Abstract
We have designed a p53 DNA binding domain that has virtually the same binding affinity for the gadd45 promoter as does wild-type protein but is considerably more stable. The design strategy was based on molecular evolution of the protein domain. Naturally occurring amino acid substitutions were identified by comparing the sequences of p53 homologues from 23 species, introducing them into wild-type human p53, and measuring the changes in stability. The most stable substitutions were combined in a multiple mutant. The advantage of this strategy is that, by substituting with naturally occurring residues, the function is likely to be unimpaired. All point mutants bind the consensus DNA sequence. The changes in stability ranged from +1.27 (less stable Q165K) to −1.49 (more stable N239Y) kcal mol−1, respectively. The changes in free energy of unfolding on mutation are additive. Of interest, the two most stable mutants (N239Y and N268D) have been known to act as suppressors and restored the activity of two of the most common tumorigenic mutants. Of the 20 single mutants, 10 are cancer-associated, though their frequency of occurrence is extremely low: A129D, Q165K, Q167E, and D148E are less stable and M133L, V203A and N239Y are more stable whereas the rest are neutral. The quadruple mutant (M133LV203AN239YN268D), which is stabilized by 2.65 kcal mol−1 and Tm raised by 5.6°C is of potential interest for trials in vivo.
Keywords: protein design/thermodynamic stability
The tumor suppressor protein p53 plays a key role in preventing the development of cancer (1, 2). The loss of p53 function, which results almost entirely from mutations within the DNA binding domain, is associated with half of all human cancers. Despite the potential therapeutic relevance, the p53 cellular stability and the mechanisms by which it is regulated are not well understood (3, 4, 5). It has been reported that p53 stability affects its functions, the most important of which are the cell-cycle arrest and apoptosis (4, 5, 6, 7). The cellular concentration of p53 protein is regulated tightly, and, in normal cells, it is kept at low levels through interactions with the Mdm2 protein. In response to stress signals, such as DNA damage, the interaction between the two proteins is compromised, which leads to a sharp increase of p53 concentration in the cell and in the half-life of the normal wild-type p53 protein (4, 5). Consequently, the accumulation of p53 protein in tumors is caused by an increase in the cellular stability of p53 (5). Several reports have highlighted the low thermal stability of p53, which may limit the cellular stability of p53 in the absence of Mdm2 (8, 9). In addition, it has been shown that some mutants of p53 have a wild-type-like conformation whereas others adopt an altered, “mutant” conformation, which is reported to affect function (10). Structural studies suggested that the mutant conformation most likely represents denatured or partially denatured states of p53, rather than an alternative conformational state (6). Although the thermodynamic stability of the full length p53 is not known, it has been demonstrated that the thermodynamic stability of wild-type p53 DNA binding domain is low and, most importantly, that the known p53 core domain “hot spots” associated with tumors are destabilized (11). Finding ways to rescue the stability and/or the function of p53, therefore, appears to be of great importance. Efforts to restore wild-type function of mutant proteins have focused on antibody or peptide binding studies derived from the C-terminal domain or p53 binding to heat shock protein dnaK (8, 9, 12). Alternatively, the introduction of basic residues in the DNA binding domain, aiming to establish new contacts between p53 and the DNA phosphate backbone, has resulted in a significant DNA binding in some frequently occurring tumorigenic mutants (12). Genetic selection of intrinsic suppressor mutations that reverse the effect of three common cancer mutations (V143A, G245S, and R249S) has been published recently (13). Gene therapy, which aims at delivering functional p53 to tumor cells to reestablish the pathway to apoptosis, has shown preliminary success (ref. 14 and references within). A more powerful way for tumor destruction may, perhaps, be a superstable p53 variant. The ultimate objective of this study was to design a superstable p53 core domain without compromising the key biological function of the protein to bind DNA.
