Abstract
Since the time of Charles Darwin, studies of interspecific hybridization have been a major focus for evolutionary biologists. Although this phenomenon has often been viewed as problematic in the fields of ecology, taxonomy and systematics, it has become a primary source of data for studies on speciation and adaptation. Effects from genetic/evolutionary processes, such as recombination and natural selection, usually develop over extended periods of time; however, they are accelerated in cases of hybridization. Interspecific hybrids exhibit novel genomes that are exposed to natural selection, thus providing a key to unravel the ultimate causes of adaptation and speciation. Here we provide firstly a historic perspective of hybridization research, secondly a novel attempt to assess the extent of hybridization among animals and thirdly an overview of the reviews and case studies presented in this theme issue.
Keywords: animal hybridization, introgression, literature review
1. Introduction
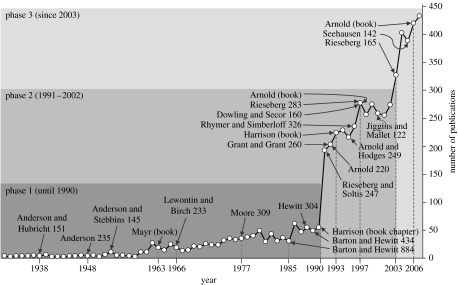
Research on interspecific hybrids has played a major role in the field of evolutionary biology, since deciphering the mechanisms preventing or allowing interbreeding of species helps to answer general questions such as: How are reproductive barriers between species evolving? Which genes contribute to reproductive isolation? Is gene flow between species (introgression) a creative, additional force in adaptation? During the past 150 years research on interspecific hybridization went through three main phases (figure 1): the first phase is characterized by the search for cases of hybridization and the attempt to develop a theoretical framework; in the second phase many examples of interspecific hybridization were explored using molecular markers; and the third and current phase is fuelled by the rapid integration of genomic and ecological methods. In the first part of this paper we will describe this development from a historical point of view. The second part will then tackle the question of how frequent hybridization occurs in animals, and test the expectation if the level of hybridization varies among taxonomic groups. The last part of the introduction will highlight some of the recent developments in the field illustrated by the contributions to this special issue.
Figure 1.
Number of publications on the topic of interspecific hybridization or introgression covering the last 70 years. Data were collected from the ISI literature databank (during December 2007). The selected key publications are highlighted and numbers with the authors indicate the number of citations (Anderson & Hubricht 1938; Anderson 1948; Anderson & Stebbins 1954; Mayr 1963; Lewontin & Birch 1966; Moore 1977; Barton & Hewitt 1985, 1989; Hewitt 1988; Harrison 1990, 1993; Rieseberg & Soltis 1991; Arnold 1992, 1997, 2006; Grant & Grant 1992; Arnold & Hodges 1995; Rhymer & Simberloff 1996; Dowling & Secor 1997; Rieseberg 1997; Jiggins & Mallet 2000; Rieseberg et al. 2003; Seehausen 2004).
2. One hundred and fifty years of hybridization research
Since Darwin first described hybrid formation in the context of speciation, interspecific hybridization has attracted the attention of many evolutionary biologists. Hybrids are formed when different species interbreed, resulting in the combination of genetic material from previously isolated gene pools. Darwin (1859) observed that:
Those forms which possess in some considerable degree the character of species, but which are so closely similar to some other forms, or are so closely linked to them by intermediate gradations, that naturalists do not like to rank them as distinct species, are in several respects the most important to us.
