Abstract
Symbioses are widespread in nature and occur along a continuum from parasitism to mutualism. Coral–dinoflagellate symbioses are defined as mutualistic because both partners receive benefit from the association via the exchange of nutrients. This successful interaction underpins the growth and formation of coral reefs. The symbiotic dinoflagellate genus Symbiodinium is genetically diverse containing eight divergent lineages (clades A–H). Corals predominantly associate with clade C Symbiodinium and to a lesser extent with clades A, B, D, F, and G. Variation in the function and interactive physiology of different coral–dinoflagellate assemblages is virtually unexplored but is an important consideration when developing the contextual framework of factors that contribute to coral reef resilience. In this study, we present evidence that clade A Symbiodinium are functionally less beneficial to corals than the dominant clade C Symbiodinium and may represent parasitic rather than mutualistic symbionts. Our hypothesis is supported by (i) a significant correlation between the presence of Symbiodinium clade A and health-compromised coral; (ii) a phylogeny and genetic diversity within Symbiodinium that suggests a different evolutionary trajectory for clade A compared with the other dominant Symbiodinium lineages; and (iii) a significantly lower amount of carbon fixed and released by clade A in the presence of a coral synthetic host factor as compared with the dominant coral symbiont lineage, clade C. Collectively, these data suggest that along the symbiotic continuum the interaction between clade A Symbiodinium and corals may be closer to parasitism than mutualism.
Keywords: cnidaria, Symbiodinium, disease, mutualistic, parasitic
The term symbiosis is inclusive of a variety of interactions that occur along a continuum that includes mutualistic, commensal, and parasitic associations (1). In commensal and mutualistic symbioses one or both partners benefit from the association, respectively, which contrasts with parasitic interactions, where one partner benefits whereas the other is harmed by the relationship (1, 2). The most well known examples of mutualistic symbioses in marine ecosystems are those between corals and photosynthetic dinoflagellates belonging to the genus Symbiodinium. Here, inorganic waste metabolites from the animal host are exchanged for organic nutrients fixed by dinoflagellate photosynthesis, primarily in the form of glycerol, that serve as respiratory substrates in the animal host that support the growth and formation of coral reefs (3, 4). Other common mutualisms include interactions between plants with mycorrihizal fungi and nitrogen-fixing bacteria (5, 6).
The dinoflagellate genus Symbiodinium is divided into eight divergent lineages, referred to as clades A–H, with each containing many subclade types based on rDNA (7–13). The diversity within the genus can be appreciated by the large number of taxa that form symbiotic interactions with Symbiodinium, including marine invertebrates from four phyla (Cnidaria: corals, jellyfish, anemones, zoanthids; Mollusca: snails and clams; Platyhelminthes: flatworms; Porifera: sponges) and the single-celled protist Foraminifera (14). Furthermore, an early phylogenetic comparison of partial 18S rDNA revealed that within the genus Symbiodinium the genetic diversity is equivalent to order-level taxonomic differences seen in other dinoflagellate groups (15).
The genetic diversity within Symbiodinium is likely to correlate with an equally diverse range of physiological properties in the host–symbiont assemblages. For example, juvenile coral hosts harboring clade C Symbiodinium have been shown to grow two to three times faster than juveniles harboring clade D (16). Clade D, however, has been shown to have a higher tolerance to thermal stress than clade C, suggesting corals harboring clade D are more resilient to coral bleaching events (17). These data point to differences in the function of these symbiotic interactions that reflect not only the type of Symbiodinium present but also the environment and developmental stage of the coral.
Understanding the functional diversity and physiological thresholds of coral–dinoflagellate symbioses is critical to predicting the fate of corals under the threat of global climate change and the increasing incidence of coral bleaching events and disease outbreaks (18, 19). In this study, we challenge the assumption that clade A and C Symbiodinium form equally mutualistic symbioses with corals by (i) evaluating the health state of corals that harbor clade A and C Symbiodinium, (ii) evaluating the phylogeny and diversity of Symbiodinium relative to free-living dinoflagellate groups, and (iii) investigating in vitro carbon fixation and release by Symbiodinium clades A and C.
