Abstract
The isomerization of chorismate to prephenate by chorismate mutase in the biosynthetic pathway that forms Tyr and Phe involves C5—O (ether) bond cleavage and C1—C9 bond formation in a Claisen rearrangement. Development of negative charge on the ether oxygen, stabilized by Lys-168 and Glu-246, is inferred from the structure of a complex with a transition state analogue (TSA) and from the pH-rate profile of the enzyme and the E246Q mutant. These studies imply a protonated Glu-246 well above pH 7. Here, several 500-ps molecular dynamics simulations test the stability of enzyme–TSA complexes by using a solvated system with stochastic boundary conditions. The simulated systems are (i) protonated Glu-246 (stable), (ii) deprotonated Glu-246 (unstable), (iii) deprotonated Glu-246 plus one H2O between Glu-246 and the ether oxygen (unstable), (iv) the E246Q mutant (stable), and (v) addition of OH− between protonated Glu-246 and the ether oxygen. In (v), a local conformational change of Lys-168 displaced the OH− into the solvent region, suggesting a possible rate-determining step that precedes the catalytic step. In a 500-ps simulation of the enzyme complexed with the reactant chorismate or the product prephenate, no water molecule remained near the oxygen of the ligand. Calculations using the linearized Poisson–Boltzmann equation show that the effective pKa of Glu-246 is shifted from 5.8 to 8.1 as the negative charge on the ether oxygen of the TSA is changed from −0.56 electron to −0.9 electron. Altogether, these results support retention of a proton on Glu-246 to high pH and the absence of a water molecule in the catalytic steps.
Keywords: allosteric enzyme/molecular dynamics/stochastic boundary condition/enzyme catalysis/protonation state
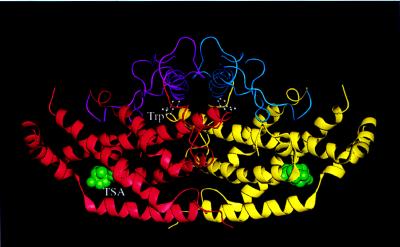
In archaebacteria, eubacteria, fungi, and other plants, chorismate mutase (chorismate pyruvate mutase, EC 5.4.99.5) (CM) catalyzes the isomerization of chorismate to prephenate. This branch of the metabolic pathway to aromatic amino acids leads to phenylalanine and tyrosine. The other branch (the conversion of chorismate to anthranilate by anthranilate synthase) leads to tryptophan (1, 2). In Saccharomyces cerevisiae, CM (Fig. 1) is a homodimer consisting of two 30-kDa polypeptide chains. This yeast CM (YCM) shows homotropic activation by the substrate chorismate, approximately 10-fold allosteric activation by tryptophan, and approximately 10-fold inhibition by tyrosine (2, 3). This 100-fold modulation of YCM activity by Trp and Tyr controls the flux of chorismate into these two branches of the biosynthesis of aromatic amino acids. The structural basis for these regulatory processes has been described recently (4, 5).
Figure 1.
Schematic representation of the dimeric structure of YCM. The allosteric domain of the left monomer (in red) is drawn in magenta and that of the right monomer (in yellow) is drawn in blue. The two effector tryptophans (in white) are in ball-and-stick form and the transition state analogues (in green) are in space-filling model. This figure, as well as Figs. 4–7 and 9, was made with quanta (Molecular Simulations, San Diego, CA) [(modified from (4)].