However, until the 1930s the common opinion was that sterility of interspecific hybrids caused the lack of evident hybridization. In the 1930s and 1940s a number of botanists started to experimentally study interspecific hybridization, either using crossing experiments (Anderson & Hubricht 1938) or field studies to estimate the rate of hybridization and to understand the consequences of interbreeding (Anderson 1948). These studies revealed that genetic information was exchanged between species (introgression) and illustrated that introgression was not rare among plants. In the 1960s Mayr (1963) suggested that interspecific hybridization among animals was of minor significance, whereas Lewontin & Birch (1966) indicated that hybridization might represent a source of novel evolutionary trajectories. Their experiments on a fruitfly (genus Dacus) suggested that the introduction of genes from another species could serve as the raw material for an adaptive evolutionary advance even though the original hybridization was disadvantageous. During the 1950s and 1960s a number of empirical cases of plant and animal hybridization led to the development of models of hybridization, which were then discussed in the seminal review by Moore (1977). Based on additional empirical evidence generated during the 1970s and early 1980s, Barton & Hewitt (1985) compiled their comprehensive, and very influential, review of instances of interspecific hybridization; they concluded that the majority of cases were explained by a balance between dispersal of parental taxa into a (usually narrow) hybrid zone and selection against hybrids. Another major milestone in the field of interspecific hybridization was presented by Harrison (1990). He concluded that although ample examples of interspecific hybridization had been reported, no single scenario or hybrid model seemed to explain all cases. Instead, the diversity in the origin, dynamics and fate of hybrids suggested that multiple evolutionary pathways were responsible for this phenomenon. Although these landmark reviews fuelled future research, a technological invention, i.e. the polymerase chain reaction, caused a revolution in the studies of hybridization. The number of publications increased fourfold to approximately 200 publications per year (figure 1). During this second phase many studies focused on detailed genetic analyses, hybrid fitness and selection in hybrid zones. The latter issue became a prominent controversy in the field; either selection against hybrids was considered to be independent of environmental factors (tension zone models), or fitness of different genotypes varies in response to environmental variation (e.g. hybrid superiority model) and thus fitness of hybrids is at least equal to that of the parental species. Although historically environmental independent scenarios have been mainly proposed by zoologists and the environmental dependent scenarios preferentially by botanists, this taxonomic division has been softened during the 1990s, and an integrative scenario was presented by Arnold (1997). Owing to the technological advances that allowed the simultaneous measurement of genetic variation at nuclear and mitochondrial loci, the issue of gene flow among species (i.e. introgression) became a major focus in the field of interspecific hybridization of animals (Dowling & Secor 1997). The third phase of hybrid research has been driven by the availability of multiple nuclear markers, i.e. microsatellite DNA, AFLP and SNP, and the first genomic studies of hybrid taxa (Rieseberg et al. 2003; Mallet 2005; Rogers & Bernatchez 2007). One of most controversial issues since the 1960s has concerned the adaptive significance of interspecific hybridization. Is hybridization a significant evolutionary mechanism which creates opportunities for adaptation and speciation, or is it just some ‘evolutionary noise’ (Arnold 1997)? Genomic studies of animal and plant systems provided evidence for the first scenario, since hybridization did indeed facilitate major ecological transitions (Rieseberg et al. 2003; Gompert et al. 2006; Rogers & Bernatchez 2007). These and other intriguing cases of interspecific hybridization have led to the development of new conceptual models on hybridization, which either focus on adaptive radiations or introgression and adaptation (Seehausen 2004; Arnold 2006).
An overview of the past 150 years of hybridization studies illustrates two aspects that are most likely apply to many other fields. Firstly, scientific progress is largely driven by technological advances. Secondly, questions raised 100 years ago are still in the focus of current research projects. The latter is most likely explained by the remaining gap in the recent integration of genetics and ecology. Although we have experienced a fruitful exchange of ideas among both fields of research, the field of molecular ecology has been mainly characterized by the application of selectively neutral markers, such as mitochondrial or microsatellite DNA, to address ecological questions. Mainly, these studies used genetic information to identify species to measure the effect of gene flow or to reconstruct phylogenetic relationships or phylogeographic patterns. However, the genetic basis of natural selection and reproductive isolation were examined only recently, and thus there are still relatively few ecological genetic analyses that have gathered data concerning functional genes, neutral loci and hybrid fitness. Yet, these few studies have illustrated, for example, how introgressive hybridization contributes to the evolution of interbreeding species (Grant & Grant 2008).
We would also like to stress two other, more general, aspects emerging from studies using literature or other databases. On the one hand, they illustrate the additive value of multiple studies of the same phenomenon over a broad taxonomic range, such as pure taxonomic or barcoding studies (Hebert et al. 2003). Thus, only the availability of focused, individual studies and structured databases allows us to extract patterns beyond the scope of single observation. On the other hand, our finding that the frequency of hybridizing taxa might represent a neutral by-product of evolutionary change is consistent with similar studies looking at the frequency of cryptic species (Pfenninger & Schwenk 2007). This study also supports the hypothesis that neutral processes are frequently underestimated in evolutionary studies (Lynch 2007).