The design of proteins with increased stability is still at a very preliminary stage, with few rational approaches reported. An empirical approach based on molecular evolution of the p53 DNA binding domain appeared to be an attractive option. This strategy has been developed from earlier studies of the effects of conversion of barnase to its homologue binase, which differs in only 17 positions in the sequence. It was found that six of the sequence differences in binase were stabilizing when substituted into barnase and that the effects of the individual substitution on stability were additive (15). Further, the thermostable mutant was fully active. A similar approach has been applied to the stabilization of antibodies (16). The semirational approach we used and present here is based largely on the information available from the p53 sequence homology database (7). Earlier phylogenetic comparison revealed a number of features common to all p53 proteins (7). The core domain of p53 contains four of the five evolutionary conserved blocks and is reported to be an archetypal domain with its unique fold, structure, function, and molecular evolution (7). Molecular evolution data had confirmed that ≈84% of missense mutation target codons are evolutionary conserved in all p53 proteins and are mapped to the core domain, where the mutational hot spots for human cancer are located (p53 database, http://www.iarc.fr). Because conserved residues in the sequence alignment most likely reflect biological selection for function, the risk of introducing changes that would be detrimental to the function by substituting residues in the human p53 protein with the most prevalent evolutionary conserved residue at a specific position should be minimal (15, 16, 18). Thus, we also may find more stable variants. In addition, we looked at the effects of known p53 suppressor mutations that have been found to restore function (13). We report here that the replacement of four residues in the sequence of the human p53 core domain enhances the thermodynamic stability of the protein from 8.6 to 11.3 kcal mol−1 whereas the function of the variant p53 core domain to bind DNA is unaltered and virtually identical to that of wild type.
MATERIALS AND METHODS
DNA Cloning and Site-Directed Mutagenesis.
The p53 DNA binding (core) domain (residues 94–312) was amplified by PCR from plasmid pT7hp53, was subcloned into the polylinker region of the expression vector pRSET(A) (Invitrogen), and was transformed into Escherichia coli DH5α as described (11). Mutations were introduced by using the QuikChange side-directed mutagenesis kit (Strategene) and were confirmed by sequencing the entire coding region for each mutant by using T7 promoter and T7 terminator primers.
Protein Expression and Purification.
The expression and purification of wild type and mutants were performed as described (11). Purified proteins were subjected to electrospray mass spectroscopy, which further confirmed the sequencing and electrophoresis data.
Differential Scanning Calorimetry.
Differential scanning calorimetry was performed on a VPDSC Microcal (Microcal, Amherst, MA), and data were analyzed as reported (11).
Equilibrium Urea Denaturation.
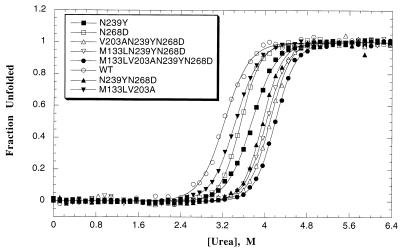
Urea-induced unfolding of p53 in 50 mM sodium phosphate and 5 mM DTT at pH 7.2 was monitored by fluorescence using an Amico-Bowman Series 2 luminescence spectrofluorimeter with excitation at 280 nm and emission scans between 300 and 370 nm as reported (11). Samples were incubated for at least 5 h at 10°C and were analyzed at the same temperature in 1-ml thermostatted cuvettes. Equilibrium measurements were made from the urea-induced denaturation curves, which exhibited a good two-state transition behavior (Fig. 1). Although wild-type p53 is reversibly denatured at 25°C, 10°C was chosen for all our studies because some destabilized mutants do not denature reversibly at elevated temperatures (11). During the measurements, the temperature was closely monitored by a thermocouple. The protein concentration of each sample was ≈2 μM. Data were analyzed as reported (11) by using the equation for a two-state model and were fitted to this equation by the program kaleidagraph (Synergy Software, Reading, PA), taking into an account the slopes of the baselines.
Figure 1.
Urea equilibrium unfolding of wild-type (WT) and mutant p53 core domain represented as fraction of unfolded protein vs. concentration of denaturant. Unfolding was monitored in 50 mM sodium phosphate (pH 7.2) and 5 mM DTT at 10°C by fluorescence using an excitation wavelength of 280 nm and normalized emission at 356 nm (11). The protein concentration was 2 μM. The fitting of the experimental data to a two-state transition equation (11) was performed by the program kaleidagraph, and the best fit is indicated by solid lines.
DNA Binding.
