Abstract
BACKGROUND & AIMS
Irritable bowel syndrome (IBS) has been associated with mucosal dysfunction,, mild inflammation, and altered colonic bacteria. We used microarray expression profiling of sigmoid colon mucosa to assess whether there are stably expressed sets of genes that suggest there are objective molecular biomarkers associated with IBS.
METHODS
Gene expression profiling was performed using Affymetrix GeneChips with RNA from sigmoid colon mucosal biopsies from 36 IBS patients and 25 healthy control subjects. RTQ-PCR was used to confirm the data in 12 genes of interest. Statistical methods for microarray data were applied to search for differentially expressed genes, and to assess the stability of molecular signatures in IBS patients.
RESULTS
Mucosal gene expression profiles were consistent across different sites within the sigmoid colon and were stable on repeat biopsy over ~3 months. Differentially expressed genes suggest functional alterations of several components of the host mucosal immune response to microbial pathogens. The most strikingly increased expression involved a yet uncharacterized gene, DKFZP564O0823. Identified specific genes suggest the hypothesis that molecular signatures may enable distinction of a subset of IBS patients from healthy controls. Using 75% of the biopsies as a validation set to develop a gene profile, the test set (25%) was correctly predicted with ~70% accuracy.
CONCLUSIONS
Mucosal gene expression analysis shows there are relatively stable alterations in colonic mucosal immunity in IBS. These molecular alterations provide the basis to test the hypothesis that objective biomarkers may be identified in IBS and enhance understanding of the disease.
INTRODUCTION
Irritable bowel syndrome (IBS) is a highly prevalent disorder affecting 10–20% of people in Western countries. It is characterized by recurrent abdominal pain associated with change in stool frequency or consistency at the time of pain, as well as alterations in bowel function. In contrast to inflammatory bowel disease (IBD), overt histological inflammation or ulceration in the intestines are not observed in IBS. The same IBS phenotype may result from different pathophysiological mechanisms1 (e.g. motor, or secretory function, or a post-inflammatory state) in these patients, even when there is a consistent bowel dysfunction e.g. diarrhea- (IBS-D) or constipation-predominant (IBS-C).
Low-grade chronic inflammation is recognized in a subgroup of patients with IBS.2 Alterations in circulating cytokines,3,4 increased mucosal permeability2 and altered colonic bacterial counts5 in IBS subgroups suggest that altered mucosal immune function may contribute to the development of IBS. However, other studies do not confirm immune activation. For example, eosinophil protein X (EPX), myeloperoxidase (MPO), tryptase, interleukin 1β or tumor necrosis factor α measured in supernatants from processed feces of patients with IBS were not elevated, in contrast to positive (IBD) controls.6 Similarly, increased CD3 lymphocytes are observed in colonic biopsies.7 or mast cells are increased in colonic or ileal biopsies in some studies;8–10 however, other studies did not confirm these findings e.g., mast cell numbers are not increased in the colonic biopsies from patients with post-dysentery IBS.10 Moreover, increases in indices of inflammation may apply only to the subgroup of patients with post-infectious IBS. Increased rectal mucosal mRNA expression of IL-1β in post-infectious IBS patients suggests there is some evidence of an inflammatory diathesis.11 From the prior literature, therefore, the pathogenetic role of inflammation and the mechanisms involved are unclear.
Our hypothesis was that differentially-expressed genes in colonic mucosal biopsies may lead to the identification of stable sets of genes that are associated with IBS. Our aims were to further understand mucosal mechanisms at the molecular level that may be associated with IBS and to determine whether expression of these markers is stable and lends itself to identifying biomarkers of the disease that would require confirmation in future studies. We performed a microarray expression profiling study of mucosal sigmoid colon biopsies that were collected as in routine clinical practice from IBS patients and healthy controls. In a subset of genes of interest, the data were confirmed by RTQ-PCR.
METHODS
PARTICIPANTS AND COLLECTION OF COLON BIOPSY SAMPLES
Our study included 36 IBS patients (21 IBS-D and 15 IBS-C) and 25 healthy controls. IBS participants were recruited by mail from an administrative database of 752 patients with IBS who reside within 150 miles radius of Rochester, MN. All patients fulfilled the Rome II criteria for IBS diagnosis12 and had undergone clinical examination and investigation to exclude other gastrointestinal disorders. Predominant bowel dysfunction was confirmed at the time of the study by means of a validated bowel symptom questionnaire.13 Healthy volunteers were recruited by public advertisement in Rochester, MN. Table 1 further describes the study cohort. The Mayo Clinic Institutional Review Board approved the study, and all participants signed informed consent.
Table 1.
Description of the study cohort.
| IBS |
||||
|---|---|---|---|---|
| Healthy controls | IBS | IBS-C | IBS-D | |
| N | 25 | 36 | 15 | 21 |
| Caucasian (n, %) | 24 (96) | 36 (100) | 15 (100) | 21 (100) |
| Gender (n, % female) | 23 (92) | 33 (92) | 15 (100) | 15 (86) |
| Age (mean ± SEM) | 39 ± 2 | 42 ± 2 | 47 ± 3 | 39 ± 3 |
| (range) | (18 – 60) | (22 – 73) | (27 – 73) | (22 – 64) |
| BMI (mean ± SEM) | 26.1 ± 1.2 | 27.4 ± 1 | 25.5 ± 1.5 | 28.8 ± 1.3 |
| (range) | (18.3 – 40.2) | (20 – 42.6) | (20.0 – 42.6) | (20.9 – 41.8) |
| Concurrent treatment | ||||
| SSRI | n=2 | n=10 | n=2 | n=8 |
| SNRI | - | n=1 | n=1 | - |
| DA | - | n=3 | n=2 | n=1 |
| TCA | - | n=2 | n=1 | n=1 |
SSRI: selective serotonin reuptake inhibitor; SNRI: serotonin and norepinephrine reuptake inhibitor; DA: dopaminergic agent (bupropion); TCA: tricyclic antidepressant
Flexible sigmoidoscopy was performed without sedation after one magnesium sulphate enema (Fleet®, CB Fleet). Using standard, large size biopsy forceps, three sigmoid colon mucosal biopsies were collected from each participant: two (10 cm apart) for microarray studies, and one for formalin-fixation, hematoxylin and eosin (H&E)–staining and examination by a single expert histopathologist (TS) who used standardized criteria.14 A fourth sigmoid colon biopsy was collected ~3 months later from 10 randomly selected subjects (5 IBS patients and 5 healthy controls) to assess stability of the molecular observations‥
ARRAY PROCESSING AND DATA PRE-PROCESSING
Colon biopsy samples were submerged in RNAlater solution (Ambion, Austin, TX) and stored at −20°C until further analysed. Tissue was homogenized in a mixer mill 501 (Retsch, Aartselaar, Belgium) in RLT cell lysis buffer (Qiagen, Hilden, Germany), followed by RNA extraction (RNeasy mini kit, Qiagen) with DNase treatment. One µg biotin-labelled total RNA was hybridised on Human Genome U133-Plus2.0 GeneChips (Affymetrix, Santa Clara, CA) according to the Affymetrix protocol. Sample processing (n=132) was performed in four batches, each of which comprised samples from both IBS and healthy subjects. Gene expression summary values for raw GeneChip data were computed using the gcRMA algorithm,15 which performs background adjustment, quantile normalization and summarization, taking guanosine-cytidine affinities into account. We used PANP for determining whether the expression of a gene exceeded background,16 and declared filtered genes when they were called present in at least 50% of the samples in one group.17 Spectral map analysis18 clearly indicated that an effect of sample processing on different batches remained after normalization. Therefore, this source of technical variation was corrected for by modeling the expression levels in function of batch of origin in a one-way ANOVA, and by using the residuals of this model for all subsequent analyses (Supplementary Figure 1; see supplemental material online at www.cghjournal.org). To avoid the potential of obtaining misleading results due to pseudoreplication,19 we averaged the expression values of the replicated samples per patient for the SAM (significance analysis of microarrays) and PAM (predictive analysis of microarrays) analyses. Replicate samples were distributed across batches. The microarray data have been deposited in the ArrayExpress Data Warehouse (http://www.ebi.ac.uk/arrayexpress/) with accession number E-TABM-176. [The data are password-protected until acceptance for publication.]