3. Extent of interspecific hybridization among animals
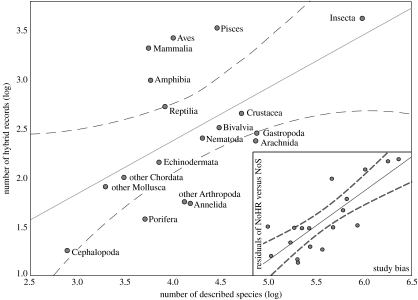
During the recent past, a number of reviews on interspecific hybridization illustrated an unexpectedly wide taxonomic range and, depending on the group of organisms, relatively high frequencies of interbreeding species, e.g. among vascular plants (25%), European butterflies (12%), fruitflies (1%), birds (10%), British ducks (25%) and European mammals (6%; Grant & Grant 1992; Mallet 2005). These figures suggest that interspecific hybridization represents a group-specific process, e.g. ducks have been impacted much more by introgression than have mammals. However, since taxonomic groups are not randomly chosen, but based on available data, they might be biased due to organismic preferences of research teams. Many diverse taxonomic groups may be under-represented in the literature and estimates of the degree to which their species participate in hybridization is thus unknown. Here we present a comprehensive analysis of hybridizing species in the animal kingdom, which includes a correction for study (i.e. organismic) bias. This allows us to test the hypothesis of differential hybridization rates among major metazoan groups. This literature survey is based on a recently developed approach to detect the rate of ‘cryptic species’ in animals (Pfenninger & Schwenk 2007).
(a) The ‘neutral hypothesis of hybridization’
We have used the publicly available literature database ‘Zoological Record’ (1947–2007). We found 21 972 publications concerning hybrid animals within the field of ecology, evolution and systematics (table 1). In order to test if the abundance of hybrids is equally distributed among various taxonomic groups, we correlated the number of hybrid records with the estimated number of described species (Baillie et al. 2004) in each taxonomic group (figure 2). A first significant correlation between both parameters indicates that the level of hybridization is predicted by the number of species per taxon (R2=0.29, p=0.02). The deviations of the regression line (residuals) are composed of differences in study intensity and taxonomic practice in the respective research community, true differences among taxonomic groups and random error. From a second correlation between all records used in our study and the number of described species per taxonomic group, we isolated the residuals as an indicator of a study bias (R2=0.34, p=0.01). Some taxa, especially vertebrates and the economically important pests and crops, have received far greater attention from taxonomists and population geneticists than most other taxa (e.g. marine invertebrates). This parameter explained a large part of the residual variation derived from the first regression (R2=0.66, p=0.00004). This pattern indicates that the rate of interspecific hybridization is relatively homogeneous among animal groups and only a few groups deviate from this pattern. Thus, the level of interspecific hybridization might represent an inherent and neutral parameter of evolutionary change. This analysis does not reveal anything about the potential significance of hybridization, or the frequency of backcrossing and introgression. However, it implies that in each taxonomic group we have a priori similar chances for gene exchange among species. We do not know how accurately the hybrid records reflect the actual number of interspecific hybrids. However, if the proportion of multiple hits in the number of hybrid records and the total number of records is roughly the same, we estimate the average hybridization rate to be approximately 1% (range 0.1–3%). This relatively low frequency of hybridization in animals is certainly at odds with previous estimates, which have predicted a frequency of approximately 10% (Mallet 2005). This discrepancy raises the question whether previous studies overestimated rates of hybridization due to taxon bias and taxonomic practice, or if a literature survey underestimates the rate since non-hybridizing taxa might on average be more frequently studied than hybridizing taxa. Future studies that focus on other kingdoms, such as plants and fungi, might allow us to better understand the potential impact of interbreeding.
Table 1.