Results
Symbiodinium Genotypes and Coral Health.
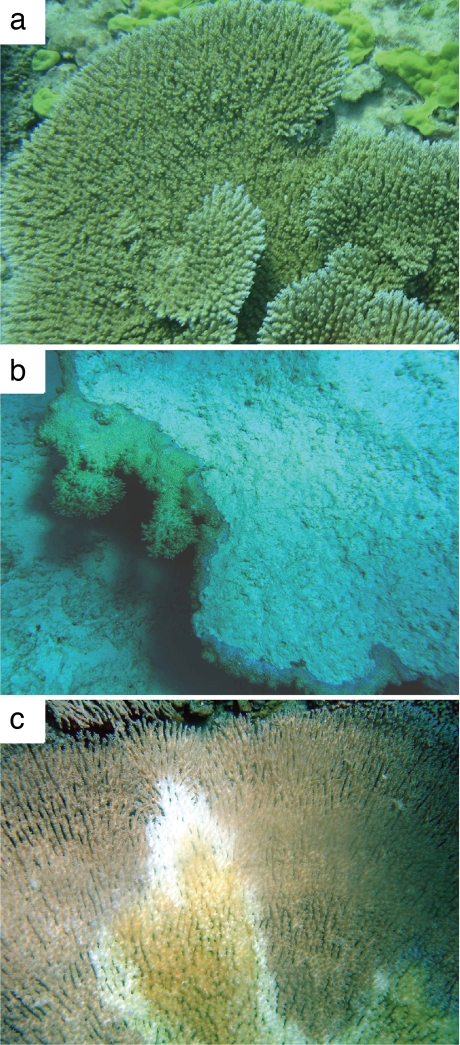
Symbiodinium belonging to clades A and C were recovered from 43 colonies of Acropora cytherea. Sixteen of 17 healthy A. cytherea colonies (Fig. 1a) harbored Symbiodinium belonging to clade C and one, to clade A. Corals displaying abnormal phenotypes (Fig. 1b) contained either clade A (n = 5) or clade C (n = 11), whereas corals showing evidence of disease (Fig. 1c) contained either clade A (n = 5), clade C (n = 5), or clade A and C (n = 1). There was a significant association between the symbiont clade and the observed health state of colonies (likelihood ratio test; 8.924, df = 2, P = 0.012), with corals hosting clade A symbionts showing a significantly higher incidence of disease or abnormal phenotypes than those hosting clade C (Fisher Exact Test; P = 0.015).
Fig. 1.
Colonies of A. cytherea sampled from Papahanaumokuakea (Northwestern Hawaiian Islands Marine National Monument) with healthy specimens (a), abnormal morphology showing evidence of past tissue loss and blue pigmentation (b), and active tissue loss phenotypes (c).
Genetic Diversity and Phylogenetic Analyses.
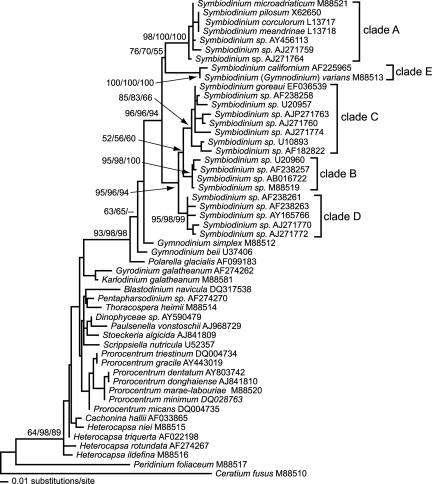
The phylogenetic analyses using maximum-likelihood, maximum-parsimony, and neighbor-joining resulted in trees with very similar topologies (maximum-likelihood shown in Fig. 2). Among the dinoflagellates, the genus Symbiodinium forms a well supported monophyletic group with clades A and E as a sister grouping to clades B, C, and D.
Fig. 2.
Maximum-likelihood phylogeny inferred by using partial nuclear SSU rDNA. Numerals at nodes indicate bootstrap analyses for maximum-likelihood (100 replicates)/maximum-parsimony (1,000 replicates)/neighbor-joining (1,000 replicates).
