage causes well-defined, biological responses (4, 5) but would have been at the limit of detection in our assays. The daily change in bulk PHYB protein thus might be functionally relevant. Light-regulated nuclear import of PHYB in Arabidopsis (20, 21) and in tobacco (19), and the interaction of PHYB with a putative basic helix–loop–helix transcription factor (18) have been reported. These data indicate that at least some components of the phototransduction cascade can be localized in the nucleus. The regulation of partitioning therefore provides an additional control mechanism, which operates on mature phyB and which might be affected by the circadian clock. However, the functional significance of the phyB localization for circadian or light responses remains to be elucidated.
Comparison to Other Circadian Systems.
phyB null mutations lengthen the circadian period in a wavelength- and irradiance-dependent manner, whereas PHYB overexpressors shorten the circadian period (10). phyB is therefore one of at least three phytochromes and two cry photoreceptors that mediate light signaling to the Arabidopsis circadian clock. The circadian regulation of PHYB shows that this gene not only encodes a bona fide circadian input photoreceptor but is also a downstream target of circadian output pathways. Recent evidence suggests that CRY genes in animals share this dichotomous relationship to the circadian system, but that their function is indispensable for circadian rhythmicity. CRY mRNA levels are circadian-regulated in Drosophila and in the mouse (12, 43, 44) but the abundance of Drosophila CRY protein (dCRY) shows no obvious rhythmicity in darkness (12), similarly to PHYB (Fig. 5). dCRY confers light sensitivity on yeast and insect cells (45) and is involved in circadian light input in the fly, though it is probably not the only photoreceptor (12). cry2 mutant mice are affected in light input (14) but it is unclear whether the mouse CRY proteins are functional photoreceptors in the circadian system. Mutant mice that completely lack CRY are not period-altered but arhythmic, as are most cells in the equivalent mutant flies (13, 15). The arhythmia probably arises because interaction with CRY regulates the availability of the TIMELESS protein (in Drosophila) or the PERIOD proteins (in mouse) in the nucleus (44, 45), where these proteins are crucial for the circadian oscillator mechanism.
Circadian input in photoautotrophic organisms typically is coupled to more than one class of photoreceptor molecule. PHYB is the first plant protein that is known to share the circadian regulation and nuclear localization of the animal CRYs, and that clearly affects circadian function. The PAS domain of PHYB might mediate interaction with PERIOD or TIMELESS homologues, if these exist in plants. These similarities are consistent with the idea that the circadian oscillator mechanism in plants is distinct from that in animals, but that it involves phytochromes in a fashion analogous to the animal CRY proteins. The apparent light-dependence of phyB nuclear translocation (19, 21) might limit the perdurance of this function in extended darkness, under which conditions many plant circadian rhythms damp out rapidly. The Arabidopsis phyB null mutant does not cause arhythmia, however, under any conditions (10, 46), suggesting that the putative oscillator function of phyB might be redundant with other phytochromes (10); in this case their removal by multiple mutations will lead to arhythmia. Alternatively, the evolutionary adoption of phytochrome as a circadian input photoreceptor may not have been independent of CRY, but rather as an addition to a CRY-dependent oscillator. Recent results have identified substrates for phytochrome kinase activity in vitro, one of which is recombinant CRY1 protein (47); some cry1 mutations also impair phytochrome responses in vivo (47). It therefore will be of great interest to determine whether phytochromes and CRYs have an obligate interaction in the circadian input pathway, or whether phytochromes have been adopted in a novel, CRY-independent mechanism for a functional circadian oscillator in plants.
Acknowledgments
We are grateful to Dr. Tim Kunkel and Prof. Eberhard Schäfer for the phytochrome and histone antibodies. This research was supported by grants from the Biotechnology and Biological Sciences Research Council (G08667) to A.J.M., the Human Frontier Science Program Organization to A.J.M. and F.N., and the Howard Hughes Medical Institute (75195-542401) and Országos Tudományos Kutatási Alapprogramok (Hungary) (T016167) and the National Committee for Technological Development (E133) to F.N. The luminescence imaging facility at Warwick was established with funding from the Biotechnology and Biological Sciences Research Council and the Gatsby Charitable Foundation and Royal Society. L.K.B.’s work was supported in part by a Joint Project grant from the Royal Society, and F.N.’s work was supported by a fellowship from the British Council.
Abbreviations
- phyA
phytochrome A
- phyB
phytochrome B
- LUC
firefly luciferase gene
- CAB
chlorophyll a/b-binding protein gene
- LD
light-dark cycle
- cry
cryptochrome
Note Added in Proof
Similar regulation has been demonstrated for the circadian clock-associated protein, CCA1 (48).