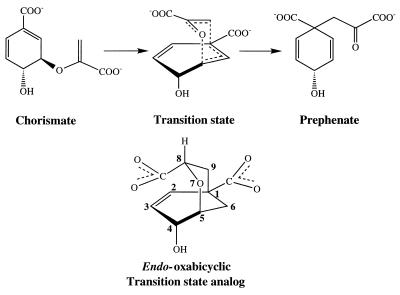
The CMs catalyze the only known Claisen rearrangement in living systems (Fig. 2). It has been shown that the enzyme preferentially binds the less stable pseudodiaxial conformation of the enolpyruvate side chain (6) (Fig. 2), which is positioned thereby for the rearrangement in the chair-like transition state (7, 8). Its complex with the active site of YCM (4) is shown in Fig. 3. The mechanistic aspects of catalysis, outlined before the structures were known, include several possibilities (9), one of which was transient protonation of the ether (leaving) oxygen of the enolpyruvate side chain, and another involved hydrogen bonds to the ether oxygen to stabilize the developing negative charge on this leaving oxygen. Indeed, when an oxabicyclic diacid transition state analogue, prepared by Bartlett and coworkers (10, 11), is bound to YCM, hydrogen bonds involving the ether oxygen are inferred from its contacts with Lys-168 and Glu-246 (4). Also, the protonated form of Glu-246 is supported by pH-rate profiles of the wild-type enzyme and the Glu-246 → Gln mutant (3, 12), and by the determination of the location (13) of the active site in YCM on the basis of homology with the P protein of the CM of Escherichia coli (14). However, in a recent computational study (15) of docking the transition state analogue to YCM, it is argued that, around the activity maximum of about pH 6.5 when Trp is present to maximize activation, Glu-246 is unprotonated, and that its interaction with the ether (leaving) oxygen is mediated by an intervening water molecule. Because of the well known uncertainty of such modeling studies in determining specific interactions, we have performed molecular dynamics studies based on the crystal structure of YCM (4) complexed with the TSA. Simulations of the active site complexed with the TSA are performed for protonated or unprotonated Glu-246, for the mutant Gln 246, and for the structure, which has a water molecule between Glu-246 and the ether (leaving) oxygen; this last configuration was suggested as the stable structure in the modeling study (15). However, there is no evidence in the dynamics studies presented here for the presence of such an intervening water molecule in any of the structures in which the TSA is bound to YCM. A simulation is also performed for a system with an ionized water molecule modeled with its OH− part in the active site at a position similar to that of the intervening water. Simulations indicate that this OH− ion is transported to solvent by a local conformational change of Lys-168. No water was observed in this position during the simulations with reactant chorismate or product prephenate. Finally, the effective pKa value of Glu-246, calculated by using the linearalized Poisson–Boltzmann method, is raised significantly so that Glu-246 is likely to retain a proton at an elevated pH.
Figure 2.
Transformation of chorismate to prephenate via proposed transition state.
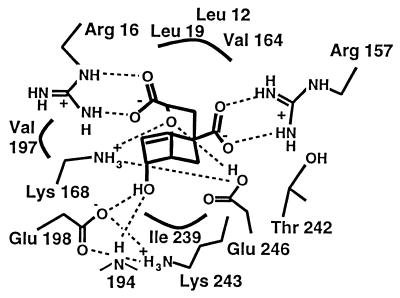
Figure 3.
Active site-binding interactions (4) [Reproduced with permission from Current Biology Ltd. (4) (Copyright 1997, Current Biology Ltd.)].
METHODS
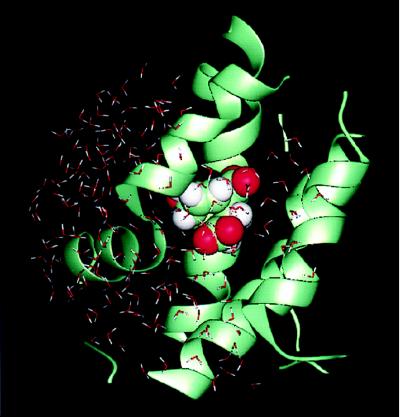
Initial coordinates (Protein Data Bank code 3csm) were those of the wild-type “super R” state bound to inhibitor plus Trp (ITRP in ref. 4), in which tryptophan and the TSA are bound to the regulatory and active sites, respectively. To reduce the necessary simulation time, in accord with many other studies of active sites, the stochastic boundary molecular dynamics method (16, 17) was used; this procedure also avoided the need to model certain loops that are not well defined in the crystal structure. The charmm program (Molecular Simulations, San Diego, CA) (18) was used for the simulation. The all-hydrogen potential function (PARAM22) (19) was used for all protein atoms, and a modified TIP3P water model (20) was used for the solvent (21). Atomic partial charges for the transition state analogue were obtained from the Merck Molecular Force Field (Merck) (22–26) implemented for molecular dynamics in the charmm program. The system was separated into a reaction zone and a reservoir region, and the reaction zone was further divided into the reaction region and the buffer region (for details of stochastic boundary molecular dynamics, see ref. 17). The reference point for partitioning the system in stochastic boundary molecular dynamics was chosen as the bridge ether oxygen of the inhibitor. The reaction region around an active site was the sphere of radius r of 14 Å, the buffer region was 14 < r <16 Å, and the reservoir region corresponded to r > 16 Å; all atoms in the reservoir region were deleted. The simulation system, shown in Fig. 4, consisted of 101 protein residues (1731 atoms), a TSA, and 193 waters. Inside the reaction region, atoms were propagated by molecular dynamics (17), whereas atoms in the buffer region were propagated by Langevin dynamics. Atoms inside the buffer region were retained by a harmonic restoring force with force constants derived from the temperature factors in the crystal structure. Water molecules were confined to the active site region by a deformable boundary (27). The friction constant in the Langevin dynamics (17) was 250 ps−1 for the protein atoms and 62 ps−1 for the water molecules. During the simulation, all of the bonds with hydrogen atoms were fixed by the SHAKE algorithm (18). A 1-fs time step was used for integration of the equations of motion during the molecular dynamic simulation. After 200 steps of minimization by using the steepest descent method and 300 steps of minimization by using the Adapted Basis Newton Raphson’s method (18), the system was equilibrated for 50 ps at 300 K. This procedure was followed by a 500-ps production run. Seven simulations were performed on the enzyme-inhibitor complex with (i) the Glu-246 protonated, (ii) the Glu-246 deprotonated, (iii) the Glu-246 replaced by Gln-246 (the E246Q mutant), (iv) a water molecule between an unprotonated Glu-246 and the ether oxygen of the TSA, (v) an ionized water molecule modeled in a position similar to that in iv, such that the H+ is attached to Glu-246 and the OH− is placed nearby, (vi) the transition state analogue modeled as the reactant chorismate, and (vii) the transition state analogue modeled as the product prephenate.