Results of database search in Zoological Record (1947–2007) for several metazoan taxa. (Information on the estimated number of currently described species for each taxon was obtained from the IUCN 2004 report; Baillie et al. 2004.)
| number of | |||
|---|---|---|---|
| taxa | hybrid records | total records | described species |
| Arthropoda | |||
| Insecta | 4291 | 427 279 | 950 000 |
| Crustacea | 452 | 91 275 | 52 000 |
| Arachnida | 242 | 58 445 | 73 000 |
| other Arthropoda | 57 | 19 361 | 13 000 |
| Chordata | |||
| Mammalia | 2094 | 180 409 | 5416 |
| ‘Amphibia’ | 999 | 33 864 | 5743 |
| ‘Reptilia’ | 536 | 58 526 | 8163 |
| Aves | 2685 | 186 024 | 9917 |
| ‘Pisces’ | 3393 | 161 095 | 28 500 |
| other Chordata | 100 | 4715 | 3025 |
| Mollusca | |||
| Cephalopoda | 18 | 10 952 | 768 |
| Bivalvia | 323 | 25 320 | 30 000 |
| Gastropoda | 289 | 34 557 | 75 000 |
| other Mollusca | 81 | 39 950 | 1950 |
| Annelida | 55 | 20 536 | 15 000 |
| Echinodermata | 144 | 16 522 | 7000 |
| Porifera | 38 | 6384 | 5000 |
| Nematoda | 253 | 31 866 | 20 000 |
Figure 2.
The log of hybrid reports as a function of the log of described species in the respective taxon. Dashed lines represent 95% confidence limits. Residuals of this regression represent either true differences of taxonomic groups or a study bias. In order to test which of the two contributes most to this deviation, we correlated the residuals to the study bias (deviations of a regression of the number of records in the database with the number of the described species). The regression of both residuals resulted in a significant relationship (inset figure).
But even if the hybridization frequency is low, we would argue that the studies of interspecific hybridization should remain a major focus of evolutionary biology for two reasons. Firstly, although the frequency of individuals which actually interbreed with other species might be rare, the occurrence of occasional hybridization events might be sufficient to cause long-lasting changes in the genomic architecture of species. Thus hybridization might be rare but if it occurs it has a major impact on the origin and fate of evolutionary lineages. Secondly, hybridization provides a natural setting to study efficiently the interaction between genetic and ecological differentiation. In an attempt to justify the study of rare phenomena, Vrijenhoek (1989) once argued: ‘Examination of unusual processes opens a window to understanding what is common and what is normal. We learn about human health by studying innumerable disorders and diseases, and we learn about the function of normal genes by studying the phenotypic consequences of mutations. To understand the functional properties of outcrossing, biparental sexuality, we must study the consequences of aberrant reproductive processes.’ This argument applies to studies of ‘rare’ hybridization as well.
4. The process and its evolutionary impact
By combining previously isolated gene pools, interspecific hybridization results in the origin of new genotypes. If this phenomenon is viewed at the level of populations, then interspecific hybridization can also result in significant shifts of allele frequencies. When compared with mutation and recombination within species (the more usual mechanisms for the origin of new genotypes), interspecific hybridization can result in rapid and long-lasting changes among interbreeding species. The evolutionary change due to hybridization can occur within one generation, thereby exposing new gene combinations to natural selection. Furthermore, by compressing the time scale over which evolutionary processes occur, and by allowing the formation of new gene combinations, interspecific hybridization enables investigations into the processes of natural selection. Owing to these properties, examples of interspecific hybridization have been described as ‘natural laboratories for evolutionary studies’ (Hewitt 1988), ‘windows on evolutionary process’ (Harrison 1990) or as ‘an ecologically dependent behavioural phenomenon, with genetic consequences’ (Grant & Grant 2008). In the following subsections, we will address several topics of current hybridization research exemplified by the contributions to this theme issue.
(a) Introgressive hybridization
During the past decades, introgressive hybridization became one of the well-studied phenomena in evolutionary biology. This development is explained by two achievements. Firstly, advances in the field of genomics have allowed a definition of introgression at an unprecedented level of detail (Arnold 2006). Secondly, there has been a simultaneous series of analyses that have addressed the possible evolutionary impact of introgression events. For example, combined analyses of mitochondrial and multiple nuclear markers have defined the parental contributions to hybrid genomes. Studies based on these technological advances supported two main conclusions: (i) introgression is much more frequent than previously thought and (ii) hybridization can play an important creative role in adaptive evolution (Arnold et al. 2008). The unambiguous detection of introgression is not trivial since processes such as lineage sorting (i.e. retention of ancestral polymorphisms) can result in identical patterns of genetic differentiation (Alves et al. 2008). However, it is even more difficult to determine the evolutionary importance of introgression among species (Futuyma & Shapiro 1995). Yet, comprehensive studies on the Darwin's finches (Geospiza) have revealed, for example, the effect of introgressive hybridization on developmental genetic pathways of phenotypic traits (i.e. beak shape; Grant & Grant 2008).