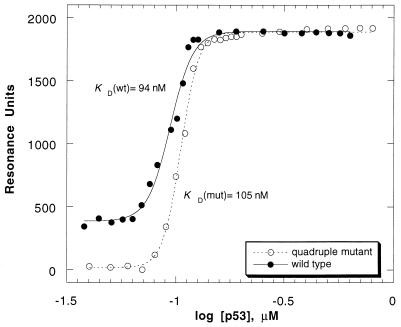
Surface plasmon resonance experiments were performed to investigate DNA binding of p53 core domain to the gadd45 promoter, which has the following sequences: Gadd45UP, 5′-biotin-GTACA GAACATGTCT AAGCATGCTG GGGAC-3′; and Gadd45LO, 3′-CATGT CTTGTACAGA TTCGTACGAC CCCTG-5′. The experiments were performed by immobilizing the gadd45 double-stranded DNA on a streptavidin-coated chip (SA5) and by using a BIAcore 2000, Pharmacia Biosensor AB. The gadd45 sequences were annealed and purified on a MonoQ column (Pharmacia) before immobilization on the chip surface. Double-stranded oligonucleotide (1 μM) was injected manually at 5 μl/min on three different flow cells. One flow cell was left empty of immobilized DNA and was used to determine background response. Background response units were substracted for determination of the binding constant. The protein was dialyzed and diluted in 50 mM Tris (pH 7.2) and 5 mM DTT to concentrations ranging from 0.03 to 1 μM. Different concentrations of p53 core domain (80 μl) were injected at a flow rate of 10 μl/min. The relative change in resonance units was monitored for 500 seconds after injection. To regenerate the surface after each sample, 10 μl of 1 M NaCl in 50 mM Tris (pH 7.2) and 5 mM DTT were injected and proved sufficient to dissociate the p53-DNA complex. The temperature was maintained at 20°C. To determine the KD of p53 binding to gadd45 promoter DNA, changes in resonance units on binding were plotted vs. protein concentration, and the data were fitted to the two-state equation (Fig. 2) (11).
Figure 2.
Binding curves of the wild-type and quadruple mutant p53 core domain to the gadd45 double-stranded DNA. The experiments were performed at 20°C in 50 mM Tris (pH 7.2) with 5 mM DTT. The protein concentration ranged from 0.03 to 1 μM whereas that of the double-stranded oligonucleotide was 1 μM. The experimental data were fitted by the program kaleidagraph, and the best fit is indicated by solid and dotted lines. The dissociation constant KD was calculated as the midpoint of the transition curve.
Solvent-Accessible Area Calculations.
The solvent accessibility and the distances between atoms were calculated by using the program what if (19).
RESULTS AND DISCUSSION
Design Strategy and Structural Analysis of the Mutants.
The amino acid sequence of human p53 protein was compared with 22 homologous proteins to identify candidates for the point mutation substitutions (7). We excluded the six mutational hot spots that occur in cancer because they involve structural and DNA binding mutants with altered stability and/or function (6, 11, 12, 13). The frequency of occurrence of the human wild-type residue and that of the variant residue(s) at randomly selected positions was calculated and used as a guideline when designing the p53 variants. The residues selected for mutation are listed in Table 1. No specific location in the structure was targeted for the replacements, and the 20 selected residues are distributed throughout the core domain. The solvent accessibility of the residues selected for mutation is variable, and nearly half of the residues were highly exposed to solvent. The hydrogen bonds made by the side chains of the residues selected for mutation in the wild-type protein are presented in Table 2. Of the 20 mutations selected, the majority occurred outside the conserved boxes, with the exception of residues Ala 129 and Met 133 (conserved box II), Arg 174 (box III) and Tyr 236 and Asn 239 (box IV) (7). In addition, the mutation N239Y was chosen based on a report that it acts as a suppressor mutation by restoring the activity when introduced as a second site to the cancer mutation G245S (13).
Table 1.