References
- 1.Kendrick R E, Kronenberg G H M. Photomorphogenesis in Plants. Dordrecht, The Netherlands: Kluwer; 1994. [Google Scholar]
- 2.Sharrock R A, Quail P H. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- 3.Reed J W, Nagpal P, Poole D S, Furuya M, Chory J. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koornneef M, Rolff E, Spruit C J P. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- 5.Wester L, Somers D E, Clack T, Sharrock R A. Plant J. 1994;5:261–272. doi: 10.1046/j.1365-313x.1994.05020261.x. [DOI] [PubMed] [Google Scholar]
- 6.Millar A J. New Phytol. 1999;141:175–197. doi: 10.1046/j.1469-8137.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- 7.Dunlap J C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 8.Yan O Y, Andersson C R, Kondo T, Golden S S, Johnson C H. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millar A J, Straume M, Chory J, Chua N-H, Kay S A. Science. 1995;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- 10.Somers D E, Devlin P F, Kay S A. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 11.Zeng H K, Qian Z W, Myers M P, Rosbash M. Nature (London) 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 12.Emery P, So W V, Kaneko M, Hall J C, Rosbash M. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 13.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager Smith K, Kay S A, Rosbash M, Hall J C. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 14.Thresher R J, Vitaterna M H, Miyamoto Y, Kazantsev A, Hsu D S, Petit C, Selby C P, Dawut L, Smithies O, Takahashi J S, Sancar A. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 15.van der Horst G T J, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker A P M, van Leenen D, et al. Nature (London) 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 16.Crosthwaite S K, Dunlap J C, Loros J J. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 17.Lagarias D M, Wu S H, Lagarias J C. Plant Mol Biol. 1995;29:1127–1142. doi: 10.1007/BF00020457. [DOI] [PubMed] [Google Scholar]
- 18.Ni M, Tepperman J M, Quail P H. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 19.Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schäfer E, Nagy F. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakamoto K, Nagatani A. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi R, Nakamura M, Mochizuki N, Kay S A, Nagatani A. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millar A J, Short S R, Chua N H, Kay S A. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowson-Day M J, Millar A J. Plant J. 1999;17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- 24.Kolar C, Fejes E, Adam E, Schäfer E, Kay S, Nagy F. Plant J. 1998;13:563–569. doi: 10.1046/j.1365-313x.1998.00048.x. [DOI] [PubMed] [Google Scholar]
- 25.Koncz C, Schell J. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 26.Clack T, Mathews S, Sharrock R A. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- 27.Millar A J, Short S R, Hiratsuka K, Chua N-H, Kay S A. Plant Mol Biol Rep. 1992;10:324–337. [Google Scholar]
- 28.Michelet B, Chua N H. Plant Mol Biol Rep. 1996;14:320–329. [Google Scholar]
- 29.Adam E, Szell M, Szekeres M, Schäfer E, Nagy F. Plant J. 1994;6:283–293. [Google Scholar]
- 30.Millar A J, Kay S A. Plant Cell. 1991;3:541–550. doi: 10.1105/tpc.3.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel T, Speth V, Buche C, Schäfer E. J Biol Chem. 1995;270:20193–20200. doi: 10.1074/jbc.270.34.20193. [DOI] [PubMed] [Google Scholar]
- 32.Harter K, Kircher S, Frohnmeyer H, Krenz M, Nagy F, Schäfer E. Plant Cell. 1994;6:545–559. doi: 10.1105/tpc.6.4.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Juez E, Nagatani A, Tomizawa K I, Deak M, Kern R, Kendrick R E, Furuya M. Plant Cell. 1992;4:241–251. [PMC free article] [PubMed] [Google Scholar]
- 34.Adam E, Kozma-Bognar L, Kolar C, Schäfer E, Nagy F. Plant Physiol. 1996;110:1081–1088. doi: 10.1104/pp.110.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somers D E, Quail P H. Plant Physiol. 1995;107:523–534. doi: 10.1104/pp.107.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Somers D E, Quail P H. Plant J. 1995;7:413–427. doi: 10.1046/j.1365-313x.1995.7030413.x. [DOI] [PubMed] [Google Scholar]
- 37.Wildermann A, Drumm H, Schäfer E, Mohr H. Planta. 1978;141:211–216. doi: 10.1007/BF00387891. [DOI] [PubMed] [Google Scholar]
- 38.Kolar C, Adam E, Schäfer E, Nagy F. Proc Natl Acad Sci USA. 1995;92:2174–2178. doi: 10.1073/pnas.92.6.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hicks K A, Millar A J, Carré I A, Somers D E, Straume M, Meeks-Wagner D R, Kay S A. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- 40.Hung C Y, Lin Y, Zhang M, Pollock S, Marks M D, Schiefelbein J. Plant Physiol. 1998;117:73–84. doi: 10.1104/pp.117.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hauser B A, Cordonnier Pratt M M, Pratt L H. Plant J. 1998;14:431–439. doi: 10.1046/j.1365-313x.1998.00144.x. [DOI] [PubMed] [Google Scholar]
- 42.Millar A J, Kay S A. Proc Natl Acad Sci USA. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyamoto Y, Sancar A. Proc Natl Acad Sci USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kume K, Zylka M J, Sriram S, Shearman L P, Weaver D R, Jin X W, Maywood E S, Hastings M H, Reppert S M. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 45.Ceriani M F, Darlington T K, Staknis D, Mas P, Petti A A, Weitz C J, Kay S A. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 46.Anderson S L, Somers D E, Millar A J, Hanson K, Chory J, Kay S A. Plant Cell. 1997;9:1727–1743. doi: 10.1105/tpc.9.10.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmad M, Jarillo J A, Smirnova O, Cashmore A R. Mol Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- 48.Green R M, Tobin E M. Proc Natl Acad Sci USA. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]