(b) Identification of hybrid classes
Owing to the application of numerous genetic techniques and the recent availability of advanced analytical tools, identification of interspecific hybrids as well as parental and backcross classes have become a relatively straightforward exercise. In particular, software based on Bayesian inference such as Structure and NewHybrids provides a powerful discriminatory tool (Excoffier & Heckel 2006; Vaha & Primmer 2006). In this theme issue, Anderson (2008) presents an extension of the frequently used software NewHybrids for the detection of hybrid classes. In addition, the application of various molecular approaches for the identification and analysis of hybridization is presented by Vigfúsdóttir et al. (2008).
(c) Prezygotic reproductive isolation
If interspecific hybridization is costly, then selection might favour the evolution of prezygotic isolating mechanisms (e.g. mating behaviours) that reduce heterospecific matings. This process will subsequently enhance reproductive isolation between species and result in reinforcement. If hybridization is less costly, or provides fitness advantages, prezygotic isolation might be weak and allow hybridization events. Even though these events might occur at a low frequency they are bound to have long-lasting consequences for the genetic architecture of populations due to gene flow among species. How behaviour of males and females determines the outcome of hybridization is illustrated in the studies of den Hartog et al. (2008), van der Sluijs et al. (2008) and Stelkens et al. (2008).
(d) Mating biology
Many studies on interspecific hybridization have focused on aspects of mating biology, since parental taxa and interspecific hybrids are often characterized by significant shifts in reproductive modes (i.e. from sexual to asexual) and ploidy levels (Schultz 1969). In this context, interspecific hybridization provides a natural setting to study the origin and genetic background of major changes in mating biology. A change in the mode of reproduction might facilitate (i) the geographical expansion of interspecific hybrids (Choleva et al. 2008), (ii) the origin of new lineages (Lampert & Schartel 2008), (iii) unexpected patterns in social insects (Feldhaar et al. 2008), or (iv) speciation (Cunha et al. 2008).
(e) Ecological differentiation and conservation genetics
One of the main topics in the field of interspecific hybridization is related to the question of whether the selection against hybrids is environmentally independent or dependent (i.e. reflecting endogenous or exogenous selection, respectively). The spatio-temporal distribution of plankton species and hybrids along environmental gradients suggests the action of exogenous selection (Petrusek et al. 2008), and the geographical distribution of species and hybrids indicating a role for man-made habitat disturbances, such as lake eutrophication (Keller et al. 2008). Anthropogenic disturbances, including the invasion of species or biomanipulation, not only have an impact on biodiversity and the ecology of systems but also on the evolutionary fate of organisms. The introgression between wild and feral domestic cats is an example of the effects that anthropomorphic habitat modifications may have (Oliveira et al. 2008).
5. Synopsis
Futuyma & Shapiro (1995) mentioned in their review of the 1993 book edited by Harrison that ‘Arbitrarily chosen molecular and morphological markers have provided abundant insight into hybrid zones, but they have not answered some of the most difficult and important questions: on what genes and characters does selection act, and what are the agents of selection? …most of the work lies ahead of us.’ Since the 1990s, we have witnessed a tremendous increase in the number of studies on interspecific hybridization, but only during the last few years has it been feasible to study simultaneously multiple functional genes and selectively neutral markers. Presently, we have the tools to examine instances of interspecific hybridization in unprecedented detail. In particular, the analyses of neutral markers and candidate genes, in combination with life-history experiments, have the potential to unravel central questions in evolutionary biology. In addition, a number of currently controversial issues in biology are related to the field of interspecific hybridization, which are as follows. (i) What are the consequences of hybridization among genetically modified organisms and wild congeners? (ii) What are the consequences of interbreeding between invasive and indigenous species? (iii) How will global climate changes contribute to species invasions and hybridization? (iv) What is the role of genetic exchange in the evolution of organisms? (v) To what extent does hybridization determine the fate of endangered species?