REAL-TIME QUANTITATIVE POLYMERASE CHAIN REACTION (RTQ-PCR)
Using banked samples that were stored in RNA later, we conducted further studies using RTQ-PCR in order to validate results from microarray analyses suggesting differential expression. Briefly, cDNA synthesis was performed using 500 ng of total RNA using random hexamer primers and Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). RTQ-PCR was performed on an ABI Prism 7900 cycler (Applied Biosystems, Foster City, CA) using the qPCR Core kit (Eurogentec, Seraing, Belgium) and validated TaqMan gene expression assays (Applied Biosystems) for the following genes: CASP1 (Hs00354836_m1), DKFZP564O0823 (Hs00209876_m1), DUOX2 (Hs00204187_m1), FCGR2A (Hs01017702_g1), LYZ (Hs00426231_m1), M160 (Hs00264549_m1), MS4A4A (Hs00254780_m1), MUC20 (Hs00416321_m1), NCF1 (Hs00165362_m1), NCF4 (Hs00241129_m1), VSIG2 (Hs00204823_m1), VSIG4 (Hs00200695_m1), and the moderately expressed reference gene SART1 (Hs00193002_m1). Serial dilutions of cDNA were used to generate standard curves of threshold cycles versus the logarithms of concentration for SART1 (reference gene) and the genes of interest.
A randomly selected subset of samples from 15 healthy controls and 30 IBS patients (15 IBS-C, 15 IBS-D) was submitted to RTQ-PCR. This selection of samples for the RTQ-PCR was done so that the analyses for each gene could be performed in one run, avoiding any possible batch effect. For each of the subjects selected, we included the two collected colon mucosal samples and analyzed both in duplicate. Thus, for each subject, a total of four data points were generated. Fold changes between IBS / controls were calculated based on the average expression value per group (IBS vs controls). Significances between IBS and healthy controls for the RTQ-PCR analyses were based on t-test statistics. The average fold change in expression levels of the genes of interest between IBS patients and healthy controls was calculated and compared to the microarray results.
ASSESSING CONCORDANCE OF REPEATED MEASUREMENTS
To quantify sample reproducibility at two sites in the sigmoid at the first biopsy, and over 2 times in a subset, we calculated concordance correlation coefficients (CCC)20 for the 1,000 most variable gene probe sets in the dataset, as well as for the set of 32 gene probe sets from the PAM analysis.
TESTING FOR DIFFERENTIALLY EXPRESSED GENES
SAM analysis (http://www-stat.stanford.edu/~tibs/SAM/) was applied21 to identify differentially expressed genes in IBS versus health. An alternative, more rigorous statistical model was also applied to the raw data, (i.e. pre-processed data of all biopsy samples before batch correction), by application of mixed ANOVA with batch and disease status as fixed effect and patient as a random effect, and with false discovery rate correction.22
DIFFERENTIATION OF MOLECULAR SIGNATURES OF IBS AND CONTROLS FROM MICROARRAYS
For identifying disease status, we applied PAM analysis.23 We, therefore, randomly divided the 61 subjects into a “training” set and a “test” set. The training set (n=45) comprised 17 healthy subjects and 28 IBS patients (16 IBS-D, 12 IBS-C); the test set (n=16) comprised the remaining 8 healthy controls and 8 IBS patients (5 IBS-D, 3 IBS-C). The samples from the test set were kept independent from the model-building step to assess the model’s predictive power, and to check for possible over-fitting.
HIERARCHICAL CLUSTERING
To identify an underlying structure in the molecular signatures, we applied hierarchical clustering (Spotfire DecisionSite 8.2 software) on a set of 16 gene probes selected in both PAM and SAM analyses, using average linkage and correlation as measures of similarity. Genes with similar expression profiles across the subjects are grouped together (X-axis) and, similarly, subjects with a similar expression profile group together (Y-axis) in a hierarchical way.
RESULTS
PARTICIPANTS
Table 1 summarizes the information on participants in the study.
HISTOLOGICAL ASSESSMENT OF MUCOSAL BIOPSIES
H&E-stained sigmoid biopsies were normal in most healthy subjects and patients with IBSC and IBS-D. One healthy subject and 2 patients with IBS-D had focal acute colitis. Melanosis coli was observed in 3 patients with IBS-C and 1 patient with IBS-D. The thickness of the subepithelial collagen layer was at the upper limit of normal (i.e., 10 µm) in 1 healthy subject, 1 patient with IBS-C, and 2 patients with IBS-D. Differences among groups were not statistically significant.
STABILITY OF MRNA EXPRESSION IN COLON MUCOSA
The concordance correlation coefficient (CCC) for repeat samples of the same partcicipant using the 1,000 most variable gene probe sets on the microarray is shown in Figure 1A. The CCC between two simultaneously collected samples as well as between two samples collected from the same person with an interval of ~3 months significantly exceeded the overall concordance. The concordance values among repeat samples did not differ between IBS patients and healthy controls. Since the overall expression profiles of sigmoid colon biopsies were relatively stable for two site and two time sample collections, we averaged the gene probe expression levels of the two collected colon samples per patient for the subsequent analyses.
Figure 1.

Concordance correlation analysis of the expression profiles of sigmoid colon samples collected from 10 individuals. The degree of similarity (concordance correlation coefficient, CCC) of the samples is indicated by color codes. The analysis included three samples for each subject: samples A and B, taken at the same time, about 10 cm apart in the sigmoid colon, and sample C collected an average of 85 days later. Blue squares indicate the CCC for samples from one individual. A thick black line distinguishes IBS and healthy subjects. Panel A (left) represents the analysis on 1,000 gene probes with the largest variation in expression within the dataset. The within-subject CCC between simultaneously collected samples (A versus B: 0.70 ± 0.03) and between samples collected with an interval of ~3 months (A/B versus C: 0.41 ± 0.03) significantly exceeded the overall concordance (0.25 ± 0.12; Mann-Whitney U test with unequal variances: respectively W = 3510, P<0.001 and W = 6744, P<0.001). Panel B (right) shows the analysis on 32 gene probes identified in the prediction analysis for microarrays. The within-subject CCC between simultaneously collected samples (A versus B: 0.76 ± 0.05) and between samples collected with an interval of ~3 months (A/B versus C: 0.67 ± 0.04) significantly exceeded the overall concordance (−0.02 ± 0.02; Mann-Whitney U test with unequal variances: respectively W = 3938, P<0.001 and W = 7683, P<0.001).
DIFFERENTIALLY EXPRESSED GENES IN IBS
Using the SAM algorithm, and at a 5% false discovery rate, 25 gene probe sets were differentially expressed between IBS and healthy persons (Table 2). These probe sets represented 20 different genes: 4 were up-regulated and 16-down regulated in IBS patients compared to healthy controls. Using the normalized raw data, the mixed ANOVA model revealed a very similar list of genes with q-values comparable to those obtained with the SAM analysis (Table 2 and Supplementary Figure 2 (see supplemental material online at www.cghjournal.org)). The differential gene expression reflected mostly subtle changes in expression levels with only a few of the significant genes with > 1.5-fold difference in expression in IBS patients compared to healthy controls (Table 2). Plots of the relative expression levels in the IBS patients and healthy persons for the individual genes are shown in Figure 2. RTQ-PCR analysis of several of the identified genes largely confirmed the microarray results with reference to fold change levels of the individual genes (Figure 3), but the same level of statistical significance was not reached.
Table 2.
Differentially expressed genes in sigmoid colon mucosal biopsies from IBS patients compared to healthy controls.
