Abstract
Normal mammalian cells arrest primarily in G1 in response to N-(phosphonacetyl)-l-aspartate (PALA), which starves them for pyrimidine nucleotides, and do not generate or tolerate amplification of the CAD gene, which confers resistance to PALA. Loss of p53, accompanied by loss of G1 arrest, permits CAD gene amplification and the consequent formation of PALA-resistant colonies. We have found rat and human cell lines that retain wild-type p53 but have lost the ability to arrest in G1 in response to PALA. However, these cells still fail to give PALA-resistant colonies and are protected from DNA damage through the operation of a second checkpoint that arrests them reversibly within S-phase. This S-phase arrest, unmasked in the absence of the G1 checkpoint, is dependent on p53 and independent of p21/waf1.
Keywords: Li-Fraumeni cells/REF52 cells/gene amplification/cell cycle arrest/G1 checkpoint
Normal mammalian cells maintain genomic integrity through controls that regulate their ability to progress through the cell cycle when they encounter environmental stress (1, 2). When these controls are deranged, the result is genomic instability, a hallmark of neoplasia. The tumor suppressor protein p53, commonly inactivated in tumors, is a major regulator of cell cycle progression in response to DNA damage or arrest of DNA synthesis (3–5). The signals generated by these stresses lead to an increase in the level of p53 protein and also stimulate its ability to activate transcription, leading to cell cycle arrest in both G1 (6–8) and G2/M (9, 10). An important transcriptional target of p53 is the cyclin-dependent kinase inhibitor p21/waf1 (11), which helps to mediate cell cycle responses to DNA damage (12–14). In primary human fibroblasts, the p53-mediated cell cycle arrest in response to DNA damage is irreversible (15). However, overexpression of p53 without DNA damage leads to reversible arrest in both G1 and G2/M (16–18).
By using p53-null mouse or human cells, or by using the E6 protein of human papilloma virus 16 to inactivate p53, it was shown that cells become permissive for CAD gene amplification and thus N-(phosphonacetyl)-l-aspartate (PALA) resistance when p53 is absent (19–21). In contrast, cells with intact p53 arrest stably, predominantly in G1, and do not form resistant colonies in PALA (19, 20). These observations reveal that PALA induces a p53-dependent cell cycle arrest in G1, thus suppressing gene amplification and genetic instability (16). In rodent cells, CAD gene amplification is the only mechanism observed for PALA resistance, but human cells become resistant to PALA through additional mechanisms including but not limited to CAD gene amplification (22).
In normal human fibroblasts deprived of pyrimidine nucleotides by PALA, wild-type p53 mediates G1 arrest through mechanisms that do not involve broken DNA (16). Paulson et al. (23) have shown that, in the absence of a functional p53 pathway, PALA treatment of tumor cells leads to DNA damage and thus to CAD gene amplification. Furthermore, in some cells the induction of p53 and associated G1 arrest are defective in response to PALA but normal in response to γ-radiation (24), suggesting that there are distinct and separable pathways of p53 activation in response to these different stimuli. In this report, we describe the responses of several cell lines to pyrimidine nucleotide depletion or radiation-induced DNA damage. All of these lines have functional, wild-type p53 and are not permissive for gene amplification. However, instead of accumulating in G1 after treatment with PALA, two of the cell lines arrest within S-phase. We suggest that activation of p53 by pyrimidine starvation causes a stable and reversible arrest in G1 and also within S-phase, thus regulating genomic stability at more than one point of the cell cycle.
MATERIALS AND METHODS
Cell Lines and Culture Conditions.
The MDAH041 post-crisis cell line, kindly provided by M. Tainsky (25), was derived from the fibroblasts of a patient with Li-Fraumeni syndrome. There is a frame shift mutation in one p53 allele at codon 184, and the normal p53 allele has been lost during in vitro propagation (20). Normal human WI38 fibroblasts were obtained from the American Type Culture Collection. TR9-7 cells, expressing wild-type p53 under the control of a tetracycline-regulated promoter, were derived from MDAH041 cells (17). Cell lines containing a neo marker were grown in the presence of G418 (600 μg/ml). Early passage p53-null and p21-null mouse embryo fibroblasts (MEFs) were provided by L. Donehower and G. Hannon, respectively. Normal human fibroblast (LF1) cells and their p21-null derivative (H07.2–1) were gifts of J. Sedivy (26). Low passage primary rat embryo fibroblast (REF) cells were provided by Andrei Gudkov (University of Illinois, Chicago) and REF52 cells were as described by Franza et al. (27). All cells were grown in 10% CO2 in DMEM, supplemented with 10% fetal bovine serum.