Although previous studies identified certain taxonomic groups that are known to have accelerated levels of interspecific hybridization, our literature survey suggests that hybridization represents a phenomenon distributed uniformly across the animal kingdom. As the historical overview, the reviews and the case studies presented in this issue demonstrate, interspecific hybridization remains a growing and exciting field in evolutionary biology.
Acknowledgments
We thank Eric Anderson, Michael Arnold, Rosemary Grant, Markus Pfenninger and Bob Vrijenhoek for their helpful comments on the earlier drafts of this manuscript. This project was partly supported by research grants (SCHW830/7) to K.S. and a PhD scholarship (Hessen State Ministry of Higher Education, Research and the Arts) to N.B.
We are also grateful to the authors, anonymous reviewers, Bob Vrijenhoek and Derek Taylor and supporters of the symposium on ‘Interspecific hybridization in animals—extent, processes and evolutionary impact’ held at the J.W. Goethe-University Frankfurt am Main, October 2006. In particular, we would like to thank Michael Arnold, Eric Anderson, Ole Seehausen, Rosemary Grant and Peter Grant for stimulating contributions and discussions. We are grateful to James Joseph (publishing editor of Phil. Trans. R. Soc. B) and DFG (4851/194/06) for their support.
Footnotes
One contribution of 16 to a Theme Issue ‘Hybridization in animals: extent, processes and evolutionary impact’.
References
- Alves P.C, Melo-Ferreira J, Freitas H, Boursot P. The ubiquitous mountain hare mitochondria: multiple introgressive hybridization in hares, genus Lepus. Phil. Trans. R. Soc. B. 2008;363:2831–2839. doi: 10.1098/rstb.2008.0053. doi:10.1098/rstb.2008.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E. Hybridization of the habitat. Evolution. 1948;2:1–9. doi:10.2307/2405610 [Google Scholar]
- Anderson E.C. Bayesian inference of species hybrids using multilocus dominant genetic markers. Phil. Trans. R. Soc. B. 2008;363:2841–2850. doi: 10.1098/rstb.2008.0043. doi:10.1098/rstb.2008.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E, Hubricht L. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am. J. Bot. 1938;25:396–402. doi:10.2307/2436413 [Google Scholar]
- Anderson E, Stebbins G.L. Hybridization as an evolutionary stimulus. Evolution. 1954;8:378–388. doi:10.2307/2405784 [Google Scholar]
- Arnold M.L. Natural hybridization as an evolutionary process. Annu. Rev. Ecol. Syst. 1992;23:237–261. doi:10.1146/annurev.es.23.110192.001321 [Google Scholar]
- Arnold M.L. Oxford series in ecology and evolution. Oxford University Press; New York, NY: 1997. Natural hybridization and evolution. [Google Scholar]
- Arnold M.L. Oxford University Press; Oxford, UK: 2006. Evolution through genetic exchange. [Google Scholar]
- Arnold M.L, Hodges S.A. Are natural hybrids fit or unfit relative to their parents? Trends Ecol. Evol. 1995;10:67–71. doi: 10.1016/S0169-5347(00)88979-X. doi:10.1016/S0169-5347(00)88979-X [DOI] [PubMed] [Google Scholar]
- Arnold M.L, Sapir Y, Martin N.H. Genetic exchange and the origin of adaptations: prokaryotes to primates. Phil. Trans. R. Soc. B. 2008;363:2813–2820. doi: 10.1098/rstb.2008.0021. doi:10.1098/rstb.2008.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie J.E.M, Hilton-Taylor C, Stuart S.N. The IUCN Species Survival Commision; Gland, Switzerland; Cambridge, UK: 2004. IUCN red list of threatened species. A global species assessment. [Google Scholar]
- Barton N.H, Hewitt G.M. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. doi:10.1146/annurev.es.16.110185.000553 [Google Scholar]
- Barton N.H, Hewitt G.M. Adaption, speciation and hybrid zones. Nature. 1989;341:497–503. doi: 10.1038/341497a0. doi:10.1038/341497a0 [DOI] [PubMed] [Google Scholar]