Corrected maximum-likelihood pairwise sequence comparisons of Symbiodinium clades A, B, C, D, and E and the dinoflagellates Gymnodinium beii, Gymnodinium simplex, Polarella glacialis, Thoracosphaera heimii, and Cachonina hallii are available in supporting information (SI) Table S1. The distances between Symbiodinium clade A and G. beii, G. simplex, and P. glacialis (0.061, 0.064, and 0.081 respectively) are less than those between clade A and the Symbiodinium clades B, C, D, and E (0.115, 0.115, 0.099 and 0.107, respectively). Similarly, the distances between clade E Symbiodinium and G. beii and G. simplex (0.091 and 0.088, respectively) are less than the distances between clade E and clades B, C, and D (0.102, 0.115, and 0.100, respectively). Further, the pairwise sequence diversity within clade C (0.051) and between different Symbiodinium clades is greater than the genetic distance between dinoflagellates that are in different orders (e.g., 0.036 between T. heimii order Thoracosphaerales and C. hallii order Peridiniales).
Because of a lower pairwise genetic distance between clade A and G. beii, G. simplex, and P. glacialis than between clade A and other Symbiodinium lineages, the Shimodaira–Hasegawa test was used. There was a significant difference in tree topologies when forcing Symbiodinium into a paraphyletic grouping by constraining clades A and clade E with the outgroup G. simplex [δ = L1 − L2 = −18.57092; P = 0.026; log-likelihood (L1) = 2866.68434].
Carbon Fixation and Release.
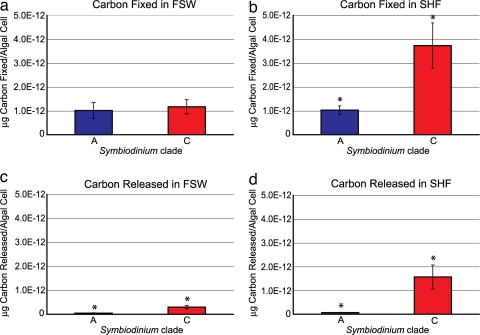
Freshly isolated symbionts (FIS) belonging to clade A and C incubated in filtered sea water (FSW) fixed similar amounts of carbon (mean value of 1.0168E-12 and 1.1698E-12, respectively; Fig. 3a). In contrast, clade C FIS incubated in synthetic host factor (SHF) fixed approximately four times more carbon than clade A FIS (mean value of 3.7344E-12 and 1.0290E-12, respectively; t test = −2.8032, P = 0.020; Fig. 3b). Similarly, clade C FIS incubated in both FSW and SHF released significantly greater amounts of carbon into the medium than clade A (FSW mean value of 2.9924E-13 and 5.0654E-14, respectively; t test = −3.3052, P = 0.008; SHF mean value of 1.56878E-12 and 6.68978E-14, respectively; t test = −2.984, P = 0.014; Fig. 3 c and d).
Fig. 3.
Mean values of carbon expressed as μg/algal cell fixed (a and b) and released (c and d) in the presence of FSW or SHF by Symbiodinium clade A (blue histograms) and clade C (red histograms). Bars in the histograms represent the standard error and an asterisk denotes values that are significantly different within a treatment.
For clade A, there was no significant difference between the amount of carbon fixed in FSW or SHF or in the amount released in FSW or SHF. In contrast, for clade C, there was a significant difference in the amount of carbon fixed in FSW to SHF (t test = −2.5834, P = 0.027) and the amount released in FSW to SHF (t test = −2.4959, P = 0.032).
Discussion
The efficiency of mutualistic symbioses depends on the host genotype, symbiont genotype, and the environment (1, 2, 20). Coral–dinoflagellate symbioses are defined as mutualistic; however, here we provide evidence that the symbiosis between a Pacific coral and the Symbiodinium clade A lineage appears as a reduction in the health state of the coral that may result from the host not receiving sufficient nutritional input from this symbiont lineage.