Figure 4.
System included in the stochastic boundary molecular dynamics simulation. The protein is in ribbon representation, the TSA is in space-filling model, and the water molecules are drawn as a stick model.
For the evaluation of the pKa, the linearized Poisson–Boltzmann equation was solved by using the finite difference method (for details, see refs. 28–31). For this part of the study, the whole YCM monomer structure was used. The protein coordinate was prepared by 200 steps of minimization by using the steepest descent method and 100 steps of minimization by using the Adapted Basis Newton Raphson’s method. The calculation, using a modified version of the uhbd program (28) that is integrated with the charmm program, was carried out on a 180 × 180 × 180 grid at an initial spacing of 3 Å and converged iteratively to 0.2 Å. Dielectric constants of 80 for solvent and 20 for the interior of the protein were used. A physiological ionic strength (145 mM), and a temperature of 298 K were chosen. The Stern (ion-exclusion) layer is 2 Å. The pKa calculation was performed by using a program developed by M. Schaefer, M. Sommer and M. Karplus (31).
RESULTS
Dynamics with Glu-246 Protonated.
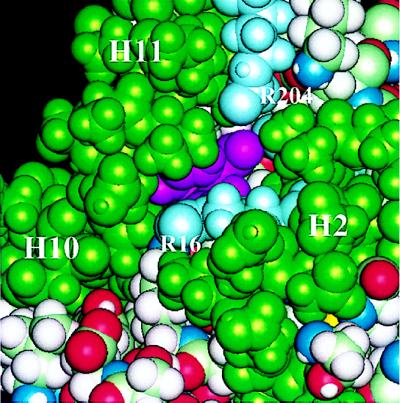
During the 500-ps simulation in the presence of the TSA and a protonated Glu-246, the TSA remained bound deeply buried within the four-helix bundle formed by H2, H8, H11, and H12, consistent with the crystal structure (4). The two carboxylate groups of the inhibitor form well defined salt bridges with Arg-16 and Arg-157. Except for the channel (the “allosteric pathway”) between H2 and H11, the inhibitor remained inaccessible to solvent. Two arginine residues, Arg-16 on H2 and Arg-204 on H11, are located near the entry of this channel. In a space-filling model (Fig. 5), one can see part of the inhibitor from outside in the direction of this channel, which may be the route for substrate binding and escape. In particular, the negatively charged inhibitor may be guided by the electrostatic field generated by the arginine residues, especially Arg-204. This channel is located furthest from the allosteric domain and facing away from the dimer interface. Inside this part of the channel, there is a hydrogen-bonding network formed by the water molecules and protein atoms (see Fig. 6).
Figure 5.
Space-filling representation of the substrate channel. The helices surrounding the channel (H2, H10, and H11) are in green. The arginine side chains of R 16 and R 204 located at the entry of the channel are in light blue, and the inhibitor is in magenta.
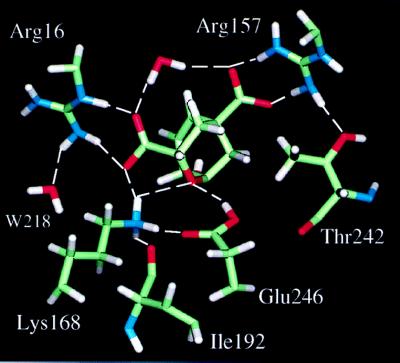
Figure 6.