Solvent accessibility, secondary structure, and frequency of the residues selected for mutation
| Residue position* | Location† | Solvent accessibility of side chain in wild type, %‡ | Frequency of residue in p53 family§
|
|
|---|---|---|---|---|
| human p53 | variant p53 | |||
| Q104 (P)(H) | End of N terminus | 56.7 | 0.2 | 0.4 (0.15) |
| A129 (S)(D)(E) | Turn between S2 and S2′ | 77.3 | 0.1 | 0.3 (0.15) (0.15) |
| M133(L) | S2′ | 0.7 | 0.15 | 0.8 |
| D148(S)(E) | Turn between S3 and S4 | 71.1 | 0.43 | 0.24 (0.1) |
| T150(P) | Turn between S3 and S4 | 64.3 | 0.43 | 0.52 |
| Q165(K) | L2 | 43.4 | 0.14 | 0.86 |
| Q167(E) | L2 | 71.2 | 0.43 | 0.52 |
| R174(K) | L2 | 57.5 | 0.81 | 0.19 |
| C182(S) | L2 | 35.3 | 0.55 | 0.36 |
| L201(P) | Turn between S5 and S6 | 89.5 | 0.73 | 0.14 |
| V203(A) | Turn between S5 and S6 | 0.7 | 0.18 | 0.68 |
| L206(S) | S6 | 47.8 | 0.55 | 0.1 |
| D228(E) | Turn between S7 and S8 | 77.5 | 0.50 | 0.5 |
| Y236(F) | End of S8 | 0.4 | 0.57 | 0.43 |
| N239(Y) | L3 | 34.5 | 0.91 | 0 |
| S260(P) | Turn between S9 and S10 | 64.9 | 0.61 | 0.22 |
| N268(D) | S10 | 13.8 | 0.52 | 0.1 |
The position according to the sequence of the human 53 protein and the Protein Data Bank (6, 7). The residues in brackets are those of the mutants; in bold are cancer mutations.
Location of the residue according to the crystal structure of the protein in the Protein Data Bank (6).
Solvent accessibility (in percent), defined as solvent accessible surface area of the amino acid residue in its parent protein calculated by whatif (19), divided by the solvent accessibility of that residue in an extended Ala-X-Ala peptide.
Relative frequency of occurrence as calculated from the alignment of 23 p53 proteins by using human p53 as the reference (7).
Table 2.
Side chains of those residues involved in hydrogen bonding
| Residue | Source atom | Target atom | Distance, Å, donor/acceptor |
|---|---|---|---|
| Asp 148 | Oδ1 | Nη2 Arg 110 | 3.28 |
| Oδ2 | Nη1 Arg 110 | 3.31 | |
| Gln 165 | Oɛ | Nδ1 His 168 | 3.16 |
| Arg 174 | Nη2 | Oɛ1 Glu 180 | 3.38 |
| Asp 228 | Oδ1 | Sγ Cys 229 | 2.79 |
| Tyr 236 | Oη | Oγ Thr 253 | 2.83 |
| Asn 268 | Nδ2 | O Arg 267 | 3.12 |
| O Phe 109 | 2.95 |
All the calculations were performed by using Collaborative Computational Project 4 (17).
Stability and Folding of p53 Mutants.
The impact of individual side chains on stability can be evaluated by analysis of mutant proteins carrying single amino acid replacements (15, 21). Table 3 summarizes the effect on the free energy of unfolding of the 20 single mutations and a number of multiple mutants of the DNA binding domain of p53. Of the 20 single mutations, N239Y and N268D have greater stability (ΔGD−NH2O = 10.1 and 9.6 kcal mol−1, respectively) than the wild-type p53 core (ΔGD−NH2O = 8.64 kcal mol−1). However, Q165K has lower stability (ΔGD−NH2O = 7.4 kcal mol−1), and the remaining mutations have only a small effect on stability.
Table 3.