Clade A Symbiodinium are rarely reported in coral hosts; however, this group has been described as fast growing and opportunistic because it is found in corals recovering from bleaching events (21, 22). Here we show that A. cytherea from Hawaii harboring clade A exhibit suboptimal health states and an increased incidence of disease as compared with corals sampled on the same reef harboring Symbiodinium clade C. Although the incidence of coral disease in Hawaii is low, mass coral mortality caused by disease is widespread in the Atlantic Caribbean, especially within the genus Acropora (23–26). Interestingly, these corals also harbor Symbiodinium belonging to clade A as the dominant symbiont type (27, 28).
The relationship between health-compromised corals and clade A symbionts may reflect a directly harmful trait of the interaction between the coral host and these symbionts or the proliferation of opportunistic symbionts in health-compromised corals. Our in vitro carbon fixation experiments suggest that clade A symbionts may not provide as much carbon to the host as clade C symbionts and thus may not meet the host's nutritional requirements. In corals that have a high autotrophic dependence, such a scenario would certainly be reflected in a reduction of fitness that could ultimately render the host more susceptible to disease. Conversely, for the symbionts, the capacity to retain carbon would increase fitness and account for the high in situ growth rates reported for this group.
The evidence presented here does not allow us to infer that there is a direct negative effect of hosting clade A symbionts for the animal host (1, 29, 30). Thus there is no direct evidence that the interaction between clade A symbionts and corals belonging to the genus Acropora is parasitic. However, it is generally believed that horizontal transmission of symbionts promotes the evolution of parasitism (31–36). Acropora acquires its symbionts via horizontal transmission (37), and the competition between different lineages of Symbiodinium found within Acropora (clade A and C co-occurring) may be driving clade A Symbiodinium toward parasitism (34–36, 38–40). Furthermore, as health-compromised corals persist in nature, this phenotype would promote an increase in abundance of clade A Symbiodinium. Interestingly, in the symbiosis between the scyphozoan Cassiopea and Symbiodinium, a shift from mutualism to parasitism was inferred from a decoupling of the symbiosis from vertical symbiont acquisition to horizontal acquisition that resulted in a reduction in fitness of the host (41). In the Cassiopea that acquired symbionts from the environment, there was a reduction in host growth rate and a proliferation leading to higher densities of Symbiodinium.
An alternative interpretation is that mutualism has never been achieved between clade A Symbiodinium and corals. The phylogenetic positioning of clades A and E is either basal (8, 10, 13, 42–47) or sister to the other Symbiodinium clades (this study and refs. 8, 10, 13, and 42–47). This positioning suggests that clade A (and E, or both) was the first group to initiate symbiosis with invertebrates, that they are divergent lineages on a different evolutionary trajectory to other Symbiodinium clades that evolved from the ancestral dinoflagellate that first formed symbiosis with invertebrates, or that a free-living state is ancestral within the genus and a symbiotic lifestyle evolved independently in the two sister lineages. The transition from free-living to mutualism in clade A may not be a completely evolved trait, whereas the sister grouping or more derived Symbiodinium clades (especially the dominant coral symbiont clade C) may have successfully achieved mutualism. It is generally believed that parasitism is the ancestral state in the evolution of endosymbiotic mutualisms and occurs during the process of endocytosis, recognition, and the adaptive evolution of both partners (1, 48–51). Clade A and clade C Symbiodinium may represent symbiont lineages characterizing these two steps in the symbiosis continuum. The phylogeny presented here places clade A Symbiodinium as sister taxa along with clade E to clades B, C, and D. Clade E Symbiodinium is extremely rare and has only been identified in the temperate soft-bodied anemone Anthopleura elegantissima (10). The rarity and absence of this Symbiodinium lineage suggests that it does not form symbioses readily, it is going extinct, or it is an opportunistic dinoflagellate that is more abundant as a free-living entity. In terms of the latter, it is important to note that the diversity of Symbiodinium in the ocean environment is not well characterized (8, 52, 53).