| Affymetrix probe set | Gene symbol | q-value SAM | q-value ANOVA | Fold change | Gene annotation |
|---|---|---|---|---|---|
| HIGHER expression in IBS patients versus controls | |||||
| 225809_at | DKFZP564O0823 | 0.018 | 0.03 | 1.41 | DKFZP564O0823 (IBS1) |
| 204687_at | DKFZP564O0823 | 0.018 | 0.01 | 1.24 | DKFZP564O0823 (IBS1) |
| 226622_at | MUC20 | 0.018 | 0.05 | 1.52 | Mucin 20 |
| 229369_at | VSIG2 | 0.023 | 0.01 | 1.20 | V-set and immunoglobulin domain containing, 2 |
| 231941_s_at | MUC20 | 0.030 | 0.07 | 1.47 | Mucin 20 |
| 200884_at | CKB | 0.039 | 0.05 | 1.29 | Creatine kinase, brain |
| LOWER expression in IBS patients versus controls | |||||
| 223655_at | M160 | 0.018 | 0.05 | 0.69 | Scavenger receptor cysteine-rich type 1 protein M160 (CD163 antigen-like 1) |
| 204787_at | VSIG4 | 0.023 | 0.05 | 0.66 | V-set and immunoglobulin domain containing, 4 |
| 211368_s_at | CASP1 | 0.030 | 0.05 | 0.75 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) |
| 205147_x_at | NCF4 | 0.030 | 0.05 | 0.70 | Neutrophil cytosolic factor 4, 40kDa |
| 1555745_a_at | LYZ | 0.030 | 0.11 | 0.48 | Lysozyme |
| 205968_at | KCNS3 | 0.039 | 0.10 | 0.66 | Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| 211367_s_at | CASP1 | 0.039 | 0.06 | 0.75 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) |
| 201762_s_at | PSME2 | 0.045 | 0.01 | 0.84 | Proteasome activator subunit 2 (PA28 beta) |
| 211366_x_at | CASP1 | 0.049 | 0.06 | 0.76 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) |
| 219607_s_at | MS4A4A | 0.056 | 0.13 | 0.74 | Membrane-spanning 4-domains, subfamily A, member 4 |
| 220085_at | HELLS | 0.058 | 0.07 | 0.82 | Helicase, lymphoid-specific |
| 1552703_s_at | COP1 | 0.080 | 0.06 | 0.81 | Caspase 1 dominant-negative inhibitor pseudo-ICE |
| 203561_at | FCGR2A | 0.080 | 0.08 | 0.77 | Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| 204023_at | RFC4 | 0.084 | 0.06 | 0.83 | Replication factor C (activator 1) 4, 37kDa |
| 206011_at | CASP1 | 0.084 | 0.11 | 0.77 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) |
| 216237_s_at | MCM5 | 0.084 | 0.08 | 0.76 | MCM5 mini-chromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) |
| 225973_at | TAP2 | 0.084 | 0.16 | 0.71 | Transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| 219759_at | LRAP | 0.084 | 0.21 | 0.39 | Leukocyte-derived arginine aminopeptidase |
| 218585_s_at | DTL | 0.084 | 0.11 | 0.79 | Denticleless homolog (Drosophila) |
The shown data are limited to gene probe sets with q<0.1 in the SAM analysis. Fold change values indicate the ratio of the mean gene expression in IBS patients compared to healthy controls.
Figure 2.
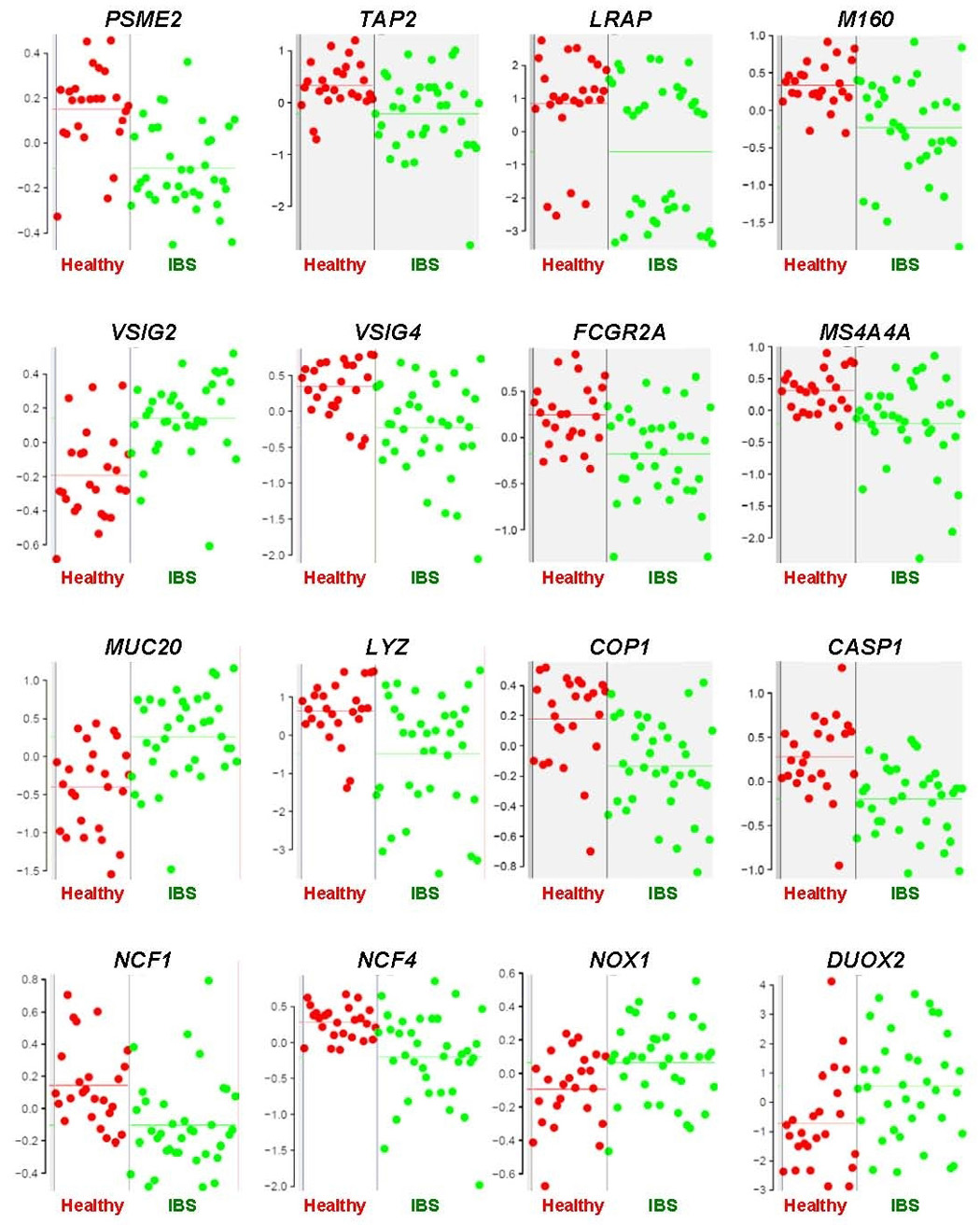
Relative expression levels of individual genes in mucosal colon samples from IBS patients versus healthy controls. Relative expression levels (y-axis) represent fluorescent signal intensity measured on the array after pre-processing of the raw data. Each individual dot represents the averaged expression value of two samples per subject (red: healthy, green: IBS). Horizontal lines indicate mean expression levels in healthy and IBS subjects.
Figure 3.
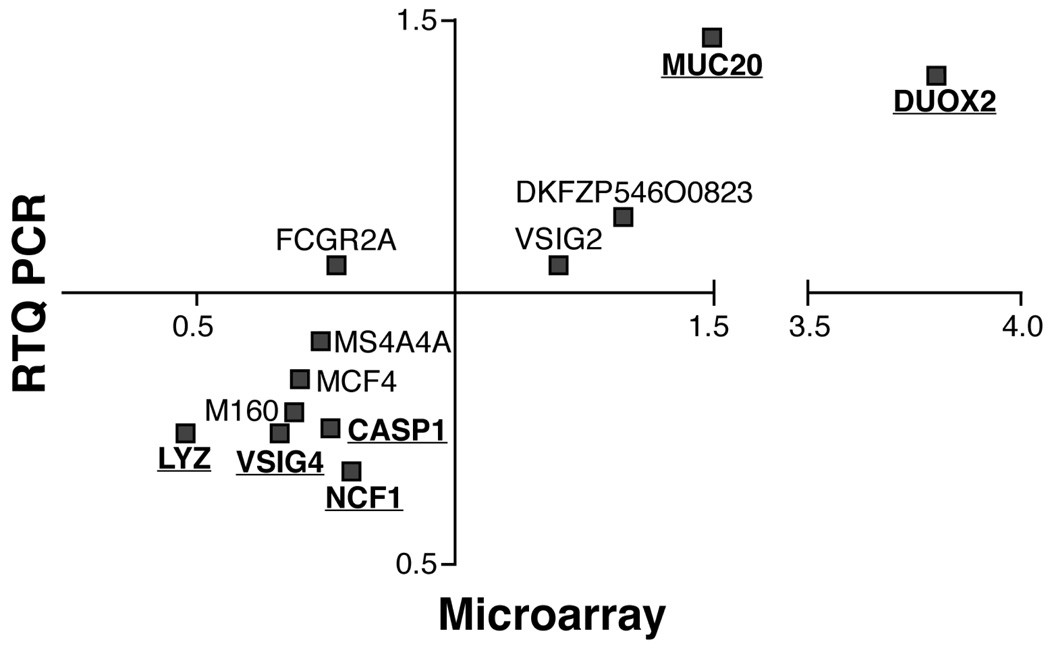
Comparison of fold changes in mRNA expression level, as measured by microarray and RTQ-PCR, between IBS patients and healthy subjects. Significant genes from the microarray study that were confirmed statistically significant (p<0.05) in RTQPCR analysis are underlined.