Changes in the free energies of urea-induced unfolding of p53 DNA binding domain
| p53 Variant | m, kcal mol−1M−1 | [Urea]50%, M | ΔGD−NH2O (kcal mol−1) | ΔΔGD−NH2O (kcal mol−1) |
|---|---|---|---|---|
| WT (2) | 2.40 ± 0.12 | 3.20 ± 0.01 | 8.64 | |
| Q104P | 2.50 ± 0.08 | 3.24 ± 0.01 | 8.75 | −0.11 |
| Q104H | 2.50 ± 0.09 | 3.29 ± 0.01 | 8.88 | −0.24 |
| A129D | 3.11 ± 0.14 | 2.94 ± 0.01 | 7.94 | 0.70 |
| A129E | 2.83 ± 0.11 | 3.06 ± 0.01 | 8.26 | 0.38 |
| A129S | 3.01 ± 0.09 | 3.13 ± 0.01 | 8.45 | 0.19 |
| M133L | 2.60 ± 0.11 | 3.31 ± 0.01 | 8.94 | −0.30 |
| D148E | 2.51 ± 0.09 | 3.04 ± 0.01 | 8.21 | 0.43 |
| D148S | 2.57 ± 0.09 | 3.28 ± 0.01 | 8.86 | −0.22 |
| T150P | 2.86 ± 0.13 | 3.17 ± 0.01 | 8.56 | 0.08 |
| Q165K | 2.89 ± 0.17 | 2.73 ± 0.01 | 7.37 | 1.27 |
| Q167E | 2.90 ± 0.07 | 3.04 ± 0.06 | 8.21 | 0.43 |
| R174K | 2.80 ± 0.09 | 3.12 ± 0.01 | 8.42 | 0.22 |
| C182S | 2.11 ± 0.10 | 3.26 ± 0.02 | 8.80 | −0.16 |
| L201P | 2.24 ± 0.09 | 3.33 ± 0.01 | 8.99 | −0.35 |
| V203A (2) | 2.40 ± 0.07 | 3.38 ± 0.01 | 9.13 | −0.49 |
| L206S | 2.75 ± 0.11 | 3.16 ± 0.01 | 8.55 | 0.10 |
| D228E | 2.40 ± 0.07 | 3.22 ± 0.01 | 8.69 | −0.05 |
| Y236F (2) | 2.74 ± 0.08 | 3.30 ± 0.01 | 8.91 | −0.27 |
| N239Y | 2.40 ± 0.07 | 3.75 ± 0.01 | 10.13 | −1.49 |
| S260P | 2.30 ± 0.11 | 3.08 ± 0.01 | 8.32 | 0.32 |
| N268D | 2.97 ± 0.06 | 3.54 ± 0.004 | 9.56 | −0.92 |
| N239YN268D | 3.03 ± 0.11 | 3.96 ± 0.01 | 10.68 | −2.04 |
| M133LV203A | 2.50 ± 0.05 | 3.44 ± 0.005 | 9.29 | −0.65 |
| M133LN239YN268D | 2.82 ± 0.10 | 4.03 ± 0.01 | 10.87 | −2.23 |
| V203AN239YN268D | 2.84 ± 0.16 | 4.09 ± 0.01 | 11.05 | −2.41 |
| M133LV203AN239YN268D | 2.73 ± 0.08 | 4.18 ± 0.01 | 11.29 | −2.65 |
The mean m value of 27 denaturation curves (at 10°C in 50 mM sodium phosphate/5 mM DTT at pH 7.2) is 2.7 ± 0.05 kcal mol−1 and was used to calculate ΔGD−NH2O and ΔΔGD−NH2O according to the equation for a two-state transition as reported (11). The errors given are the standard errors, and the number of measurements is shown in parentheses. ΔGD−ND = ΔGD−NH2O − m [D] whereas ΔΔGD−N[D]50% = 〈m〉Δ[D]50%, where m is the slope of the transition from native to denatured state, [D] is the concentration of the denaturant, [D]50% is the concentration of denaturant at which 50% of the protein is unfolded, 〈m〉 is the average value of m for mutant and wild-type proteins, and Δ[D]50% = [D]50%wt − [D]50%mutant.
Stabilizing Mutations.
The most stable mutants are N239Y, N268D, V203A, and M133L, which stabilized the p53 DNA binding domain by 1.5, 0.9, 0.5, and 0.3 kcal mol−1, respectively. Of interest, the two most stabilizing mutations, N239Y and N268D, are known to act as second-site suppressor mutations capable of restoring the wild-type function to two of the most common p53 cancer mutations, namely G245S and V143A, respectively.
Asn 239 is 4 Å away from Cys 238 (one of the four ligands for the zinc atom) and ≈6 Å away from the zinc. In the wild-type structure of the p53 core domain–DNA consensus complex, the side chain of Asn 239 has a solvent accessibility surface area of 44 Å2 and, although it is ≈4 Å away from the O2P and O5 of the Gua13, it is not close enough to be able to make hydrogen bonds with the DNA. It is 3–3.5 Å away from Cys 242, which is also a ligand for the zinc. Its closest neighbors within 4.5 Å are Leu 137, Cys 238, Ser 240, Val 274, Cys 275, and Ala 276, with which it forms hydrophobic interactions. Depending on the rotamer formed, the substitution of Asn to Tyr in the p53 core–consensus complex increases the solvent accessibility of the residue from 44 to 77 Å2. Although it is suggested that the Tyr residue may come closer to the phosphodiester backbone of the DNA in the core–DNA complex and may influence the DNA binding (13), our equilibrium thermodynamic data (no DNA present) demonstrated that this substitution clearly has a major stabilizing effect on the structure of this domain.