Hydrogen-bonding network observed inside the substrate channel.
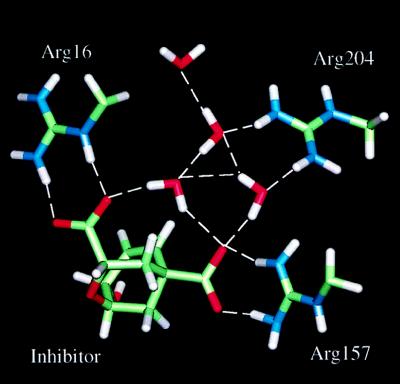
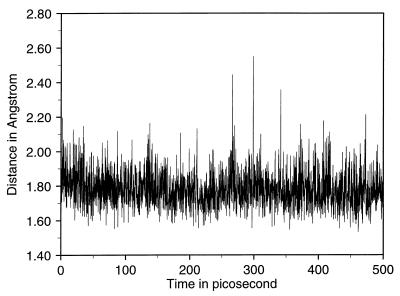
An essential result of the simulation is the stability of the interactions between the ether bridge oxygen atom of the inhibitor and the polar side chains of Lys-168 and Glu-246. The key residue Glu-246 remains completely solvent inaccessible, as does the ether bridge oxygen; the waters nearest to the active site are separated from Glu-246 by the side chain of Lys-168. A water molecule interacts with the guanidinium group of Arg-16 (see Fig. 7). It was suggested in previous work (4) that the side chain of Glu-246 is probably protonated, based on the distance of about 3.1 Å (4) between its carboxylate group and the ether oxygen observed in the crystal structure. The present simulation clearly demonstrates that, when the proton is added to the carboxylate group of Glu-246, the structural integrity is well maintained. Also, Glu-246 remains hydrogen-bonded to Lys-168, which donates a good hydrogen bond to one of the carboxylate groups of the inhibitor (see Fig. 7). The side chain of Lys-168 is within hydrogen-bonding distance of the ether oxygen and of the main chain carbonyl group of Ile 192. Fig. 8 shows the hydrogen bond distance as a function of time for the interaction between the ether oxygen and the proton on the carboxylate oxygen of Glu-246 that is distal to Lys-168. A simulation in which the proton is added to the carboxylate oxygen atom of Glu-246 proximal to Lys-168 resulted in a flip of the side chain of Glu-246. Thus, the protonation of the oxygen distal to Lys-168 is more favorable.
Figure 7.
Interactions between the inhibitor and the groups in the active site.
Figure 8.
Hydrogen-bonding distance as a function of time for the proton on Glu-246 and the ether oxygen of the inhibitor.
Dynamics with Glu-246 Unprotonated.
A 500-ps dynamics simulation with the Glu-246 anion resulted in a severe distortion of the active site structure. The side chain of Glu-246, which remained hydrogen-bonded to Lys-168, was shifted to a distance of about 4.5 Å from the ether oxygen. This result is inconsistent with the x-ray crystal structure (4), in which the distance between the carboxylate of the Glu-246 side chain and the ether bridge oxygen is only about 3.1 Å. The Lys-168 side chain also moved away from its position in the crystal structure, and the hydrogen bond between Lys-168 and the carboxylate group of the inhibitor that interacts with Arg-16 was weakened (i.e., the distance increased).
Dynamics with a Gln-246 Mutant.
When the E246Q mutant was simulated for 500 ps, the structure was stable. One hydrogen of the amide group of the glutamine side chain makes a hydrogen bond to the main chain carbonyl of Thr-242, whereas the other hydrogen of this amide group makes the expected hydrogen bond to the ether oxygen of the inhibitor. There were no significant changes in the structure.
Dynamics with a Water Molecule Between Unprotonated Glu-246 and the Ether Oxygen.
To examine the possibility, suggested by the recent docking study (15), that the Glu-246 side chain interacts with the bridge oxygen through a water molecule, a 500-ps simulation was performed in which a water molecule was modeled into the space between the side chain of Glu-246 and the ether oxygen (as in Fig. 6a of ref. 15). The results show that the presence of a water molecule in this position has a strong effect on the binding mode of the inhibitor. Because of the size of the water molecule, the inhibitor was displaced in the early stage of simulation so that it could no longer make good hydrogen bonds to Arg-16 and Arg-157. Near the middle of the simulation (about 200 ps from the beginning of the production run), the water dissociated from the ether oxygen, though it remained bound to Glu-246. As a result, the inhibitor shifted back toward its original position and regained the hydrogen bonds to Arg-16 and Arg-157. Moreover, even though the water was modeled as hydrogen bonded to both Glu-246 and the ether oxygen, its oxygen atom is surrounded by hydrophobic groups including the methyl groups of Thr-242, Leu-19, Val-164, and a methyl group from the inhibitor. This environment, plus the lack of space, appear to make it unfavorable for the water to stay at the modeled position.