PALA Selections.
PALA (NSC224131), an inhibitor of the aspartate transcarbamylase activity of the multifunctional CAD enzyme, was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute, Bethesda, MD. Treatments of cells with PALA were as described by Agarwal et al. (28). For each cell line or strain, the ID50 was determined and selections of 2 × 105 cells per 10-cm plate were done in the presence of 10% (vol/vol) dialyzed fetal calf serum, using PALA at 3 × ID50 (29). The selective medium was changed every 4–5 days until colonies of 100–200 cells were observed, typically in 3–4 weeks.
Western and Northern Analyses.
Total cellular protein was isolated by lysing cells in 20 mM Tris⋅HCl, pH 7.5, 2% (wt/vol) SDS, 2 mM benzamidine, and 0.2 mM phenylmethanesulfonyl fluoride. Protein concentrations were determined by the Bradford method (Bio-Rad). Proteins (25 μg/lane) were separated by SDS-10% PAGE and electroblotted to polyvinylidene difluoride membranes (Stratagene; ref. 30). After transfer, the gels were stained with Coomassie blue (Sigma) to verify equal loading. The membranes were probed with mAb DO-1 (Santa Cruz Biotech), directed against p53, and bound antibody was detected by enhanced chemiluminescence (Amersham). Northern analysis: Total RNA was extracted with the Trizol reagent (GIBCO/BRL) as specified by the manufacturer. Analysis was performed as described by Sambrook et al. (31).
Cell Cycle Analysis.
The DNA content of trypsinized cells was determined by staining with propidium iodide, using a Cycletest kit (Becton Dickinson). Cell cycle distribution was determined by using a FACScan instrument (Becton Dickinson), cell fit software, and an HP340 Series 9000 Workstation (Hewlett–Packard). Dead cells were gated out by using pulse processing.
Measurement of DNA Synthesis.
For tritium labeling, C11 and MDAH041 cells were grown in 6-well plates and treated with PALA at 3 × ID50. After 3 days, 5 μCi of 3H-labeled thymidine was added to each well, and the cells were incubated for 2 hr more. After a trichloracetic acid wash, 0.5 ml of 2 M perchloric acid was added to each well at 60°C for 30 min. The resulting solution was transferred to scintillant and 3H incorporation was determined. The residue was dissolved in alkali for protein determination. For BrdUrd incorporation, cells grown on coverslips were treated with PALA (3 × ID50) and were incubated for an additional 4 hr in medium containing 10 μM BrdUrd. The incorporation of BrdUrd into DNA was determined by immunostaining (Amersham).
RESULTS
Li-Fraumeni Cell Lines in Which Wild-Type p53 Has Been Restored Have Many Properties of Normal Fibroblasts.
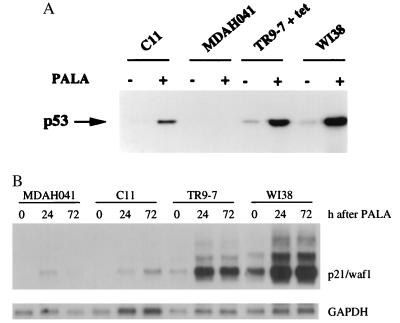
Previously, we established tetracycline-regulated expression of wild-type p53 in the p53-null MDAH041 cell line (17). Clone TR9-7 undergoes reversible growth arrest when wild-type p53 is overexpressed (in the absence of tetracycline), indicating that the pathway mediating this response to p53 is intact. We have now restored p53 expression in MDAH041 cells in a different way by introducing a wild-type p53 minigene under the control of the normal p53 promoter. One clone (C11) expresses a low level of p53 constitutively, and the p53 can be induced and activated by treatment with PALA (Fig. 1) or adriamycin (data not shown). The level of p53 is lower in C11 than in the normal fibroblast cell strain WI38, whereas the level in TR9-7 cells plus tetracycline is similar to that of WI38 cells (Fig. 1). After transient transfection of C11 cells with a construct in which the expression of luciferase is regulated by a p53-responsive element (32, 33), luciferase was induced by 5- to 10-fold upon treatment with PALA, γ-radiation, UV radiation, or adriamycin (data not shown), demonstrating that the p53 transgene is functional. In parallel experiments, p53-null MDAH041 cells did not express luciferase before or after treatment (data not shown). We also analyzed the expression of a p53 target gene, p21/waf1, in C11, TR9-7, MDAH041, and WI38 cells. PALA treatment induced p21 mRNA in both TR9-7 and WI38 cells (Fig. 1B). In contrast, the induction of p21 in C11 cells was not efficient after PALA treatment and was similar to that in MDAH041 cells. These observations suggest that the levels of p53 in C11 cells are below the threshold required for efficient induction of p21.