The majority of the genes identified in the colonic mucosa play a role in the immune response or the host defense against microbial invasion. A detailed description of the individual genes and their potential role in IBS is provided as Supplementary Discussion (see supplemental material online at www.cghjournal.org). Briefly, at least three genes with significantly lower expression levels in IBS patients play an essential role in the pathway of antigen processing and presentation by the major histocompatibility I complex (MHC-I). These genes are: PSME2 (proteasome activator subunit 2, PA28 beta), TAP2 (transporter 2, ATP-binding cassette, subfamily B), and LRAP (leukocyte-derived arginine aminopeptidase).
Six other significantly altered genes participate in the immune response. In IBS patients, there was a higher expression of VSIG2 (a V-set and immunoglobulin domain containing protein) and MUC20 (mucin 20). In contrast, IBS patients had lower expression of VSIG4, FCGR2A (CD32, encoding immunoglobulin Fc receptors), MS4A4A (encoding a homologue of the β subunit of immunoglobulin receptors), and M160 (CD163 molecule-like 1).
A third set of genes, all involved in the host defense response to pathogens in the colon, are expressed at significantly lower levels in IBS relative to healthy controls. These are: lysozyme (LYZ), an anti-microbial agent whose natural substrate is the bacterial cell wall peptidoglycan; and cysteine protease caspase-1 (CASP1) and its neighbouring gene on chromosome 11q, caspase-1 dominant negative inhibitor (COP1). These enzymes are involved in the proteolytic cleavage of precursor proteins leading to the synthesis of IL-1β and IL-18, both of which are important in antimicrobial defense.
Finally, the expression of multiple members of the family of NOX/DUOX oxidase genes that are responsible for the generation of an oxidative burst of superoxide as part of nonspecific host defense against microbial organisms, was either decreased (NCF1, NCF4) or increased (NOX1, DUOX2). The expression of dual oxidase 2 (DUOX2) in the sigmoid colon mucosal biopsies of IBS patients was increased on average 3.8-fold, the largest observed fold-change of all genes.
CONFIRMATION OF MICROARRAY ANALYSIS USING RTQ-PCR
We selected 12 genes that were identified from the microarray data analysis for confirmatory analysis by validated fluorogenic TaqMan gene expression assays-on-demand (Applied Biosystems). Normalisation of the TaqMan assay results was done relative to the control SART1 gene, because this gene was found earlier to be stable and is also moderately expressed in colon samples.24 Of the 12 genes of interest, 11 showed a change in gene expression that was in the same direction (up or down) in IBS patients (i.e., in Figure 3, they appear in the left-lower or the right-upper quadrants of the graph). Note that the level of fold change also appears to be consistent between the microarray and the RTQ-PCR as there is almost a linear relation between both analyses (apart from the DUOX2 gene that shows a clearly larger fold change in microarray analysis as compared to RTQ-PCR). Significant differences (p<0.05) between IBS and healthy subjects were confirmed in 6 out of the 12 genes, and these represented the genes with the largest fold change values. Overall, our data show substantial concordance between Affymetrix microarray and TaqMan data when comparing the fold change in expression level between IBS patients and healthy subjects (Figure 3).
UP REGULATION OF A NOVEL GENE IDENTIFIED IN IRRITABLE BOWEL SYNDROME
Two of the most significantly up regulated probe sets in colon mucosal biopsies of IBS patients (Table 2 and Figure 4A) both represent a gene that is annotated in the public sequence databases as DKFZP564O0823. Although little is known about this gene, in silico analysis demonstrates that this gene encodes a predicted plasma membrane protein of 310 amino acids (Figure 4B). The gene is mainly expressed in colon and placenta, but it is also found in many other tissues. Amino acid sequence alignment of the human, mouse, and rat homologs demonstrate a highly conserved (94–95%) sequence in the transmembrane and intracellular regions but less homology in the extracellular region (51% sequence identity).
Figure 4.
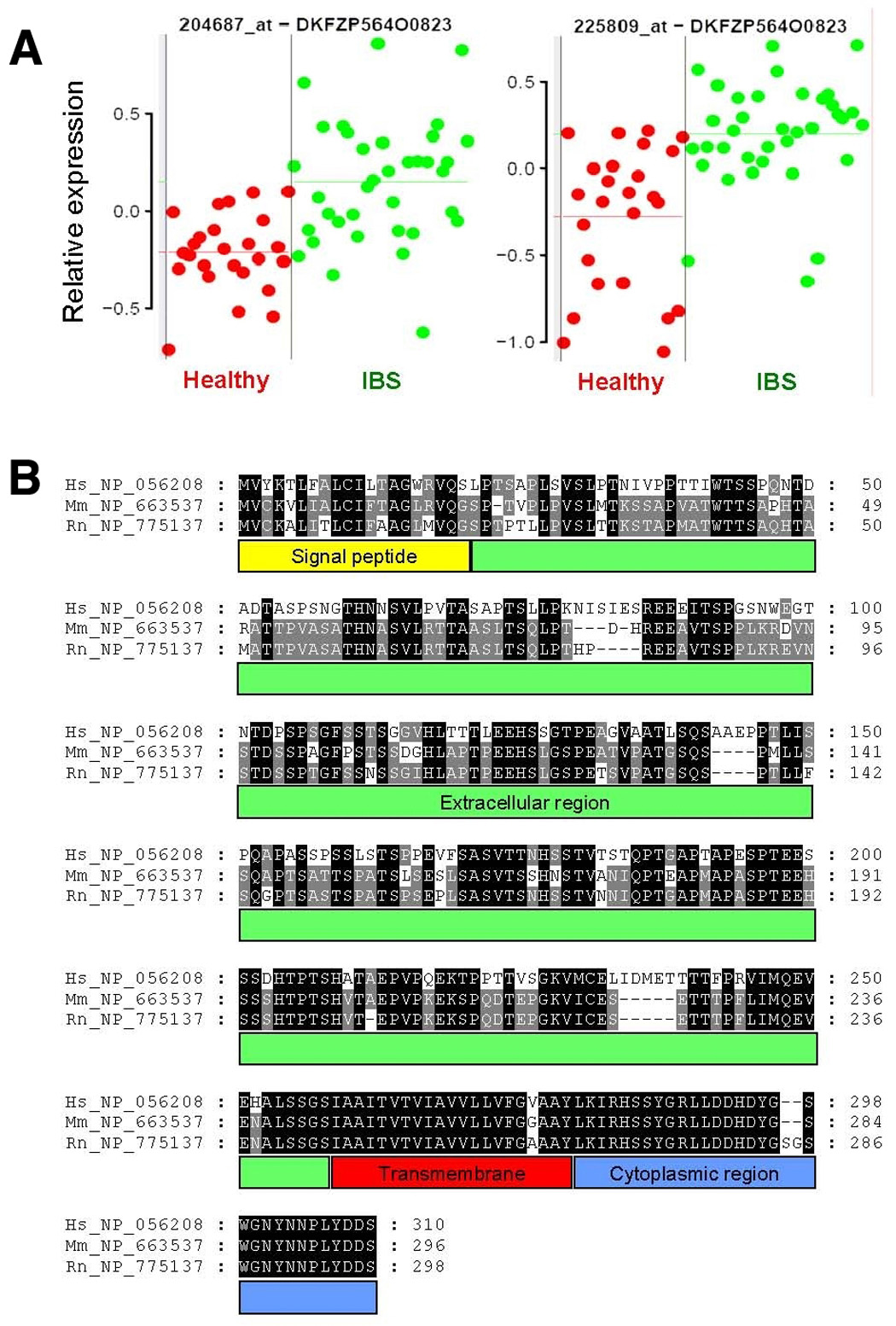
Gene expression of DKFZP564O0823 (IBS1) and comparative sequence analysis. (A) Gene expression of two probe sets on the GeneChip that encode for DKFZP564O0823 (IBS1) in mucosal biopsies from colon of healthy (red) and IBS (green) subjects. Each dot represents the average of two samples from one individual. Relative expression levels (y-axis) represent fluorescent signal intensity measured on the array after pre-processing of the raw data. Horizontal lines indicate mean expression levels in healthy and IBS subjects, respectively. (B) Comparative protein sequence analysis of human DKFZP564O0823 (Hs_NP_056208) and its mouse (Mm_NP_663537) and rat (Rn_NP_775137) homologs. Identical amino acids over different species are highlighted with a black or grey background. Protein domains are indicated below the sequence.