- Choleva L, Apostolou A, Ráb P, Janko K. Making it on their own: sperm-dependent hybrid fishes (Cobitis) switch the sexual hosts and expand beyond the ranges of their original sperm donors. Phil. Trans. R. Soc. B. 2008;363:2911–2919. doi: 10.1098/rstb.2008.0059. doi:10.1098/rstb.2008.0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C, Doadrio I, Coelho M.M. Speciation towards tetraploidization after intermediate processes of non-sexual reproduction. Phil. Trans. R. Soc. B. 2008;363:2921–2929. doi: 10.1098/rstb.2008.0048. doi:10.1098/rstb.2008.0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. John Murray; London, UK: 1859. The origin of species by means of natural selection or the preservation of favoured races in the struggle for life. [PMC free article] [PubMed] [Google Scholar]
- den Hartog P.M, Slabbekoorn H, ten Cate C. Male territorial vocalizations and responses are decoupled in an avian hybrid zone. Phil. Trans. R. Soc. B. 2008;363:2879–2889. doi: 10.1098/rstb.2008.0046. doi:10.1098/rstb.2008.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling T.E, Secor C.L. The role of hybridization and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 1997;28:593–619. doi:10.1146/annurev.ecolsys.28.1.593 [Google Scholar]
- Excoffier L, Heckel G. Computer programs for population genetics data analysis: a survival guide. Nat. Rev. Genet. 2006;7:745–758. doi: 10.1038/nrg1904. doi:10.1038/nrg1904 [DOI] [PubMed] [Google Scholar]
- Feldhaar H, Foitzik S, Heinze J. Lifelong commitment to the wrong partner: hybridization in ants. Phil. Trans. R. Soc. B. 2008;363:2891–2899. doi: 10.1098/rstb.2008.0022. doi:10.1098/rstb.2008.0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma, D. & Shapiro, L. 1995 Book review: hybrid zones. In Hybrid zones and the evolutionary process (ed. R. G. Harrison), Evolution49, 222–226.
- Gompert Z, Fordyce J.A, Forister M.L, Shapiro A.M, Nice C.C. Homoploid hybrid speciation in an extreme habitat. Science. 2006;314:1923–1925. doi: 10.1126/science.1135875. doi:10.1126/science.1135875 [DOI] [PubMed] [Google Scholar]
- Grant B.R, Grant P.R. Fission and fusion of Darwin's finches populations. Phil. Trans. R. Soc. B. 2008;363:2821–2829. doi: 10.1098/rstb.2008.0051. doi:10.1098/rstb.2008.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.R, Grant B.R. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. doi:10.1126/science.256.5054.193 [DOI] [PubMed] [Google Scholar]
- Harrison, R. G. 1990 Hybrid zones: windows on evolutionary process. In Oxford surveys in evolutionary biology, vol. 7 (eds D. J. Futuyma & J. Antonovics), pp. 69–128. Oxford, UK: Oxford University Press.
- Harrison R.G. Oxford University Press; New York, NY: 1993. Hybrid zones and the evolutionary process. [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. doi:10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G.M. Hybrid zones-natural laboratories for evolutionary studies. Trends Ecol. Evol. 1988;3:158–167. doi: 10.1016/0169-5347(88)90033-X. doi:10.1016/0169-5347(88)90033-X [DOI] [PubMed] [Google Scholar]
- Jiggins C.D, Mallet J. Bimodal hybrid zones and speciation. Trends Ecol. Evol. 2000;15:250–255. doi: 10.1016/s0169-5347(00)01873-5. doi:10.1016/S0169-5347(00)01873-5 [DOI] [PubMed] [Google Scholar]
- Keller B, Wolinska J, Manca M, Spaak P. Spatial, environmental and anthropogenic effects on the taxon composition of hybridizing Daphnia. Phil. Trans. R. Soc. B. 2008;363:2943–2952. doi: 10.1098/rstb.2008.0044. doi:10.1098/rstb.2008.0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert K.P, Schartl M. The origin and evolution of a unisexual hybrid: Poecilia formosa. Phil. Trans. R. Soc. B. 2008;363:2901–2909. doi: 10.1098/rstb.2008.0040. doi:10.1098/rstb.2008.0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R.C, Birch L.C. Hybridization as a source of variation for adaptation to new environments. Evolution. 1966;20:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. doi:10.2307/2406633 [DOI] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl Acad. Sci. USA. 2007;104:8597–8604. doi: 10.1073/pnas.0702207104. doi:10.1073/pnas.0702207104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. doi:10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Mayr E. Belknap Press of Harvard University Press; Cambridge, MA: 1963. Animal species and evolution. [Google Scholar]