There is strong support for a monophyletic grouping of Symbiodinium; however, the lower genetic distances between clade A Symbiodinium and the Symbiodinium outgroups Gymnodinium sp. and P. glacialis (G. simplex being free-living) relative to other Symbiodinium clades suggests a nonconstant molecular clock within the genus. DNA evolution rates are known to be variable between mutualists and nonmutualists of the same organism (54). Furthermore, faster DNA evolution rates have been identified in aphid bacterial endosymbionts (genus Buchnera) than in free-living relatives (55), and in fungi found in mutualisms with liverworts (56). Indeed, clade A Symbiodinium has been shown to have a slower rate of DNA evolution than other Symbiodinium clades (57), which is consistent with rates expected of a nonmutualistic lineage. Also, the bootstrap support for the Symbiodinium outgroups presented here and in another comprehensive phylogenetic study of the genus in relation to free-living pelagic symbionts using partial large subunit rDNA (58), both show very little support for the Gymnodinium species. Clearly, further analysis of these dinoflagellates is required to better understand the evolution of symbiosis within the grouping.
Genetic diversity within the genus Symbiodinium has previously been reported as comparable to that seen within different orders of dinoflagellates (15). Here, we show that this order-level genetic diversity actually exists within a single lineage of Symbiodinium, clade C. Such high levels of genetic divergence within the Symbiodinium genus supports the idea that the group contains members with highly diverse functions and physiologies, some of which may provide them with the capacity to form symbiotic interactions with coral. Based on the data presented here, a conservative approach would classify clade lineages of Symbiodinium into different families when comparing the amount of pairwise sequence diversity between other dinoflagellates. Parasitic and mutualistic symbiont lineages have been shown to group together in other interactions (59–62), and different genetic varieties of the same symbiont can be either mutualistic or harmful to the host. For example, the interactions between the actinomcyete fungi Frankia and Casuarina plant species can be both beneficial or antagonistic (63), mycorrhizal–plant interactions can be mutualistic or antagonistic (64), and the bacterial group Vibrionaceae that contains both free-living and symbiotic varieties that form symbioses with marine hosts is either cooperative or parasitic (65).
The amount of diversity in clade A and clade C is very different (58) with clade C containing many more types (21, 58). An opportunistic symbiotic lifestyle in clade A could potentially explain the discrepancy in diversity within these clades. If clade A exists primarily in the free-living environment and only occasionally infects a host colony, then diversification within the clade may be restricted because of the need to occupy two compartments, the host and the ocean. Clade C Symbiodinium (and to a lesser extent other clade lineages) associate with a diverse range of hosts, many that use vertical transmission of symbionts, providing them with more, highly specialized environments (coral and other host organisms) that would promote the evolution of multiple specialized types. This mechanism has been suggested as a factor that differentiates the genetically diverse genus Symbiodinium compared with other less diverse pelagic symbionts (58).
Further evidence that supports clade A Symbiodinium as adapted more toward a free-living dinoflagellate comes from the history of culturing Symbiodinium. Clade A Symbiodinium is easily cultured and outcompetes other clades even when the starting culture obtained from a host species contains predominantly clade C and undetectable clade A symbionts (8, 27). In addition, clade A and B Symbiodinium have been cultured from the ocean environment with a group of isolates within clade A suggested to be nonsymbiotic (53). In contrast, clade C is extremely difficult to culture and may represent an obligate symbiont adapted to survive only in the highly specialized environment of the animal host cell.
In this study we also show that clade A Symbiodinium releases very little carbon that can be used for host nutrition, the defining factor in coral-dinoflagellate mutualism (4), and there is no increase in the amount of carbon fixed in the presence of SHF. This finding is in contrast to the common coral symbiont, clade C, which both increases carbon fixation and release in the presence of SHF. Recently, Loram et al. (66) showed functional variability in the symbiosis between the giant sea anemone, Condylactis gigantea, and clades A and B Symbiodinium. In this association, clade A was shown to be more beneficial to the animal host than clade B. Considering the diversity and biology of the these host taxa, Symbiodinium belonging to clade A is likely to have a different interaction with soft-bodied anthozoans than with the Scleractinia. The most obvious differences between the two interactions are the formation of the calcium carbonate skeleton in the Scleractinia and the persistence of anemones when devoid of their dinoflagellate symbionts to rely on heterotrophy to survive, which is not possible for coral (67).
Taken together, we suggest that Symbiodinium clade A is less beneficial to corals than other Symbiodinium lineages and may be more representative of a parasitic than a mutualistic symbiont, whereby the animal host does not receive sufficient nutritional input from the dinoflagellate symbiont, a circumstance that ultimately renders the coral more susceptible to disease and mortality.