MUCOSAL EXPRESSION IN COLON AND A HYPOTHETICAL MOLECULAR SIGNATURE OF IBS
We assessed the ability of the colonic mucosal molecular expression panel to differentiate IBS from health. Using PAM analysis on a “training” set of 45 subjects (75% of the entire cohort, including both IBS and healthy subjects’ biopsies), we obtained a 32 gene probe set signature (Figure 5) with average cross-validation misclassification rate of 22%. Thus, 13/17 healthy and 22/28 IBS were correctly classified using this molecular signature. As a first step to explore the hypothesis that this molecular expression signature would be valid, the molecular signature was then applied to the independent “test” set of 16 participants (other 25% of participants who were not in the validation set). PAM analysis showed that the molecular signature correctly predicted diagnosis of 75% of the participants, with an equally accurate prediction of IBS or health status. The misclassification rates were similar for the “training” (22%) and the “test” sets (25%) of biopsies, suggesting that over-fitting was not an issue.
Figure 5.
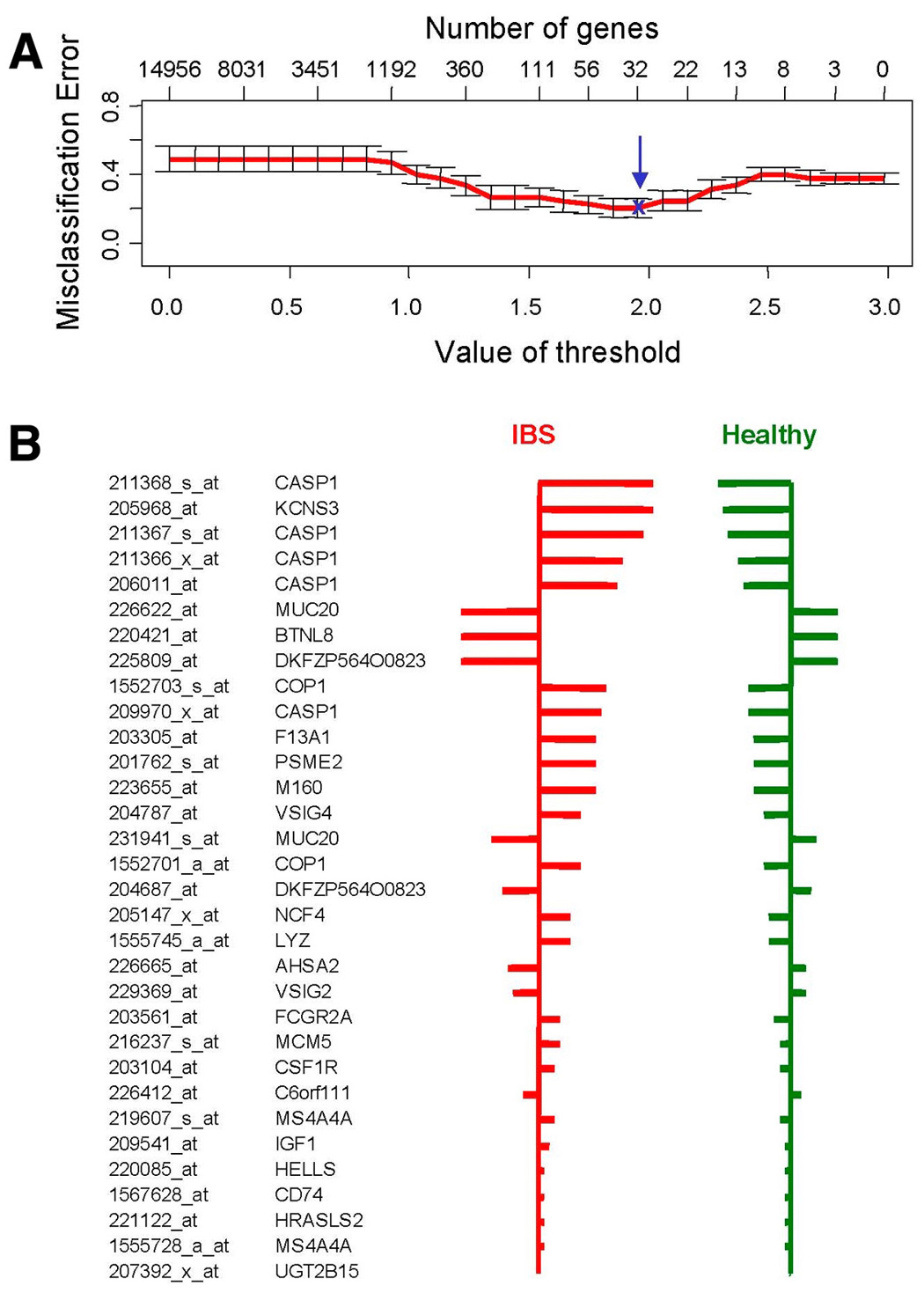
Predictive Analysis of Microarrays (PAM) used as a classification method to predict IBS disease status. First, the optimal number of genes to accurately predict IBS disease status was determined (upper panel). This was done by assessing the lowest misclassification error, using cross-validation on the samples of the training set. A set of 32 gene probe sets provided the best predictive power (blue arrow), corresponding with a threshold value (delta) of 2.0. The lower panel shows these 32 gene probe sets with their relative importance for the classification, indicated by the width of their respective bars.
As a second step to validate this signature of 32 gene probes, we determined its reproducibility by calculating the CCC (Figure 1B). The within subject reproducibility of samples significantly exceeded the overall concordance between participants suggesting that the molecular signature of 32 gene probe sets is a robust measure that is specific for the examined subject. The molecular signature was also stable over time.
Finally, in order to facilitate the generation of hypotheses for future studies that would aim at confirming the validity of such a molecular expression profile to differentiate IBS and health, we attempted to reduce the number of probe sets in the molecular signature. This was achieved through selection of gene probes that were identified in common by PAM and SAM analyses of the samples of the training set only. The resulting 16 probe sets, representing 11 different genes, were then used, in an unsupervised classification method (hierarchical clustering), that included all 61 subjects of the study (Figure 6). The first two levels of hierarchy (Y-axis) separated two groups of subjects, largely corresponding to the group of IBS patients and the healthy individuals of the training set. Thus, 25/28 IBS patients and 14/17 health controls were correctly classified. In the test set, 11 of 16 were correctly classified (69%), with positive and negative predictive values of 75% and 63%, respectively. These results were very similar to those of the PAM analysis using all 32 gene probe sets, suggesting that it would be reasonable to use the 16 probe set representing 11 genes rather than the 32 probe set in future studies attempting to validate the current findings.. Additional mixed ANOVA analyses ruled out possible interacting effects of gender or concomitant drug therapy on the findings of this study (see also Figure 6).
Figure 6.