- Moore W.S. An evaluation of narrow hybrid zones in vertebrates. Quart. Rev. Biol. 1977;52:263–277. doi:10.1086/409995 [Google Scholar]
- Oliveira R, Godinho R, Randi E, Alves P.C. Hybridization versus conservation: are domestic cats threatening the genetic integrity of wildcats (Felis silvestris silvestris) in Iberian Peninsula? Phil. Trans. R. Soc. B. 2008;363:2953–2961. doi: 10.1098/rstb.2008.0052. doi:10.1098/rstb.2008.0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrusek A, Seda J, Macháček J, Ruthová Š, Šmilauer P. Daphnia hybridization along ecological gradients in pelagic environments: the potential for the presence of hybrid zones in plankton. Phil. Trans. R. Soc. B. 2008;363:2931–2941. doi: 10.1098/rstb.2008.0026. doi:10.1098/rstb.2008.0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenninger M, Schwenk K. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol. Biol. 2007;7:121. doi: 10.1186/1471-2148-7-121. doi:10.1186/1471-2148-7-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhymer J.M, Simberloff D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 1996;27:83–109. doi:10.1146/annurev.ecolsys.27.1.83 [Google Scholar]
- Rieseberg L.H. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 1997;28:359–389. doi:10.1146/annurev.ecolsys.28.1.359 [Google Scholar]
- Rieseberg L.H, Soltis D.E. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Plants. 1991;5:65–84. [Google Scholar]
- Rieseberg L.H, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. doi:10.1126/science.1086949 [DOI] [PubMed] [Google Scholar]
- Rogers S.M, Bernatchez L. The genetic architecture of ecological speciation and the association with signatures of selection in natural lake whitefish (Coregonus sp. Salmonidae) species pairs. Mol. Biol. Evol. 2007;24:1423–1438. doi: 10.1093/molbev/msm066. doi:10.1093/molbev/msm066 [DOI] [PubMed] [Google Scholar]
- Schultz R.J. Hybridization, unisexuality and polyploidy in the teleost Poeciliopsis (Poeciliidae) and other vertebrates. Am. Nat. 1969;103:605–619. doi:10.1086/282629 [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. doi:10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Stelkens R.B, Pierotti M.E.R, Joyce D.A, Smith A.M, van der Sluijs I, Seehausen O. Disruptive sexual selection on male nuptial coloration in an experimental hybrid population of cichlid fish. Phil. Trans. R. Soc. B. 2008;363:2861–2870. doi: 10.1098/rstb.2008.0049. doi:10.1098/rstb.2008.0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaha J.P, Primmer C.R. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol. Ecol. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. doi:10.1111/j.1365-294X.2005.02773.x [DOI] [PubMed] [Google Scholar]
- van der Sluijs I, Van Dooren T.J.M, Hofker K.D, van Alphen J.J.M, Stelkens R.B, Seehausen O. Female mating preference functions predict sexual selection against hybrids between sibling species of cichlid fish. Phil. Trans. R. Soc. B. 2008;363:2871–2877. doi: 10.1098/rstb.2008.0045. doi:10.1098/rstb.2008.0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigfúsdóttir F, Pálsson S, Ingólfsson A. Hybridization of glaucous gull (Larus hyperboreus) and herring gull (Larus argentatus) in Iceland: mitochondrial and microsatellite data. Phil. Trans. R. Soc. B. 2008;363:2851–2860. doi: 10.1098/rstb.2008.0042. doi:10.1098/rstb.2008.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek R.C. Genetic and ecological constraints in the origins and establishment of unisexual vertebrates. In: Dowley R.M, Bogart J.B, editors. Evolution and ecology of unisexual vertebrates. New York State Museum; Albany, NY: 1989. pp. 24–31. [Google Scholar]