Figure 1.
Induction of p53 and p21/waf1 by PALA. (A) Cells were treated for 4 days with 0.5 mM PALA, and p53 was detected by Western analysis, using the DO-1 antibody, which was detected by enhanced chemiluminescence. (B) Levels of p21 mRNA. Total RNA was separated by electrophoresis in an agarose gel and transferred to a nylon membrane. p21 mRNA was detected with a specific human cDNA probe. Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) served as loading control.
However, TR9-7 cells plus tetracycline and C11 cells retain many other p53-dependent responses of normal human fibroblast cell strains, in contrast with their p53-null MDAH041 parents. Similarly, the rat cell line REF52, which has wild-type p53, retains many normal responses characteristic of REF cell strains. For example, none of these cell lines exhibit gene amplification in selective concentrations of PALA (refs. 29 and 34 and data presented below), TR9-7 (plus tetracycline), and REF52 cells arrest normally at the G1/S boundary in response to DNA damage (see below), and TR9-7 and C11 cells retain the normal p53-dependent arrest at G2/M in response to inhibition of DNA synthesis (5).
p53-Mediated S-Phase Arrest in Response to Pyrimidine Nucleotide Starvation.
Normal human fibroblasts arrest stably in PALA and are capable of resuming growth weeks after exposure to the drug, whereas cells lacking p53 lose viability within days. Furthermore, PALA resistance, undetectable in normal human fibroblasts, occurs in p53-null cells at a frequency of ≈10−5 (19, 20). Like WI38 cells, both TR9-7 cells plus tetracycline and C11 cells arrested stably, remained attached to the culture dishes and remained nearly constant in number for 4–6 weeks in the presence of PALA (data not shown). These cells also failed to generate PALA-resistant colonies (Table 1). As expected, MDAH041 cells were killed rapidly by PALA and gave rise to resistant colonies at a high frequency (Table 1). Thus, the formation of PALA-resistant colonies is p53-dependent in cell lines derived from MDAH041.
Table 1.
Growth properties, p53 status, and frequencies of PALA-resistant colonies
| Cell line or strain | p53 Status | ID50 for PALA, μM | Frequency* of PALA-resistant colonies at 3 × ID50 |
|---|---|---|---|
| MDAH041 | null | 75 | 5 × 10−4 |
| C11 | wt | 60 | <8 × 10−7 |
| TR9–7 | wt | 40 | <4 × 10−7 |
| WI38 | wt | 20 | <10−8 |
| REF52 | wt | 10 | <10−8 |
| REF | wt | 10 | <10−8 |
| MEF | wt | 17 | <10−7 |
| p53-null MEFs | null | 17 | 4.7 × 10−4 |
| p21/waf1-null MEFs | wt | 17 | <4 × 10−6 |
wt, wild type.
Cumulative estimate from several independent experiments.