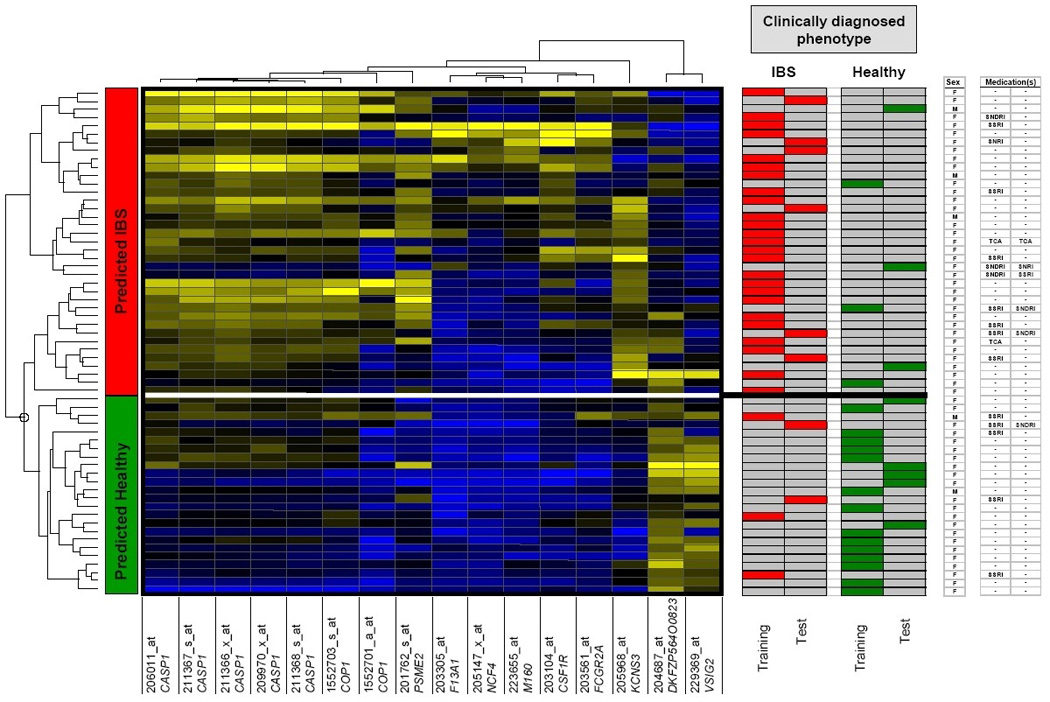
Hierarchical clustering analysis. Clustered display of heat map with hierarchical clustering of 16 probe sets and samples using average linkage and correlation as similarity measures. Heat map colors represent relative expression levels on a color gradient scale ranging from blue to black to yellow (high - intermediate - low expression). This color scale was maximized for each individual probe set over all the samples (i.e., the sample with the highest expression is blue; the sample with the lowest expression is yellow). The white horizontal line indicates IBS disease status as predicted by the molecular signature. The right panel of the figure shows the clinical diagnosis in the subjects assigned to the training or the test sets, the gender of the subjects and concomitant drug treatment. (M: male; F: female; SSRI: selective serotonin reuptake inhibitor; SNRI: serotoninnorepinephrine reuptake inhibitor; SNDRI: serotonin-norepinephrine-dopamine reuptake inhibitor; TCA: tricyclic antidepressant)
DISCUSSION
This study demonstrates the differential expression of several genes in the colonic mucosa of IBS patients. Many of these genes are directed towards the host defense mechanisms against microbiological pathogens; the well-established properties of the genes and their potential role in IBS are described in the Supplementary Discussion (see supplemental material online at www.gastrojournal.org).25–38 A pictorial summary of these genes is provided in Figure 7. Our study does not allow us to determine whether the observed differential expression is the cause or the consequence of IBS. The altered gene expression may have only been observed in a subset of IBS patients, and may reflect heterogeneity of the molecular mechanisms occurring with a common symptom phenotype. On the other hand, it is striking that most of the genes with significant differential expression are involved in the host response to intraluminal antigen or bacterial invasion or the resulting effects on immune responses. This degree of mechanistic specificity in the identified genes suggests that it is unlikely that the differential expression observed represents false positive or chance associations. Thus, the associations were demonstrated using two independent statistical approaches. Moreover, significant differences (p<0.05) between IBS and healthy subjects were confirmed by RTQ-PCR for 6 out of the 12 genes, and these represented the genes with the largest fold change values. Statistical significance was not achieved for genes with a more subtle fold change difference. This is not really surprising; although the TaqMan technology used in RTQ-PCR has some advantages with regard to sensitivity compared to microarrays (i.e., genes with low expression can be analyzed using RTQ-PCR where microarray technology may fail), it is clear that relatively larger intra-group variations are often found with TaqMan assays as compared to the microarray analyses. This can be explained, in part, by the fact that the TaqMan method only normalizes the gene expression data versus a single reference gene (SART1 in our assay), whereas the microarray method allows normalization of the expression level of each individual gene against all other genes on the microarray. This means that even relatively small gene expression variations of the SART1 gene will introduce noise (and variation) in the normalized expression level of the genes of interest using the RTQ-PCR technology. In general, the data generated by TaqMan assays largely confirm the microarray data with regard to the fold change levels of the individual genes, but the same level of statistical significance was not reached using RTQ-PCR, most likely because the TaqMan technology does not allow discrimination of subtle differences in gene expression.
Figure 7.
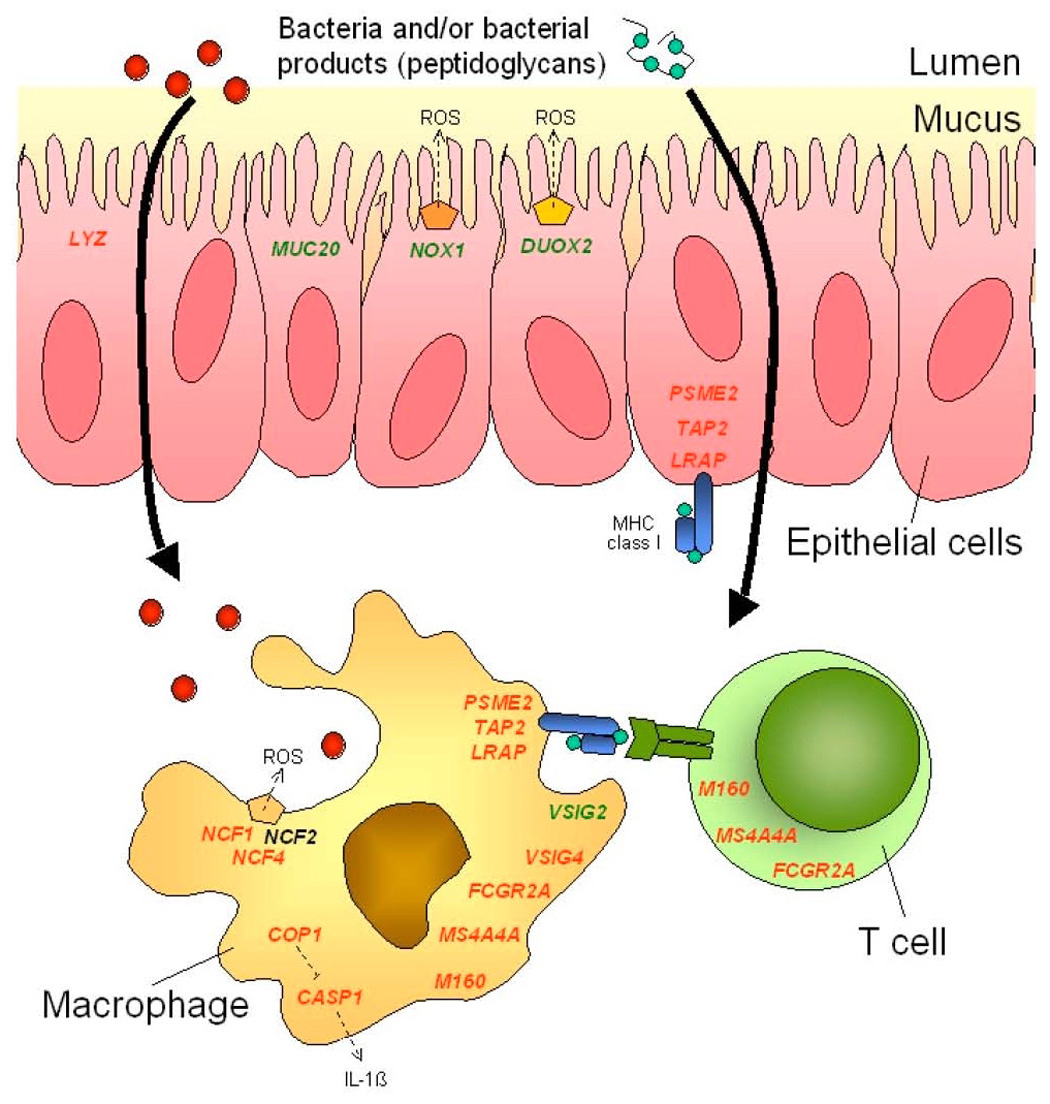
Pictorial summary of differentially expressed genes in colonic mucosa of IBS patients relative to healthy controls. Genes are color-coded according to increased (green) or decreased (red) expression in cellular elements of the colonic mucosa from IBS patients versus healthy controls. Protein complexes responsible for oxidative burst are shown as pentagon shapes. ROS: reactive oxygen species. NCF2 expression was unchanged and is shown in black.
The magnitude of the fold increase in expression of the different genes in the mucosa of patients with IBS ranges from 1.2 to 1.52; this is consistent with the subtle degree of immune activation measured cytologically or with functional studies e.g. IL-1β expression.11 We would have found the data far less believable if the fold increases were greater given observations in the literature, using complementary techniques to evaluate inflammation or immune activation. It is also intriguing that a gene involved in mucin production (e.g., MUC20) is up-regulated, and this may correspond to the frequently encountered observation of excessive mucus passage in patients with IBS. The genes demonstrating lower expression in IBS, which range from 0.48 to 0.83, also reflect the fact that IBS is not associated with clinically overt evidence of defective barrier function or immune response to enteric antigens. Thus, given the importance of these mechanisms to control bacterial and toxin invasion, the magnitude of change appears to reflect the observation that IBS patients are not more vulnerable to superinfection or to significant inflammation or immune activation that might result from an unchecked microbial interaction with the local immune system.