Methods
Symbiodinium Genotypes and Coral Health.
Colonies of A. cytherea (1–2 m in size) were sampled from French Frigate Shoals in the Northwestern Hawaiian Islands Marine National Monument, Papahanaumokuakea, during September 2005, May 2006 and September 2007. Approximately 2 cm2 of coral was removed from each colony with a hammer and chisel and stored at −20°C in DMSO preservation buffer (68). Each coral sampled was photographed and categorized as (i) healthy (Fig. 1a), (ii) displaying abnormal phenotypes consisting of pronounced blue pigmentation and evidence of past tissue loss (Fig. 1b), or (iii) suffering from a disease causing active tissue loss (Fig. 1c).
DNA from the stored coral samples was extracted by using a modified cetyltrimethylammonium bromide (CTAB) protocol (69). Briefly, the tissue was incubated in 500 μl of CTAB buffer [100 mM Tris·Hcl, 1.4 M NaCl, 20 mM EDTA, 2% (wt/vol) CTAB, 2% (vol/vol) 2-mercaptoethanol, 1% (wt/vol) polyvinylpyrrolidone, pH 8.0] at 65°C for 30 min. An equal amount of phenol/chloroform/isoamyl alcohol (25:24:1) was added, and the contents of the tube were mixed and centrifuged at 15,000 × g for 20 min. The aqueous phase was removed, and an equal volume of chloroform/isoamyl alcohol (24:1) was added, thoroughly mixed, and centrifuged at 15,000 × g again for 20 min. The DNA was precipitated from the resulting aqueous phase by the addition of 1/2 volume of 5 M NaCl and an equal volume of isopropanol and incubated for 1 h at −80°C. The DNA was pelleted by centrifugation at 15,000 × g for 30 min, washed twice with 70% ethanol, and resuspended in sterile water.
The nuclear small subunit (SSU) rDNA was amplified from DNAs extracted from Symbiodinium by using the primers ss5z and ss3z (70). The PCR products were digested by using TaqI (New England Biolabs) for 2 h at 65°C, and the clade of Symbiodinium present in each A. cytherea colony was identified based on the patterns of restriction fragment length polymorphisms (RFLPs) in the digests visualized on 2% agarose gels under UV illumination.
Genetic Diversity and Phylogenetic Analyses.
Symbiodinium and other dinoflagellate nuclear SSU rDNA sequences were obtained from GenBank. The analysis is restricted to only five clades of Symbiodinium, which represents available SSU rDNA data for the genus. A concatenated alignment of 459-nt characters consisting of two regions within the SSU rDNA was constructed and manually edited by using the software MacVector 8.0.2. The DNA alignment was tested against 56 models of DNA substitution in the program PAUP*, version 4.0b10, and the best-fit model was selected by using the program Modeltest, version 3.7 (71). The Hierarchical Likelihood Ratio Tests and the Akaike Information Criterion both selected the TrN+I+G as the best fit model of DNA evolution with stationary base frequencies of A = 0.2659, C = 0.1978, G = 0.2422, and T = 0.2941, proportion of invariable sites = 0.2485, and a gamma distribution shape parameter of 0.6227. Pairwise distances and phylogenies were inferred from the alignment by using PAUP*, version 4.0b10 (72). For corrected maximum-likelihood pairwise genetic distances, the distance showing the greatest amount of genetic diversity when comparing a Symbiodinium clade was used. The maximum-likelihood phylogeny was constructed by using the results from Modeltest, and the Kimura two-parameter model (73) was used to infer the neighbor-joining tree. For maximum-parsimony, the minimum F value was incorporated as the character-state optimization and gaps in the sequence were treated as a fifth base. Midpoint grouping was used in the maximum-likelihood and neighbor-joining trees, and Peridinium foliaceum and Ceratium fusus were used as outgroups in the maximum-parsimony tree. The validity of node placement within each phylogeny was tested by using bootstrap analyses (74) for 1,000 replicates for neighbor-joining and maximum-parsimony trees and 100 replicates for maximum-likelihood analysis.