The frequency of occurrence of Asn 268 throughout the sequences of the 23 p53 homologous proteins is 0.52 as compared with 0.1 for the Asp. In the p53 core-DNA complex, the side chain of Asn 268 is 22 Å2 solvent accessible. The side chain of this residue makes hydrogen bonds with Arg 267 and Phe 109. Asn 268 is in strand 10 and is ≈2.8–3.4 Å away from Ile 255 (located in strand 9). The close proximity of these two residues results in good hydrogen bonding that bridges the two sheets of the β-sandwich and may contribute to the stability of the protein (6, 13).
Val 203 is in the turn between strands 5 and 6, and it is completely buried. Replacement of Val by Ala leads to a modest increase in thermostability, which is difficult to explain on structural grounds. Likewise, Met 133 is located in strand 2′ and is completely buried. Its frequency of occurrence is 0.15 whereas that of the mutant residue (Leu) is 0.8. Other neighbors within 4.5 Å with which it forms weaker interactions are Thr 125, Phe 270, Cys 124, Cys 141, and Val 143. The close inspection of the structure does not provide clues as to why this variant should have a modest increase in stability.
Destabilizing Mutations.
In this study, the most deleterious mutation in the p53 core domain was that at position 165 and involved substitution of Gln to Lys (Q165K). The penalty for this substitution in terms of free energy of unfolding was 1.27 kcal mol−1. The Gln side chain is well exposed to the solvent and forms a strong hydrogen bond with His 168. Its other neighbors within 4.5 Å are Pro 250 and Arg 249. The last two residues may indirectly affect the critical DNA contact made by Arg 248 and, therefore, may destabilize the DNA binding domain. Arg 249 is one of the six hot spots, and it is classed as a structural mutation. Together with its neighbor Arg 248 (a DNA contact mutant and also a hot spot), each of these two residues destabilize the structure by 1.95 kcal mol−1 (11).
The other destabilizing mutation is that of Ala 129. Replacement with Asp or Glu destabilized the protein by 0.7 and 0.4 kcal mol−1, respectively. Similarly, D148E, Q167E and S260P decreased the stability by 0.4, 0.4, and 0.3 kcal mol−1, respectively. The side chain of Asp 148 forms hydrogen bonds with the side chain of Arg 110 (Table 2).
Neutral Mutations.
The majority of single mutants (ΔΔG = −0.27 to +0.22 kcal·mol−1) were neutral. L201P has 0.35 kcal mol−1 higher energy of unfolding than the wild-type but was not selected for the multiple mutant because of possible complications that can arise from proline substitution (21).
Multiple Mutants and Additivity Effect.
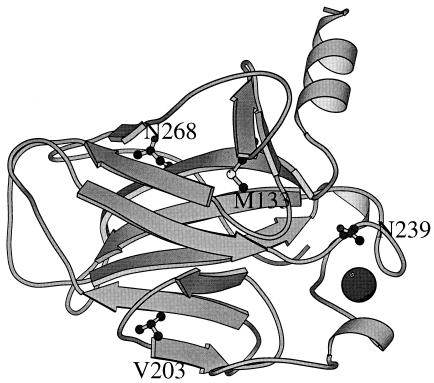
Four mutations were selected to construct the quadruple mutant, namely, M133L, V203A, N239Y, and N268D. Their location in the structure is depicted in Fig. 3. The total stability gained is 2.65 kcal mol−1, with Asn to Tyr substitution at position 239 contributing ≈50% to the stability of the quadruple mutant. To examine whether the effects of the four mutations are additive, we have constructed five different p53 core domain variants (M133LV203A, N239YN268D, M133LV203AN239Y, V203AN239YN268D, and M133LV203AN239YN268D) and have analyzed their free energies of unfolding. The data showed near additivity of changes in stabilization energies of the mutants. In all five cases, the sum of the free energies of unfolding of the individual component mutants is very close to the free energies of unfolding for the combined mutants. This is not unexpected because these residues are far apart (11–24 Å) from each other in the wild-type structure and do not interact directly (15, 18, 23). The free energy of stabilization achieved for various proteins by multiple mutations are similar, 3–4 kcal mol−1 of stabilization from 6–7 substitutions (14, 24, 25). Our data complement previous data on additivity and confirm its general applicability to a wide range of different proteins (15, 18, 23). The apparent Tm for irreversible thermal denaturation by differential scanning calorimetry for some of the variants followed the same trend as the reversible urea-induced thermodynamic stability data (11). The Tm values were 42°C for the wild-type p53, 46.2°C for M133LN239YN268D, and 47.2°C for the quadruple mutant, respectively.