It is intriguing that the measured differences in gene expression in sigmoid colon mucosal biopsies are stable over time. The genes that are differentially expressed may control mechanisms involved in colonic mucosal defense of patients with IBS. This observation, as well as the fact that the fold differences are observed relative to healthy controls who received the same bowel preparation, suggest that the observations are not the result of artifact, such as the bowel preparation, or problems with the assays. Rather, they are consistent with the hypothesis that they may represent biological changes in IBS. These data complement the literature2–11 documenting a component of immune activation in the mucosa of IBS patients. Inflammation may be associated with increased intestinal permeability,2 which may result in colonic secretion. In addition to disturbances of visceral perception or motility in IBS,1 there is evidence of altered colonic mucosal immune function documented in peripheral blood and colonic mucosa.39 Thus, IBS patients displayed an increased frequency of peripheral blood CD4+ and CD8+ T cells expressing the gut homing integrin β7, increased lamina propria CD8+ T cells in ascending colon biopsies, and increased expression of the ligand for integrin β7, mucosal addressin cell adhesion molecule-1+, on endothelial cells of ascending colon biopsies as compared with control subjects. These abnormalities were also observed in biopsies from patients with ulcerative colitis.39
Whereas, earlier studies tested the hypothesis that local colonic immune activation occurs in IBS patients and this was demonstrated by studies of known biomarkers, our study was based on an unsupervised analysis of the expression data of thousands of gene probes present on the microarray. Our study identified the differential expression of molecules associated with mucosal immune mechanisms that had not previously been identified among genes involved in the local immune function. Further studies are required to understand these alterations in mucosal expression of genes associated with mucosal immune function. However, the study leads to testable hypotheses of potential mechanisms involved in immune function in IBS.
The microarray analysis showed the IBS patients’ mucosa expressed a gene, DKFZP564O0823; it is important to note that the RTQ-PCR also showed a small-fold increase in expression that was not significant. The function of this gene is not completely understood. The DKFZP564O0823 mouse homolog, known as RIKEN cDNA 9130213B05, expresses a cell surface glycoprotein precursor. Two publications have reported on the involvement of the rat homolog of this gene in resistance to apoptosis in rat prostate which led to the name, rat gene prostatic androgen-repressed message-1 (Parm-1).40,41 Although no reports on the human DKFZP564O0823 gene have been published, Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) contains experimental microarray data showing its altered expression, and its potential role in inflammation and immune responses. More specifically, DKFZP564O0823 expression was increased in primary colon endothelial cells on treatment with TNFα, as compared to exposure to interferon-gamma or interleukin-4. The latter two are primarily associated with T helper cell subsets, whereas TNFα is a pleiotropic cytokine with a critical function in both inflammatory and immunological responses. Because these data on colon endothelial cells form part of a large study, the publication on these experiments discussed only well-known genes but did not discuss DKFZP564O0823.42
In another study, DKFZP564O0823 gene expression was assessed in Jurkat CD4+ T cells following induction of the Nef protein from the simian immune deficiency virus (SIV).43 The Nef protein is expressed early in SIV (and HIV) infections, and down-regulates MHC-I molecules from the cell surface, thereby facilitating immune evasion. The microarray experiment revealed that, among many other well-characterized genes, DKFZP564O0823 expression is upregulated by SIV-Nef. Thus, if this novel gene functions in IBS as it is proposed to function in these two studies in the literature, it would also be consistent with up-regulation of immune response mechanisms in the colon of IBS patients compared to controls.
We conducted further analyses of the molecular signatures of gene expression in the colon mucosa to provide preliminary data that form the basis for hypotheses generation. The present IBS patient cohort shows 75% specificity of the molecular signature based on a set of 32 gene probes, which shows stability of differential expression on repeat testing and a consistent message in the differential expression of the host responses to intraluminal antigen or bacterial invasion or the resulting effects on immune functions. It is clear that the hypothesis requires further testing with larger numbers of subjects, including positive controls suffering other gastrointestinal diseases such as IBD or diarrhea due to small bowel diseases.
The weaknesses and limitations in our study are the relatively small sample size and the absence of a positive control. Moreover, despite all our efforts to avoid false discovery results and over-fitting models, the precautions taken cannot guarantee the validity of our pilot study. Indeed, previous reports on molecular signatures from microarray data have shown the pitfalls and problems with interpretation and urged authors to validate such results by several - preferably completely independent - teams.44 Nevertheless, an important dimension of our study is the confirmation, using a complementary technique, RTQ-PCR, of the main findings on 6 of the 12 genes of interest identified on microarray. Although the level of significance achieved with RTQ-PCR is less impressive, we believe that this difference most likely reflects the lower sensitivity of RTQ-PCR for subtle differences in gene expression.
In summary, the current data on molecular signatures, which have been shown to be stable over three months, lead to hypotheses that there are biomarkers suggesting immune activation or other mechanisms associated with the interaction between the human (host) and colonic content. These hypotheses are testable in future studies. Our pilot study provides the basis for selection of gene probe sets for those studies and for calculating the requisite sample sizes based on the fold differences in the expression of genes of interest.
Supplementary Material
Acknowledgements
Dr M Camilleri is supported in part by grants DK 54681 and 02638 (studies in irritable bowel syndrome) and by RR024150 (Mayo Clinic CTSA) from the National Institutes of Health. This work was supported by a research grant of Johnson & Johnson Pharmaceutical Research & Development. The authors thank Debra TePoel, R.N., study coordinator, and Steven Osselaer for excellent information technology support.
Grant support information:
This work was supported by a research grant of Johnson & Johnson Pharmaceutical Research & Development.
Non-Standard Abbreviations used
- ANOVA
analysis of variance
- CCC
concordance correlation coefficient
- H&E
hematoxylin and eosin
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- PAM
predictive analysis of microarrays
- RTQ-PCR
real-time quantitative polymerase chain reaction
- SAM
significance analysis of microarrays
- SIV
simian immune deficiency virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest disclosure: A provisional patent application has been filed by the authors’ employers, Mayo Clinic and Janssen Pharmaceutica n.v., for the use of the discovered molecular signatures to diagnose IBS. The following authors were employees of Johnson & Johnson Pharmaceutical Research & Development, a division of Janssen Pharmaceutica n.v. at the time when the study was performed and the manuscript drafted: Jeroen Aerssens, Willem Talloen, Leen Thielemans, Hinrich W. H. Göhlmann, Ilse Van den Wyngaert, Theo Thielemans, and Bernard Coulie. All other authors declare no competing financial interests.
NOTE: This is not a clinical trial and therefore details consistent with CONSORT guidelines are not provided. However, the authors are happy to provide other information that might be requested.