Dynamics with an Ionized Water Molecule Between Glu-246 and the Ether Oxygen.
The uncatalyzed conversion of chorismate to prephenate involves no solvent H2O/D2O isotope effect, and a substantially asymmetric transition state: kH/kT is 1.15 ± 0.01 (equilibrium: 1.43) for C5—O bond breaking and 0.99 ± 0.01 (equilibrium: 0.78) for bond making at C9 (32). Catalysis by the E. coli CM (ECM)-T protein shows a solvent H2O/D2O effect of 2.2 on kcat but no effect (1.0) on Km; moreover, catalysis by ECM-T at low substrate concentrations (Vmax/Km conditions) is insensitive to H–T substitution at C5 or C9, thus implying a rate-determining step (RDS) before the chemical transformation (32). Catalysis by the Bacillus subtilis CM enzyme (BCM) shows no solvent H2O/D2O effect; moreover, catalysis of BCM at high substrate concentration (Vmax conditions) indicates that koff for prephenate is almost identical to kcat, suggesting that under Vmax conditions the loss of prephenate is the RDS (the “on” rate constant for prephenate is almost the same as kcat/Km for chorismate) (33).
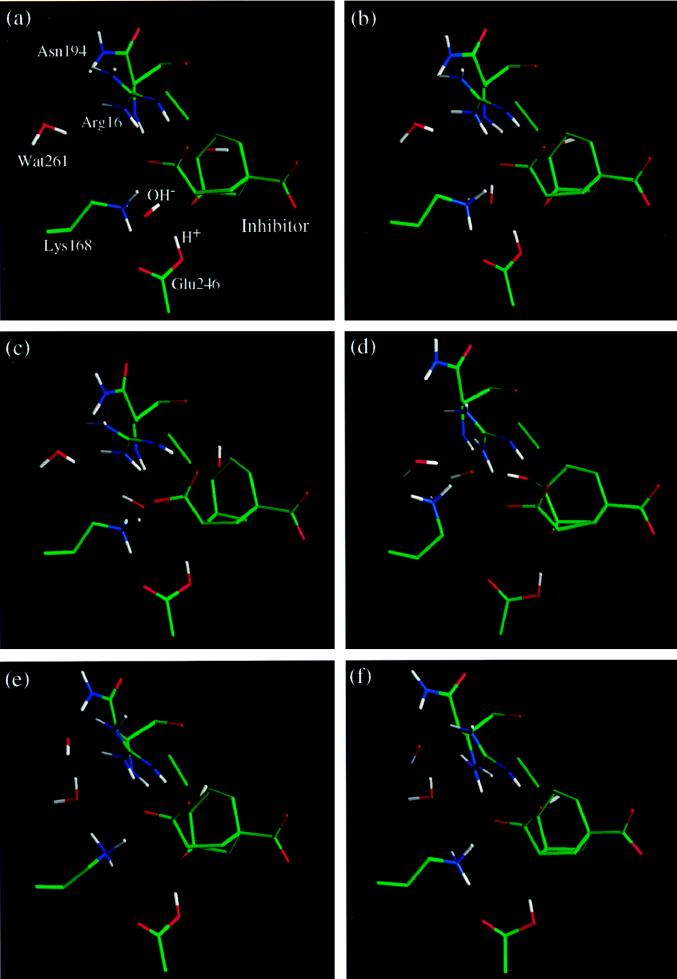
A model for the proposed RDS that precedes the chemical transformation in YCM is examined by introducing an OH−, presumably from an ionized water molecule that has protonated Glu-246. A 500-ps simulation was carried out, starting with an OH− ion in a position similar to that of the intervening water in the previous simulation (see Fig. 9a). At the beginning of the simulation, the OH− ion is hydrogen bonded to Lys-168. Around 50 ps, the OH− ion moves out into the solvent as a result of a local conformational change of the side chain of Lys-168 (Fig. 9 b–d). Although Lys-168 acts as a gate in this process, this lysine residue is bulky enough that no water molecules enter the active site even in the most open conformation involving the complex of the enzyme with TSA, OH−, and solvent. In fact, once the OH− ion is displaced (Fig. 9 d and e), the side chain of Lys-168 quickly regains its original conformation in the enzyme–TSA complex (Fig. 9 e and f). Therefore, one might expect a solvent D2O effect in YCM (Lys-168 and Glu-246) like that observed (9) for the ECM-T (Lys-37 and Glu-86), but not found (33) in BCM, which has bidentate Arg bound to the ether oxygen. In the remaining part of the active site, no large conformational change is observed. Although the transport of the OH− ion is facilitated mainly by the motion of Lys-168, other groups also contribute. In particular, the guanidinium group of Arg-16, the main chain amide group of Asn-94, and the hydroxyl group on the TSA provide favorable interactions together with Lys-168 as the OH− ion slides out (Fig. 9d). The hydroxyl group of the TSA also undergoes a displacement, but quickly recovers its original conformation (Fig. 9 c–e).