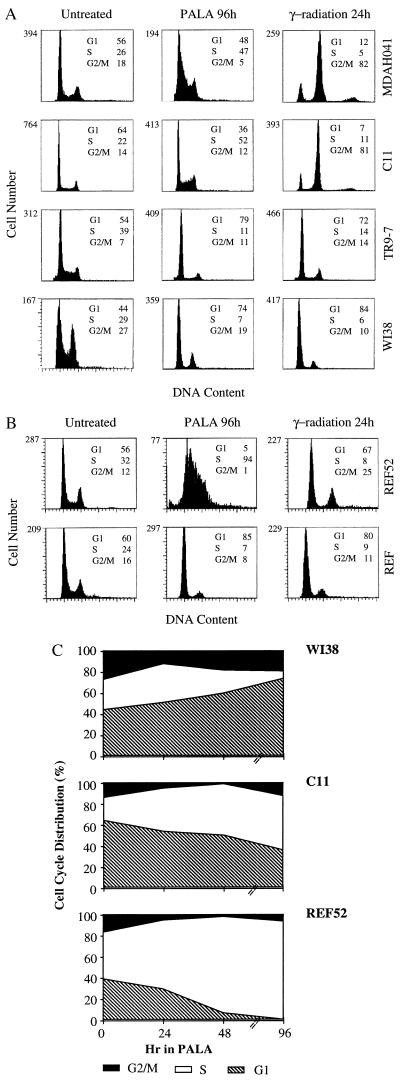
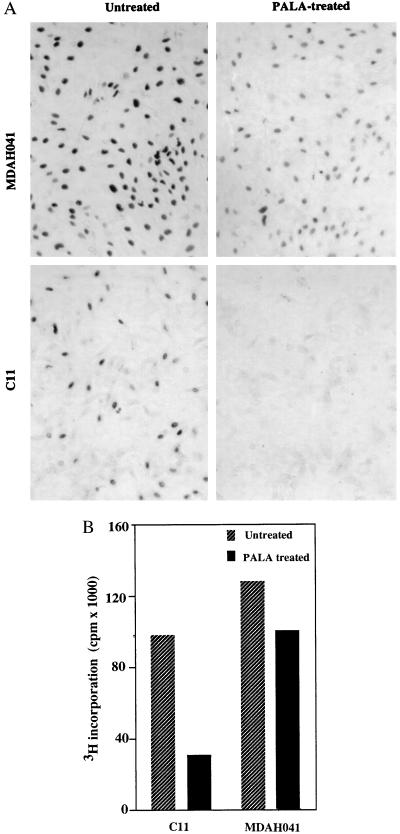
To compare their cell cycle responses, exponentially growing cells were treated with 3 × ID50 of PALA for 24, 48, or 96 hr and analyzed for DNA content. The majority of WI38 cells and TR9-7 cells plus tetracycline accumulated in G1 (2 N DNA content) and G2/M (4 N DNA content), with only 7% or 11%, respectively, in S-phase after 96 hr (Fig. 2A). In contrast, the S-phase population was much higher (52%) for C11 cells treated with PALA for 96 hr (Fig. 2A). Approximately, one-third of PALA-treated C11 cells have a 2 N content of DNA, possibly because their G1 checkpoint control is only partially effective. A similar analysis revealed virtually no G1 arrest in REF52 cells; almost all (94%) accumulated in S-phase 96 hr after PALA treatment (Fig. 2B and ref. 34). The response of a REF cell strain (Fig. 2B) was similar to that of normal human WI38 fibroblasts (Fig. 2A). To analyze whether cells arrested in S-phase make DNA, C11 and MDAH041 cells were treated with PALA for 3 days and pulse-labeled with BrdUrd for 4 hr or [3H]thymidine for 2 hr. PALA-treated C11 cells exhibited much reduced BrdUrd staining (Fig. 3A) and a 3-fold decrease in the uptake of thymidine (Fig. 3B) as compared with untreated cells. These observations reveal that the cells arrested in S-phase synthesize very little DNA. Similar observations were made in PALA-treated REF52 cells (34). In contrast, PALA-treated MDAH041 cells continue to synthesize DNA (Fig. 3), indicating that, when their deoxynucleotide pools are unbalanced by PALA in the absence of p53, these cells do enter S-phase and continue to make new DNA under suboptimal conditions, which may lead to DNA damage.
Figure 2.
Cell cycle distributions of human (A) and rat (B) cells. After treatment with PALA (3 × ID50) or γ-radiation (5 Gy), the cells were sorted after staining with propidium iodide. The percentages of cells in G1, S, and G2/M are indicated. (C) Cell cycle distributions as a function of time of treatment with PALA.
Figure 3.
DNA synthesis in PALA-treated cells. (A) BrdUrd labeling of PALA-treated MDAH041 and C11 cells. Cells grown on coverslips were treated with PALA (3 × ID50) for 3 days and pulse-labeled for 4 hr. After staining with anti-BrdUrd antibody, the cells were photographed (×57). (B) [3H]thymidine labeling of PALA-treated C11 and MDAH041 cells. Cells were treated with PALA (3 × ID50) for 3 days and pulse-labeled for 2 hr. The incorporation of 3H was normalized to the total amount of protein in each well.
PALA-Induced S-Phase Arrest Is Reversible and Protective.