Figure 3.
Diagram of p53 DNA binding domain produced by molscript (22) showing only the position of the four mutations used to design the quadruple mutant.
Effect of Mutations within the p53 Core Domain on DNA Binding Function.
The DNA binding affinity of all mutants and the wild-type protein was tested by surface plasmon resonance (26). The apparent binding constants for the quadruple mutant (KD = 105 nM) and the wild-type p53 core domain (KD = 94 nM) were nearly identical (Fig. 2). Though quantitative results here are given only for the quadruple mutant and the wild-type p53 core domain, the rest of the data, which are not shown here, confirmed that all mutants are capable of binding to double-stranded DNA consensus sequence at 20°C. It is worth emphasizing that further studies to test the in vivo activities of the quadruple p53 core domain variant but within a full length p53 protein are currently under consideration.
Point Mutations Implicated in Cancer and p53 Stability.
The thermodynamic stability of the p53 core domain is relatively low (ΔGD−NH2O = 6 kcal mol−1 at 25°C) (11). At physiological temperature, the stability of the wild-type p53 core is estimated to be 2–3 kcal mol−1 (A. N. Bullock and A.R.F., unpublished data). Cancer-associated mutations in p53 have been reported to destabilize the structure and to cause an increased tendency to aggregation (6, 8, 11). The notion that tumorigenic variants of p53 are denatured at least partially was supported by comprehensive antibody studies in which antibodies specific for the native and denatured states of p53 were used (6, 12, 27).
Ten point mutations in this study have been reported to occur in tumors (17). The frequency of occurrence of these tumorigenic mutations within the core domain is extremely low (0.01–0.08%) and may, therefore, be insignificant (e.g., mutants arising from preexisting genetic instabilities). From the 10 cancer-associated mutants, only 1 (Q165K) was destabilized significantly. Compared with some of the most frequently observed cancer-associated hot spots, for example R175H or C242S (ΔGD−NH2O = 3.0 and 2.9 kcal mol−1) (11) or that of the temperature-sensitive mutant V143A (ΔGD−NH2O = 3.0 kcal mol−1) (P.V.N. and A.R.F., unpublished data), the destabilization of Q165K (ΔGD−NH2O = 1.3 kcal mol−1) appears to be moderate. The second most destabilizing substitution was A129D (ΔGD−NH2O = 0.7 kcal mol−1). The nonconservative amino acid replacement of Ala to Asp alters the charge of the variant p53 protein, which may indeed result not only in functional aberration (28) but also in thermodynamic destabilization. On the other hand, we have found cancer-associated mutants that bind DNA and are not destabilized at all as judged from the urea-induced equilibrium unfolding and DNA binding studies. For example, M133L and V203A are indeed marginally more stable than the wild-type p53. It is plausible that some of the tumor mutations could be naturally occurring examples of polymorphism and may not necessarily be the cause of cancer. Notably, the degree of destabilization of cancer-associated mutations is within the observed changes reported for other protein-engineered model systems (15, 20, 21). The results shown here demonstrate that the key DNA binding function of the p53 core domain can tolerate a considerably extended range of sequence changes within the sequence homology map, which represents the molecular evolution of this domain across 23 different species. Because the severity of disruption caused by tumorigenic mutations is extremely varied (11), a more biophysical studies on thermodynamic stability coupled with biological activity data are needed before we understand the intricate nature of such mutations on molecular level and consider potential ways to restore their natural function.
One of the most exciting recent developments in the fight against cancer is p53 gene therapy. A superstable p53 variant might be a more potent and promising tool for tumor destruction. The quadruple mutant we have designed, which has notably improved stability while retaining wild-type DNA binding function, appears to be an initial candidate for preliminary in vivo trials.