Figure 9.
Time evolution of the displacement of OH− ion from the active site mediated by a local conformational change of Lys-168. The nearest water molecule to the active site, Wat261, is indicated explicitly.
This striking result enlarges the possible role of the protonation of Glu-246. It reveals that the ionization of a water molecule and the displacement of the resulting hydroxide anion from the active site could provide the proton to Glu-246, and also, at least in part, could contribute to the RDS. These results provide an alternative to the proposed conformational change of the enolpyruvoyl side chain of the substrate as the RDS step (33) at low substrate concentrations.
Dynamics with Reactant Chorismate or Product Prephenate.
The simulations with the reactant chorismate or the product prephenate, modeled from the transition state analogue, show that both systems are stable. The overall geometry and interactions are close to those observed in the crystal structure of the transition state analogue. Most importantly, no water molecule enters into the active site, including the vicinity of the ether oxygen. In the complexes of the enzyme fragment with either chorismate or prephenate, certain side chains, such as that of Lys-16, which makes direct contacts with the pyruvate side chain of the substrate, have larger atomic fluctuations than in the transition state analogue complex. This result is due partly to the increased flexibility of the pyruvate side chain of the substrate. The greater rigidity of the system when the TSA is present, as compared with the reactant or product, is in accord with the concept that the substrate binds tightly to the enzyme in the transition state. However, the transition state analogue is not ideal because of the lack of the stretched bonds and the developing charges of the true transition state (11). Hence, the TSA is bound more strongly than chorismate or prephenate by only a factor of about 20 to 30: the Ki is 3 μM for TSA and 70 μM for prephenate, and the Km for chorismate is 100 μM for the BCM (33). It is expected that the real transition state would bind more tightly.
Calculation of pKa Values of Glu-246.
To complement the dynamic simulation, the pKa values of Glu-246 have been calculated under various conditions by using the linearized Poisson–Boltzmann equation. The pKa value of Glu-246 is mainly affected by a few important features in the active site. They are the overall low dielectric environment in the active site because of the exclusion of the bulk water, the negatively charged bridge oxygen, and the positively charged Lys-168 side chain. The first two features increase the pKa and the third one decreases the pKa. The effective pKa value of Glu-246 is 5.8 (to be compared with the intrinsic pKa of 4.4) if the bridge oxygen is modeled as an ether oxygen in TSA, and it increases to 8.1 if the partial charge on the bridge oxygen is increased from −0.56 as in TSA to −0.9 as a mimic of the actual transition state. These calculations are consistent with the results of the dynamic simulation, i.e., Glu-246 is protonated under conditions corresponding to catalysis in the active site of YCM.
DISCUSSION
The results of the simulations indicate that a protonated Glu-246 leads to a stable structure in which Glu-246 is hydrogen bonded to the ether oxygen, consistent with the x-ray results (4). The simulation for the complex in which Glu-246 is unprotonated and that for the complex in which there is a water molecule between Glu-246 and the ether oxygen show structural distortions that are inconsistent with the x-ray results (4). The simulation with an ionized water molecule in the active site result in a quick displacement of OH− ion into the solvent. Also, the mutant Glu-246Gln is stable over a 500-ps simulation. No water is observed in the active site in the simulations of enzyme-reactant (chorismate) and enzyme-product (prephenate) complexes. The effective pKa value of Glu-246 is calculated to be shifted significantly upward under catalytic conditions with a developing negative charge on the ether oxygen.