To study the reversibility of the S-phase growth arrest, C11 cells, TR9-7 cells plus tetracycline, and REF52 cells were treated with 3 × ID50 of PALA for 4 or 15 days and then replated in normal medium for analysis of colony formation. All the cell lines with wild-type p53 survived exposure to PALA, in contrast to the p53-null MDAH041 cells (Table 2). All the human cell lines with p53 survived better than either of the rat cell types. To assess whether the cell cycle arrest protects against mutagenic DNA damage upon treatment with PALA, we examined the status of the single-copy X-linked hypoxanthine phosphoribosyl transferase (HPRT) gene, whose inactivation can be selected for with 6-thioguanine (6TG). Cells were treated either with selective concentrations of PALA (3 × ID50) or with nonselective concentrations (1 × ID50). In the latter situation, REF52 cells become permissive for CAD gene amplification, probably due to the damage caused when DNA is replicated from highly unbalanced pools of deoxynucleoside triphosphates (34). No 6TG-resistant colonies were observed when the cells were either untreated or treated with PALA at 3 × ID50, whereas 6TG-resistant colonies appeared at a frequency of ≈10−5 when REF52 cells were treated with PALA at 1 × ID50 (Table 3). When normal, p53-null or p21-null MEFs were treated with PALA at 3 × ID50 for 4 days and then subjected to selection with 6TG, resistant colonies were observed with the p53-null MEFs (frequency ≈3 × 10−5) but not with p21-null or normal MEFs (Table 3). These results show that when cells can arrest protectively in the presence of PALA, the HPRT locus is not mutated, and that HPRT mutations do arise at a high frequency when exposure to PALA does not lead to protective arrest.
Table 2.
Recovery of cells after PALA treatment
| Cell line or strain | Percentage of cells that form colonies after PALA treatment
|
|
|---|---|---|
| 4 days | 15 days | |
| MDAH041 | <1 | <1 |
| C11 | 82 | 63 |
| TR9-7 | 86 | 70 |
| WI38 | 62 | 43 |
| REF | 20 | nd |
| REF52 | 19 | nd |
| MEF | 28 | nd |
| p53-null MEF | <1 | nd |
| p21/waf1-null MEF | 27 | nd |
The values are normalized for the plating efficiency of each cell line without treatment. Cells treated with 3 × ID50 of PALA were replated into normal medium containing 1 mM uridine, to allow rapid re-establishment of pyrimidine nucleotide pools. Colonies containing 30 or more cells were fixed and counted after 10–15 days. nd, not determined.
Table 3.
Mutation of the HPRT locus in PALA-treated cells
| Treatment | Frequency of 6-thioguanine-resistant colonies
|
|||
|---|---|---|---|---|
| REF52 | MEF | p53-null MEF | p21/waf1-null MEF | |
| None | <2 × 10−6 | <2 × 10−6 | <2 × 10−6 | <2 × 10−6 |
| PALA (3 × ID50) | <2 × 10−6 | <2 × 10−6 | 1.2 × 10−5 | <2 × 10−6 |
| PALA (1 × ID50) | 1.6 × 10−5 | nd | nd | nd |
Cells treated with PALA for 4 days were then placed in normal medium containing 1 mM uridine and selected with 6TG (1 μg/ml). Colonies with 30 or more cells were counted after 15 days. nd, not determined.
p53-Mediated S-Phase Arrest and Genomic Stability Are Independent of p21/waf1.
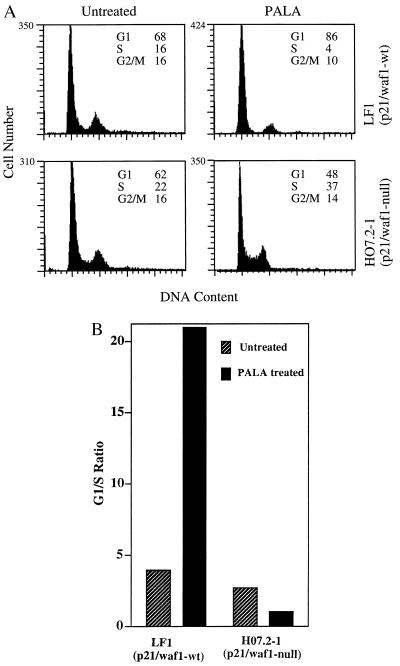
Since REF52 and C11 cells do not generate PALA-resistant colonies despite loss of the G1 checkpoint and since p21/waf1 is known to regulate the G1/S transition, we investigated whether p21 plays a role in regulating S-phase arrest, using a human cell strain in which p21 has been inactivated by homologous recombination (H07.2–1; ref. 26) and p21-null MEFs. Normal parental human LF1 cells and p21-null H07.2–1 cells were treated with 50 μM PALA (ID50 = 15 μM) for 3 days, and the cell cycle distributions were determined. The normal LF1 cells arrested in G1, with an increase of more than 5-fold in the G1/S ratio (Fig. 4). In contrast, the p21-null H07.2–1 cells accumulated in S-phase, as reflected by the change in G1/S ratio from 2.8 in untreated cells to 1.2 in PALA-treated cells (Fig. 4). Similar observations were made in p21-null MEFs (data not shown). Deng et al. (13) also found an increase in the number of S-phase cells in PALA-treated p21-null MEFs and a decrease in S-phase cells in PALA-treated normal MEFs.