REFERENCES
- 1.Camilleri M. Mechanisms in IBS: something old, something new, something borrowed. Neurogastroenterol Motil. 2005;17:311–316. doi: 10.1111/j.1365-2982.2004.00632.x. [DOI] [PubMed] [Google Scholar]
- 2.Spiller RC. Infection, immune function, and functional gut disorders. Clin Gastroenterol Hepatol. 2004;2:445–455. doi: 10.1016/s1542-3565(04)00159-4. [DOI] [PubMed] [Google Scholar]
- 3.Dinan TG, Quigley EM, Ahmed SM, Scully P, O'Brien S, O'Mahony L, O'Mahony S, Shanahan F, Keeling PW. Hypothalamic-pituitary-gut axis dysregulation in irritable bowel syndrome: plasma cytokines as a potential biomarker? Gastroenterology. 2006;130:304–311. doi: 10.1053/j.gastro.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 4.O'Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O'Sullivan GC, Kiely B, Collins JK, Shanahan F, Quigley EM. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 5.Malinen E, Rinttila T, Kajander K, Matto J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 6.Lettesjo H, Hansson T, Peterson C, Ung KA, Ringstrom G, Abrahamsson H, Simren M. Detection of inflammatory markers in stools from patients with irritable bowel syndrome and collagenous colitis. Scand J Gastroenterol. 2006;41:54–59. doi: 10.1080/00365520510023909. [DOI] [PubMed] [Google Scholar]
- 7.Chadwick VS, Chen W, Shu D, Paulus B, Bethwaite P, Tie A, Wilson I. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology. 2002;122:1778–1783. doi: 10.1053/gast.2002.33579. [DOI] [PubMed] [Google Scholar]
- 8.Weston AP, Biddle WL, Bhatia PS, Miner PB., Jr Terminal ileal mucosal mast cells in irritable bowel syndrome. Dig Dis Sci. 1993;38:1590–1595. doi: 10.1007/BF01303164. [DOI] [PubMed] [Google Scholar]
- 9.Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, Pasquinelli G, Morselli-Labate AM, Grady EF, Bunnett NW, Collins SM, Corinaldesi R. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 10.Wang LH, Fang XC, Pan GZ. Bacillary dysentery as a causative factor of irritable bowel syndrome and its pathogenesis. Gut. 2004;53:1096–1101. doi: 10.1136/gut.2003.021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gwee KA, Collins SM, Read NW, Rajnakova A, Deng Y, Graham JC, McKendrick MW, Moochhala SM. Increased rectal mucosal expression of interleukin 1beta in recently acquired post-infectious irritable bowel syndrome. Gut. 2003;52:523–526. doi: 10.1136/gut.52.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson WG, Longstreth GF, Drossman DA, Heaton KW, Irvine EJ, Muller-Lissner SA. Functional bowel disorders and functional abdominal pain. Gut. 1999;45 Suppl.2:II43–II47. doi: 10.1136/gut.45.2008.ii43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talley NJ, Phillips SF, Wiltgen CM, Zinsmeister AR, Melton LJ., 3rd Assessment of functional gastrointestinal disease: the bowel disease questionnaire. Mayo Clin Proc. 1990;65:1456–1479. doi: 10.1016/s0025-6196(12)62169-7. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins D, Balsitis M, Gallivan S, Dixon MF, Gilmour HM, Shepherd NA, Theodossi A, Williams GT. Guidelines for the initial biopsy diagnosis of suspected chronic idiopathic inflammatory bowel disease. The British Society of Gastroenterology Initiative. J Clin Pathol. 1997;50:93–105. doi: 10.1136/jcp.50.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. Dept. of Biostatistics Working Papers, Working Paper 1, Johns Hopkins University; 2004. [Google Scholar]
- 16.Warren P, Bienkowska J, Martini P, Jackson J, Taylor D. PANP - a New Method of Gene Detection on Oligonucleotide Expression Arrays. 2006 Submitted to RECOMB, Available from: http://people.brandeis.edu/~dtaylor/PANP/
- 17.McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wouters L, Göhlmann HW, Bijnens L, Kass SU, Molenberghs G, Lewi PJ. Graphical exploration of gene expression data: a comparative study of three multivariate methods. Biometrics. 2003;59:1131–1139. doi: 10.1111/j.0006-341x.2003.00130.x. [DOI] [PubMed] [Google Scholar]
- 19.Hurlbert SH. Pseudoreplication and the design of ecological field studies. Ecological Monographs. 1984;54:187–211. [Google Scholar]
- 20.Lin LIK. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 21.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camilleri M, Andrews CN, Bharucha AE, Carlson PJ, Ferber I, Stephens D, Smyrk TC, Urrutia R, Aerssens J, Thielemans L, Goehlmann H, Van Den Wyngaert I, Coulie B. Alterations in expression of p11 and SERT in mucosal biopsy specimens of patients with irritable bowel syndrome. Gastroenterology. 2007;132:17–25. doi: 10.1053/j.gastro.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preckel T, Fung-Leung WP, Cai Z, Vitiello A, Salter-Cid L, Winqvist O, Wolfe TG, Von Herrath M, Angulo A, Ghazal P, Lee JD, Fourie AM, Wu Y, Pang J, Ngo K, Peterson PA, Fruh K, Yang Y. Impaired immunoproteasome assembly and immune responses in PA28-/- mice. Science. 1999;286:2162–2165. doi: 10.1126/science.286.5447.2162. [DOI] [PubMed] [Google Scholar]
- 26.Keusekotten K, Leonhardt RM, Ehses S, Knittler MR. Biogenesis of functional antigenic peptide-transporter TAP requires assembly of pre-existing TAP1 with newly synthesized TAP2. J Biol Chem. 2006;281:17545–17551. doi: 10.1074/jbc.M602360200. [DOI] [PubMed] [Google Scholar]
- 27.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 28.Kim JK, Choi EM, Shin HI, Kim CH, Hwang SH, Kim SM, Kwon BS. Characterization of monoclonal antibody specific to the Z39Ig protein, a member of immunoglobulin superfamily. Immunol Lett. 2005;15:153–161. doi: 10.1016/j.imlet.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Unkeless JC. Function and heterogeneity of human Fc receptors for immunoglobulin G. J Clin Invest. 1989;83:355–361. doi: 10.1172/JCI113891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higuchi T, Orita T, Nakanishi S, Katsuya K, Watanabe H, Yamasaki Y, Waga I, Nanayama T, Yamamoto Y, Munger W, Sun HW, Falk RJ, Jennette JC, Alcorta DA, Li H, Yamamoto T, Saito Y, Nakamura M. Molecular cloning, genomic structure, and expression analysis of MUC20, a novel mucin protein, up-regulated in injured kidney. J Biol Chem. 2004;279:1968–1979. doi: 10.1074/jbc.M304558200. [DOI] [PubMed] [Google Scholar]
- 31.Druilhe A, Srinivasula SM, Razmara M, Ahmad M, Alnemri ES. Regulation of IL-1beta generation by Pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 2001;8:649–657. doi: 10.1038/sj.cdd.4400881. [DOI] [PubMed] [Google Scholar]
- 32.Kikuchi H, Hikage M, Miyashita H, Fukumoto M. NADPH oxidase subunit, gp91-phox homologue, preferentially expressed in human colon epithelial cells. Gene. 2000;254:237–243. doi: 10.1016/s0378-1119(00)00258-4. [DOI] [PubMed] [Google Scholar]
- 33.Kuwano Y, Kawahara T, Yamamoto H, Teshima-Kondo S, Tominaga K, Masuda K, Kishi K, Morita K, Rokutan K. Interferon-gamma activates transcription of NADPH oxidase 1 gene and upregulates production of superoxide anion by human large intestinal epithelial cells. Am J Physiol Cell Physiol. 2006;290:C433–C443. doi: 10.1152/ajpcell.00135.2005. [DOI] [PubMed] [Google Scholar]
- 34.Kawahara T, Kuwano Y, Teshima-Kondo S, Takeya R, Sumimoto H, Kishi K, Tsunawaki S, Hirayama T, Rokutan K. Role of nicotinamide adenine dinucleotide phosphate oxidase 1 in oxidative burst response to Toll-like receptor 5 signaling in large intestinal epithelial cells. J Immunol. 2004;172:3051–3058. doi: 10.4049/jimmunol.172.5.3051. [DOI] [PubMed] [Google Scholar]
- 35.El Hassani RA, Benfares N, Caillou B, Talbot M, Sabourin JC, Belotte V, Morand S, Gnidehou S, Agnandji D, Ohayon R, Kaniewski J, Noel-Hudson MS, Bidart JM, Schlumberger M, Virion A, Dupuy C. Dual oxidase2 is expressed all along the digestive tract. Am J Physiol Gastrointest Liver Physiol. 2005;288:G933–G942. doi: 10.1152/ajpgi.00198.2004. [DOI] [PubMed] [Google Scholar]
- 36.Harper RW, Xu C, Eiserich JP, Chen Y, Kao CY, Thai P, Setiadi H, Wu R. Differential regulation of dual NADPH oxidases/peroxidases, Duox1 and Duox2, by Th1 and Th2 cytokines in respiratory tract epithelium. FEBS Lett. 2005;579:4911–4917. doi: 10.1016/j.febslet.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL. Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J. 2003;17:1502–1504. doi: 10.1096/fj.02-1104fje. [DOI] [PubMed] [Google Scholar]
- 38.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 39.Ohman L, Isaksson S, Lundgren A, Simren M, Sjovall H. A controlled study of colonic immune activity and beta7+ blood T lymphocytes in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3:980–986. doi: 10.1016/s1542-3565(05)00410-6. [DOI] [PubMed] [Google Scholar]
- 40.Bruyninx M, Hennuy B, Cornet A, Houssa P, Daukandt M, Reiter E, Poncin J, Closset J, Hennen G. A novel gene overexpressed in the prostate of castrated rats: hormonal regulation, relationship to apoptosis and to acquired prostatic cell androgen independence. Endocrinology. 1999;140:4789–4799. doi: 10.1210/endo.140.10.7097. [DOI] [PubMed] [Google Scholar]
- 41.Cornet AM, Hanon E, Reiter ER, Bruyninx M, Nguyen VH, Hennuy BR, Hennen GP, Closset JL. Prostatic androgen repressed message-1 (PARM-1) may play a role in prostatic cell immortalisation. Prostate. 2003;56:220–230. doi: 10.1002/pros.10254. [DOI] [PubMed] [Google Scholar]
- 42.Sana TR, Janatpour MJ, Sathe M, McEvoy LM, McClanahan TK. Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine. 2005;29:256–269. doi: 10.1016/j.cyto.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Ndolo T, George M, Nguyen H, Dandekar S. Expression of simian immunodeficiency virus Nef protein in CD4+ T cells leads to a molecular profile of viral persistence and immune evasion. Virology. 2006;353:374–387. doi: 10.1016/j.virol.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Ioannidis JP. Microarrays and molecular research: noise discovery? Lancet. 2005;365:454–455. doi: 10.1016/S0140-6736(05)17878-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