The pH-activity profile for the wild-type Glu-246 YCM (12) shows a maximum at about pH 6.5 and more than half-maximal activity between pH values of 5 and 8.5. The assignment (12) of a high pKa for Glu-246 is strengthened by comparison of the pH-activity profile of the Glu-246 enzyme with that of the Glu-246 to Gln mutant. In this mutant (E246Q), the high end of the pH profile is extended to about 10.5, probably reflecting the pKa of Lys-168 or some other residues. Although the simulations need to be confirmed by more quantitative studies, such as free energy simulations, the results reported here are in accord with the proposal that a protonated Glu-246, with an increased pKa, is involved in the catalytic mechanism. The suggestion (15) of an active site water as the mediator between Glu-246 and the ether oxygen is not supported by the present results.
Although the structural and dynamic results give information about active site binding, the details of rate enhancement by the CM enzymes, typically by factors of 106 to 107 over the rate of the uncatalyzed reactions (34, 35), have yet to be analyzed qualitatively. Effects include (36) (i) the role of selection of the less stable pseudodiaxial conformer (about 10% of the total) of chorismate, (ii) the role of specific electrostatic stabilization of the ether oxygen (e.g., Arg-90 in BCM), including other hydrogen bonding and general electrostatic effects that promote the charge separation in the transition state (Arg-90 and Glu-78 in BCM), and (iii) the creation of a strongly hydrophobic environment. These three effects also apply to the P protein of ECM and to YCM, where Lys-39 and Lys-168, respectively, stabilize the ether oxygen, along with Gln 88 and a protonated Glu-246, respectively. Moreover, the charge separation as the transition state develops is also probably aided by Glu-52 in ECM and Glu-198 in YCM.
It is rare that the chemical transformation is the RDS in enzyme catalysis. For the BCM (36, 37), the RDS occurs before the chemical transformation at low substrate concentration (32, 33) (Vmax/Km conditions) and probably involves a change in the binding step or a conformational change that occurs before the chemical rearrangement. At high substrate concentrations (Vmax conditions), the departure of the product prephenate is the rate-limiting step (33). Although no sequence homology exists between the B. subtilis enzyme and E. coli (or yeast) enzyme, the similarity of kinetic parameters suggests functional similarity (32). Both substrate binding and product leaving are expected to show large solvent compensation effects involving ΔH‡ and ΔS‡ (38–40), and these thermodynamic parameters may depend substantially on temperature. Thus, it may be difficult to compare these thermodynamic parameters obtained from log k versus 1/T plots via the Eyring equation from the different CM enzymes (41, 42) and especially to compare these parameters with those for the uncatalyzed reaction (41, 42). Molecular mechanics studies of these solvent-sensitive stages of the reaction are needed to aid in an understanding of the rate-sensitive steps in the mechanism.
Acknowledgments
This work was supported by grants GM06920 (W.N.L.) and GM30804 (M.K.) from the National Institutes of Health. J.M. was the recipient of a postdoctoral fellowship from Burroughs Wellcome Fund, Program in Mathematics and Molecular Biology, in the early part of this study. He is now a postdoctoral fellow of the National Institutes of Health.
ABBREVIATIONS
- CM
chorismate mutase
- YCM
yeast chorismate mutase
- TSA
transition state analogue
- RDS
rate-determining step
- ECM
Escherichia coli chorismate mutase
- BCM
Bacillus subtilis chorismate mutase
References
- 1.Weiss U, Edwards J M. The Biosynthesis of Aromatic Amino Compounds. New York: Wiley; 1980. [Google Scholar]
- 2.Braus G H. Microbiol Rev. 1991;55:349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidheini T, Mosch H U, Evans J N, Braus G. Biochemistry. 1990;29:3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- 4.Sträter N, Schnappauf G, Braus G, Lipscomb W N. Structure. 1997;5:1437–1452. doi: 10.1016/s0969-2126(97)00294-3. [DOI] [PubMed] [Google Scholar]