Figure 4.
Cell cycle distributions of p21/waf1-null human cells. (A) Three days after treatment with PALA (50 μM, 3 × ID50), the cells were sorted after staining with propidium iodide. The percentages of cells in G1, S, and G2/M are indicated. (B) G1/S ratios for untreated and PALA-treated normal and p21-null human cells.
To assess whether the loss of p21/waf1 affects the ability of cells to become resistant to PALA, we exposed 2 × 106 p53-null, p21-null, or normal MEFs to 50 μM PALA (3 × ID50) for 4 weeks. Colonies were observed for the p53-null MEFs (frequency ≈1 × 10−5) but not for the p21-null or normal MEFs (Table 1). To see whether p21-null MEFs were protected from DNA damage, these cells were treated with PALA for 4 days and then selected with 6TG, with p53-null cells as a control. 6TG-resistant colonies were observed for the p53-null cells, whereas no colonies were seen for the p21-null cells (Table 3), showing that the HPRT gene had not been mutated.
Protective Arrest in Response to PALA Is Independent of the γ-Radiation-Induced G1 Checkpoint.
Irradiation of exponentially growing cells (5 Gy) led to the arrest of WI38, TR9-7 (plus tetracycline), and REF52 cells in G1 and G2/M 24 hr later (Fig. 2 A and B). In contrast, similar treatment of MDAH041 cells led to a drastic decrease in the G1 population, with a corresponding increase in cells in G2/M after 24 hr (Fig. 2A). Surprisingly, C11 cells and MDAH041 cells behaved similarly in this experiment, showing a transient arrest in G2/M with no evidence of accumulation in G1. Therefore, although the nonpermissive cell lines C11 and REF52 show a very similar accumulation in S-phase when treated with PALA, they respond quite differently to γ-radiation. We conclude that the regulation of cell cycle checkpoint functions under conditions of nucleotide starvation is different and separable from the regulation induced by γ-irradiation.
DISCUSSION
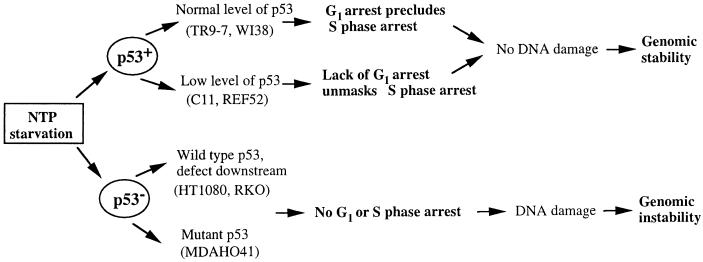
p53 plays a major role in maintaining genomic stability (reviewed in refs. 1, 2, 35, and 36) and cells lacking functional p53 readily become aneuploid and generate PALA-resistant colonies (19, 20). In normal human fibroblasts, failure to acquire PALA resistance is correlated with protective G1 arrest in response to nucleotide depletion (16) or γ-irradiation (15). Inactivation of p53 leads to loss of the G1 checkpoint and acquisition of the ability to generate PALA-resistant colonies. Our new observations suggest that, in addition to the G1 checkpoint, p53-mediated S-phase arrest is sufficient to suppress both PALA resistance and PALA-induced mutation of the HPRT locus (Fig. 5).
Figure 5.
p53-Dependent effects of PALA-induced pyrimidine nucleotide starvation.
Levels of p53 May Determine Whether Arrest Occurs in G1 or S-Phase in Response to PALA or γ-Irradiation.
C11 cells expressing wild-type p53 failed to arrest in G1 in response to PALA but did arrest stably and reversibly in S-phase. However, TR9-7 cells, derived from the same parental cells as C11, showed a phenotype very similar to that of WI38 cells. TR9-7 and WI38 cells express similar levels of p53, whereas C11 cells express a lower level, suggesting that a threshold amount of p53 may be required to achieve efficient G1 arrest in PALA and that a lower amount of p53 is sufficient for S-phase arrest in PALA. Since C11 cells continued to enter S-phase after γ-irradiation, whereas TR9-7 and WI38 cells arrested efficiently in G1, G1 arrest after γ-radiation also may depend on the level of p53 expression. Therefore, different p53-dependent cell cycle checkpoint responses seem to be triggered at different levels of p53, consistent with the previous observation that the level of p53 determines whether cells arrest or die by apoptosis (37).