Carbon Fixation and Release.
FIS were obtained from the jellyfish Cassiopeia sp. (n = 6) collected from the Hilton Hawaiian Village lagoon in Waikiki and from the coral Pocillopora damicornis (n = 6) collected from the reef surrounding the Hawaiian Institute of Marine Biology in Kaneohe Bay, Oahu. Symbiodinium were isolated from Cassiopeia sp. by homogenizing tentacles in 2 ml of FSW and from P. damicornis by blasting the tissue off the skeleton with a dental water pick. The FIS were collected by centrifugation at 5,000 × g at 24°C for 5 min, and the FIS pellets were washed free of host tissues by repeated resuspension in FSW and centrifugation (×3). The final FIS pellets were resuspended in 1.5 ml of FSW, and the number of cells were assessed with a Neubauer haemocytometer and a light microscope (four per sample) and expressed per ml. The clade of Symbiodinium harbored by each host was confirmed by using SSU rDNA RFLP as described above.
The total carbon fixed and released by FIS incubated in FSW and a SHF was assessed by using NaH14CO3. Six replicate experiments for each treatment, for each clade were performed with FIS isolated from a different animal. FIS (200 μl at 4 × 106 cells/ml) were mixed with an equal volume of FSW or SHF and incubated in the light (300 μmol quanta·m−1·s−1) for 1 h at 26°C in the presence of 1 μCi of NaH14CO3 (added in 10 μl of MilliQ water before the 1-h incubation). The SHF consisted of 1.24 μM aspartic acid, 15.15 μM glutamic acid, 3.61 μM serine, 0.92 μM histidine, 7.13 μM glycine, 2.11 μM arginine, 3.13 μM taurine, 10.41 μM alanine, 0.97 μM tyrosine, 1.73 μM methionine, 5.93 μM valine, 2.32 μM phenylalanine, 2.51 μM isoleucine, 2.43 μM leucine, and 0.64 μM asparagine, dissolved in distilled water with pH 8.3 and adjusted to a salinity 33 parts per thousand (75).
After incubation, the tubes were mixed by vortexing, and two 50-μl samples were removed to assess total 14C fixed. FIS were then pelleted at 5,000 × g for 5 min, and two 50-μl samples of the supernatant were placed in scintillation vials to calculate the amount of carbon released by the FIS. The unincorporated 14C was removed from the samples by acidification by adding 100 μl of 0.1 M HCl to each scintillation vial and incubating it at room temperature for 1 h. Biodegradable counting scintillant (BCS) (4 ml; Amersham Biosciences) was added to each scintillation vial, and the amount of radioactivity was assessed with a Beckman LS 3801 scintillation counter. The total 14C fixed and released was calculated and expressed as counts per FIS and as a ratio of counts fixed and released relative to the FSW control. An estimate of the absolute amount of 14C fixed or released was calculated by using the specific activity of the label (NaH14CO3) expressed in cpm/fmol and the following equation [cpm × (cpm/fmol −1) × 10].
Supplementary Material
Acknowledgments.
We thank B. Wheeler, I. Baums, J. Salerno, and A. Mayfield for assistance in the field and use of the coral images and M. Craig, S. Karl, R. Toonen, J. Felsenstein, X. Pochon, and two anonymous reviewers for insights that helped improve the manuscript. Coral samples were collected in the Northwestern Hawaiian Islands Marine National Monument under a permit DLNR.NWH106R006 issued to R.D.G. The collections were implemented on a National Oceanic and Atmospheric Administration cruise and supported through funding from The National Marine Sanctuary program and Hawaii Institute of Marine Biology Reserve Partnership (memorandum of agreement 2005–008/66882). This is Hawaii Institute of Marine Biology contribution 1307.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801328105/DCSupplemental.