- 5.Sträter N, Hakansson K, Schnappauf G, Braus G, Lipscomb W N. Proc Natl Acad Sci USA. 1996;93:3330–3334. doi: 10.1073/pnas.93.8.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sogo S G, Widlanski T S, Hoare J H, Grimshaw C E, Berchtold G A, Knowles J R. J Am Chem Soc. 1984;106:2701–2703. [Google Scholar]
- 7.Copley S D, Knowles J R. J Am Chem Soc. 1985;107:5306–5308. [Google Scholar]
- 8.Copley S D, Knowles J R. J Am Chem Soc. 1987;109:5008–5013. [Google Scholar]
- 9.Guilford W J, Copley S D, Knowles J R. J Am Chem Soc. 1987;109:5013–5019. [Google Scholar]
- 10.Bartlett P A, Jackson C R. Biopolymers. 1985;97:843–865. [Google Scholar]
- 11.Mader M M, Bartlett P A. Chem Rev (Washington, DC) 1997;97:1281–1301. doi: 10.1021/cr960435y. [DOI] [PubMed] [Google Scholar]
- 12.Schnappauf G, Sträter N, Lipscomb W N, Braus G H. Proc Natl Acad Sci USA. 1997;94:8491–8496. doi: 10.1073/pnas.94.16.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue Y, Lipscomb W N. Proc Natl Acad Sci USA. 1995;92:10595–10598. doi: 10.1073/pnas.92.23.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee A Y, Karplus A P, Ganem B, Clardy J. J Am Chem Soc. 1995;117:3627–3628. [Google Scholar]
- 15.Lin S L, Xu D, Li A, Rosen M, Wolfson H J, Nussinov R. J Mol Biol. 1997;271:838–845. doi: 10.1006/jmbi.1997.1168. [DOI] [PubMed] [Google Scholar]
- 16.Brooks C L, III, Brunger A, Karplus M. Biopolymers. 1985;24:843–865. doi: 10.1002/bip.360240509. [DOI] [PubMed] [Google Scholar]
- 17.Brooks C L, III, Karplus M. J Mol Biol. 1989;208:159–181. doi: 10.1016/0022-2836(89)90093-4. [DOI] [PubMed] [Google Scholar]
- 18.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 19.MacKerell, A. D., Jr., Bashford, D., Bellott, M., Dunbrack, R. L., Jr., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Gao, H., Ha, S., et al. (1998) J. Phys. Chem., Series B, 3586–3616. [DOI] [PubMed]
- 20.Jorgensen W L. J Am Chem Soc. 1981;103:335–340. [Google Scholar]
- 21.Neria E, Fisher S, Karplus M. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 22.Halgren T A. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 23.Halgren T A. J Comput Chem. 1996;17:520–552. [Google Scholar]
- 24.Halgren T A. J Comput Chem. 1996;17:553–586. [Google Scholar]
- 25.Halgren T A, Nachbar R B. J Comput Chem. 1996;17:587–615. [Google Scholar]
- 26.Halgren T A. J Comput Chem. 1996;17:616–641. [Google Scholar]
- 27.Brooks C L, III, Karplus M. J Chem Phys. 1983;79:6312–6325. [Google Scholar]
- 28.Davis M E, Madura J D, Luty B A, McCammon J A. Comput Phys Commun. 1991;62:187–197. [Google Scholar]
- 29.Honig B, Nicholls A. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 30.Bashford D, Karplus M. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 31.Schaefer M, Sommer M, Karplus M. J Phys Chem. 1997;101:1663–1683. [Google Scholar]
- 32.Addadi L, Jaffe E K, Knowles J R. Biochemistry. 1983;22:4494–4501. doi: 10.1021/bi00288a022. [DOI] [PubMed] [Google Scholar]
- 33.Gray J V, Eren D, Knowles J R. Biochemistry. 1990;29:8872–8878. doi: 10.1021/bi00489a051. [DOI] [PubMed] [Google Scholar]
- 34.Gorisch H. Biochemistry. 1978;17:3700–3705. doi: 10.1021/bi00611a004. [DOI] [PubMed] [Google Scholar]
- 35.Andrews P R, Smith G D, Young I G. Biochemistry. 1973;12:3492–3498. doi: 10.1021/bi00742a022. [DOI] [PubMed] [Google Scholar]
- 36.Chook Y M, Gray J V, Ke H, Lipscomb W N. J Mol Biol. 1994;240:476–500. doi: 10.1006/jmbi.1994.1462. [DOI] [PubMed] [Google Scholar]
- 37.Chook Y M, Ke H, Lipscomb W N. Proc Natl Acad Sci USA. 1993;90:8600–8603. doi: 10.1073/pnas.90.18.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hins H J, Shiao D F, Sturtevant J M. Biochemistry. 1971;10:1347–1352. doi: 10.1021/bi00784a012. [DOI] [PubMed] [Google Scholar]
- 39.Sturtevant J M. Proc Natl Acad Sci USA. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naghibi H, Tamura A, Sturtevant J M. Proc Natl Acad Sci USA. 1995;92:5597–5599. doi: 10.1073/pnas.92.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kast P, Asif-Ullah M, Hilvert D. Tetrahedron Lett. 1996;37:2691–2694. [Google Scholar]
- 42.Galopin C C, Zhang S, Wilson D B, Ganem B. Tetrahedron Lett. 1996;37:8675–8678. [Google Scholar]