REF52 cells arrest in G1 in response to irradiation but not PALA treatment, whereas C11 cells fail to arrest in G1 in response to either stimulus. Consistent with these results, Chen et al. (37) have described cell types in which the accumulation of p53 and G1 arrest are defective in response to PALA but normal in response to γ-radiation. It is possible that p53 is modified differentially in response to these two forms of stress, for example, by phosphorylation at different sites, leading to activation of different components downstream of p53 and thus to distinct biological outcomes.
p21/waf1-Independent Inhibition of DNA Synthesis by p53 When Cells Are Starved for Nucleotides Protects Them from DNA Damage.
Although both MDAH041 and C11 cells do incorporate thymidine or BrdUrd when arrested in S-phase by PALA, the incorporation is much lower in C11 cells. This result suggests that p53 inhibits ongoing DNA synthesis in cells arrested in S-phase by PALA. S-phase arrest in PALA-treated cells involves the p53-dependent inhibition of DNA synthesis, which prevents the breakage that would occur if DNA were synthesized from highly unbalanced deoxynucleoside triphosphate pools. In PALA, REF52 cells lack both the G1 and G2/M checkpoints and initially traverse through S-phase (34). In the next cycle, cells begin to accumulate within S and stay there for a long time. In S-phase-arrested REF52 cells, as in C11 cells, DNA synthesis is inhibited, as revealed by reduced uptake of BrdUrd (34). Furthermore, when cells lacking functional p53 attempt to propagate, most of them undergo mitotic catastrophe and cell death (5). However, rare cells overcome the effect of PALA by increasing CAD activity (22). The reversibility of arrest of cells with wild-type p53 in PALA indicates strongly that it has occurred in the absence of DNA damage, since only one double strand break may be enough to cause permanent growth arrest in human fibroblasts (15, 38). DNA damage, assayed by the generation of resistance to 6TG, is also inhibited in the S-phase-arrested cells, again showing that p53 inhibits the generation of DNA damage in these cells.
A role for p53 in blocking DNA synthesis has not yet been reported. We and others have shown that overexpression of p53 in the absence of stress does not hinder progression through S-phase but rather causes arrest in either G1 or G2 (17, 18). p53 may not be modified appropriately to carry out its S-phase checkpoint function under the conditions explored previously. Alternatively, a role for p53 in S-phase progression may only become evident when S-phase is prolonged, as in the presence of PALA. Whatever mechanism p53 uses to block progression through S-phase, p21/waf1 is not required, since both C11 cells lacking p21 expression and p21-null MEFs do not arrest in G1 in PALA (ref. 13 and the present study), do accumulate stably in S-phase, and do not form PALA-resistant colonies (the present study). Brown et al. (26) have recently found that deletion of the p21/waf1 gene by homologous recombination in normal human cells does not alter the p53-mediated regulation of genomic stability, and we find that the same p21/waf1-null cells do arrest in S-phase when treated with PALA. In summary, p53-dependent suppression of the genesis of PALA-resistant cells can occur at two points in the cell cycle, and one of these points is independent of p21/waf1.
Physiological Importance of p53-Dependent Protective Responses to the Inhibition of DNA Synthesis.
The S-phase arrest may be important under several different physiological situations. For example, when conditions for growth are unfavorable or when cells are exposed to a stress, DNA may be damaged if cells attempt to replicate it. A p53-mediated blockade of DNA synthesis may prevent the generation and propagation of additional DNA damage. After arrest, the cells may either repair the damaged DNA or undergo apoptosis before entering the next cell cycle. p53-dependent checkpoints in S-phase and G2/M also may help to protect cells in which a mutation has inactivated a protein required for G1 arrest, such as Rb. Multiple checkpoints in different parts of the cell cycle are likely to be important in realizing the full ability of p53 to maintain genomic stability. In this regard, we have recently found that the protective function of p53 in cells arrested in S-phase also includes the ability to inhibit their entry into mitosis. p53-null cells treated with hydroxyurea continue to attempt mitosis, whereas the same cells with p53 restored do not (5).