Acknowledgments
Special thanks to Drs. Mark Bycroft, Yu Wai Chen, Mauricio Mateu, Laura Itzhaki, and Alex Bullock for their help. This work was supported by grant from the Cancer Research Campaign of the United Kingdom. P.V.N. is a recipient of a Post-Doctoral Fellowship from the Medical Research Council of Canada, and J.H. holds a Kekulé scholarship from Fonds der Chemischen Industrie, Germany.
References
- 1.Lane D P. Nature (London) 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Prives C. Cell. 1994;78:543–546. doi: 10.1016/0092-8674(94)90519-3. [DOI] [PubMed] [Google Scholar]
- 3.Kubbutat M H G, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 4.Kubbutat M H G, Vousden K H. Mol Med Today. 1998;4:250–256. doi: 10.1016/s1357-4310(98)01260-x. [DOI] [PubMed] [Google Scholar]
- 5.Lane D P. Nature (London) 1998;394:616–617. doi: 10.1038/29166. [DOI] [PubMed] [Google Scholar]
- 6.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 7.Soussi T, May P. J Mol Biol. 1996;260:623–637. doi: 10.1006/jmbi.1996.0425. [DOI] [PubMed] [Google Scholar]
- 8.Hansen S, Hupp T R, Lane D P. J Biol Chem. 1996;271:3917–3924. doi: 10.1074/jbc.271.7.3917. [DOI] [PubMed] [Google Scholar]
- 9.Hansen S, Lane D P, Midgley C A. J Mol Biol. 1998;275:575–588. doi: 10.1006/jmbi.1997.1507. [DOI] [PubMed] [Google Scholar]
- 10.Milner J. Nat Med. 1995;1:879–880. doi: 10.1038/nm0995-879. [DOI] [PubMed] [Google Scholar]
- 11.Bullock A N, Henckel J, DeDecker B S, Johnson C M, Nikolova P V, Proctor M R, Lane D P, Fersht A R. Proc Natl Acad Sci USA. 1997;94:14338–14342. doi: 10.1073/pnas.94.26.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieczorek A M, Waterman J L F, Waterman M J F, Halazonetis T D. Nat Med. 1996;2:1143–1146. doi: 10.1038/nm1096-1143. [DOI] [PubMed] [Google Scholar]
- 13.Brachmann R K, Yu K, Eby Y, Pavletich N P, Boeke J D. EMBO J. 1998;17:1847–1859. doi: 10.1093/emboj/17.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phelan A, Ellit G, O’Hare P. Nat Biotechnol. 1998;16:440–443. doi: 10.1038/nbt0598-440. [DOI] [PubMed] [Google Scholar]
- 15.Serrano L, Day A G, Fersht A R. J Mol Biol. 1993;233:305–312. doi: 10.1006/jmbi.1993.1508. [DOI] [PubMed] [Google Scholar]
- 16.Steipe B, Schiller B, Plückthun A, Steinbacher S. J Mol Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- 17.Collaborative Computational Project 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 18.Pantoliano M W, Whitlow M, Wood J F, Dodd S W, Hardman K D, Rollence M L, Bryan P N. Biochemistry. 1989;28:7205–7213. doi: 10.1021/bi00444a012. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard, S. J. & Thornton, J. M. (1993) naccess 2.1.
- 20.Sauer T R, Lim W A. Curr Opin Struct Biol. 1992;2:46–51. [Google Scholar]
- 21.Fersht A R, Serrano L. Curr Opin Struct Biol. 1993;3:75–83. [Google Scholar]
- 22.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 23.Wells J A. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- 24.Shih P, Kirsch J F. Protein Sci. 1995;4:2063–2072. doi: 10.1002/pro.5560041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X J, Baase W A, Shoichet B K, Wilson K P, Matthews B W. Protein Eng. 1995;8:1017–1022. doi: 10.1093/protein/8.10.1017. [DOI] [PubMed] [Google Scholar]
- 26.Kretschman E, Raether H. Z Naturforsch. 1968;23:2135. [Google Scholar]
- 27.Gannon J V, Greaves R, Iggo R, Lane D P. EMBO J. 1990;9:1595–1602. doi: 10.1002/j.1460-2075.1990.tb08279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menon A G, Anderson K M, Riccardi V M, Chung R Y, Whaley J M, Yandell D W, Farmer G E, Freiman R N, Li F P, Barker D F, et al. Proc Natl Acad Sci USA. 1990;87:5435–5439. doi: 10.1073/pnas.87.14.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]