Acknowledgments
We thank Michael Tainsky, Larry Donehower, Greg Hannon, and John Sedivy for MDAH041, p53-null MEFs, p21/waf1-null MEFs, and p21-null human cells, respectively. This work was supported by Grant GM49345 from the National Institutes of Health. W.R.T., a fellow of the National Cancer Institute of Canada, was supported by funds from the Canadian Cancer Society.
ABBREVIATIONS
- PALA
N-(phosphonacetyl)-l-aspartate
- REF
rat embryo fibroblast
- HPRT
hypoxanthine phosphoribosyl transferase
- MEF
mouse embryo fibroblast
- 6TG
6-thioguanine
References
- 1.Di Leonardo A, Linke S P, Yin Y, Wahl G M. Cold Spring Harbor Symp Quant Biol. 1993;58:655–667. doi: 10.1101/sqb.1993.058.01.073. [DOI] [PubMed] [Google Scholar]
- 2.Chernova O B, Chernov M V, Agarwal M L, Taylor W R, Stark G R. Trends Biochem Sci. 1995;20:431–434. doi: 10.1016/s0968-0004(00)89094-5. [DOI] [PubMed] [Google Scholar]
- 3.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 5.Taylor, W. R., Agarwal, M. L., Agarwal, A., Stacey, D. W. & Stark, G. R. (1998) Oncogene, in press. [DOI] [PubMed]
- 6.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 7.Fu L, Minden M D, Benchimol S. EMBO J. 1996;15:4392–4401. [PMC free article] [PubMed] [Google Scholar]
- 8.Hupp T R, Sparks A, Lane D P. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 9.Ceraline J, Deplanque G, Duclos B, Limacher J M, Vincent F, Goldblum S, Bergerat J P. Bull Cancer (Paris) 1997;84:1007–1016. [PubMed] [Google Scholar]
- 10.Schwartz D, Almog N, Peled A, Goldfinger N, Rotter V. Oncogene. 1997;15:2597–2607. doi: 10.1038/sj.onc.1201436. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 12.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 13.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 14.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Nature (London) 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 15.Di Leonardo A, Linke S P, Clarkin K, Wahl G M. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 16.Linke S P, Clarkin K C, DiLeonardo A, Tsou A, Wahl G M. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal M L, Agarwal A, Taylor W R, Stark G R. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart N, Hicks G G, Paraskevas F, Mowat M. Oncogene. 1995;10:109–115. [PubMed] [Google Scholar]
- 19.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 20.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 21.White A E, Livanos E M, Tlsty T D. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 22.Smith K A, Chernova O B, Groves R P, Stark M B, Martinez J L, Davidson J N, Trent J M, Patterson T E, Agarwal A, Duncan P, et al. Proc Natl Acad Sci USA. 1997;94:1816–1821. doi: 10.1073/pnas.94.5.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulson T G, Almasan A, Brody L L, Wahl G M. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C-Y M, Hall I, Lansing T J, Gilmer T M, Tlsty T D, Kastan M B. Cancer Res. 1996;56:3659–3662. [PubMed] [Google Scholar]
- 25.Bischoff F, Yim S O, Pathak S, Grant G, Siciliano M J, Giovanella B C, Strong L C, Tainsky M A. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 26.Brown J P, Wei W, Sedivy J M. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 27.Franza B, Jr, Maruyama K, Garrels J I, Ruley H E. Cell. 1986;44:409–418. doi: 10.1016/0092-8674(86)90462-9. [DOI] [PubMed] [Google Scholar]
- 28.Agarwal M L, Agarwal A, Taylor W R, Wang Z-Q, Wagner E F, Stark G R. Oncogene. 1997;15:1035–1041. doi: 10.1038/sj.onc.1201274. [DOI] [PubMed] [Google Scholar]
- 29.Perry M E, Commane M, Stark G R. Proc Natl Acad Sci USA. 1992;89:8112–8116. doi: 10.1073/pnas.89.17.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 31.Sambrook J, Frisch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 34.Chernova O B, Chernov M V, Ishizaka Y, Agarwal M L, Stark G R. Mol Cell Biol. 1998;18:536–545. doi: 10.1128/mcb.18.1.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Wahl G M, Linke S P, Paulson T G, Huang L-C. Cancer Surv. 1997;29:183–219. [PubMed] [Google Scholar]
- 37.Chen X, Ko L J, Jayaraman L, Prives C. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 38.Huang L-C, Clarkin K C, Wahl G M. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]