References
- 1.Paracer S, Ahmadjian V. Symbiosis: An Introduction to Biological Associations. 2nd Ed. New York: Oxford Univ Press; 2000. [Google Scholar]
- 2.Douglas A. Host benefit and the evolution of specialization in symbiosis. Heredity. 1998;81:599–603. [Google Scholar]
- 3.Muscatine L. Glycerol excretion by symbiotic algae from corals and tridacna and its control by the host. Science. 1967;156:516–519. doi: 10.1126/science.156.3774.516. [DOI] [PubMed] [Google Scholar]
- 4.Muscatine L, Porter J. Reef corals: Mutualistic symbioses adapted to nutrient-poor environments. Bioscience. 1977;27:454–460. [Google Scholar]
- 5.Smith S, Read D. Mycorrhizal Symbiosis. San Diego: Academic; 1997. [Google Scholar]
- 6.Sprent J, Sprent P. Nitrogen-Fixing Organisms: Pure and Applied Aspects. London: Chapman & Hall; 1990. [Google Scholar]
- 7.Rowan R, Powers D. A molecular genetic classification of zooxanthellae and the evolution of animal-algal symbioses. Science. 1991;251:1348–1351. doi: 10.1126/science.251.4999.1348. [DOI] [PubMed] [Google Scholar]
- 8.Carlos A, Baillie B, Kawachi M, Maruyama T. Phylogenetic position of Symbiodinium (Dinophyceae) isolates from tridacnids (Bivalvia), cardiids (Bivalvia), a sponge (Porifera), a soft coral (Anthozoa), and a free-living strain. J Phycol. 1999;35:1054–1062. [Google Scholar]
- 9.LaJeunesse T, Trench R. Biogeography of two species of Symbiodinium (Freudenthal) inhabiting the intertidal sea anemone Anthopleura elegantissima (Brandt) Biol Bull. 2000;199:126–134. doi: 10.2307/1542872. [DOI] [PubMed] [Google Scholar]
- 10.LaJeunesse T. Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: In search of a “species” level marker. J Phycol. 2001;37:866–880. [Google Scholar]
- 11.Pawlowski J, et al. Molecular identification of algal endosymbionts in large miliolid Foraminifera: 2 dinoflagellates. J Eukaryotic Microbiol. 2001;48:368–373. doi: 10.1111/j.1550-7408.2001.tb00326.x. [DOI] [PubMed] [Google Scholar]
- 12.Pochon X, Pawlowski J, Zaninetti L, Rowan R. High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Marine Biol. 2001;139:1069–1078. [Google Scholar]
- 13.Pochon X, LaJeunesse T, Pawlowski J. Biogeographic partitioning and host specialization among foramniferan dinoflagellate symbionts (Symbiodinium; Dinophyta) Marine Biol. 2004;146:17–27. [Google Scholar]
- 14.Stat M, Carter D, Hoegh-Guldberg O. The evolutionary history of Symbiodinium and scleractinian hosts - Symbiosis, diversity, and the effect of climate change. Perspect Plant Ecol Evol System. 2006;8:23–43. [Google Scholar]
- 15.Rowan R, Powers D. Ribosomal RNA sequences and the diversity of symbiotic dinoflagellates (zooxanthellae) Proc Natl Acad Sci USA. 1992;89:3639–3643. doi: 10.1073/pnas.89.8.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little A, van Oppen J, Willis B. Flexibility in algal endosymbioses shapes growth in reef corals. Science. 2004;304:1492–1494. doi: 10.1126/science.1095733. [DOI] [PubMed] [Google Scholar]
- 17.Rowan R. Thermal adaptations in reef coral symbionts. Nature. 2004;430:742. doi: 10.1038/430742a. [DOI] [PubMed] [Google Scholar]
- 18.Hoegh-Guldberg O. Climate change, coral bleaching and the future of the world's coral reefs. Marine Freshwater Res. 1999;50:839–866. [Google Scholar]
- 19.Hughes T, et al. Climate change, human impacts, and the resilience of coral reefs. Science. 2003;301:929–933. doi: 10.1126/science.1085046. [DOI] [PubMed] [Google Scholar]
- 20.Verghese S, Misra A. Frankia-actinorhizal symbiosis with special reference to host-microsymbiont relationship. Curr Sci. 2002;83:404–408. [Google Scholar]
- 21.LaJeunesse T. “Species” radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene–Pliocene Transition. Mol Biol Evol. 2005;22:570–581. doi: 10.1093/molbev/msi042. [DOI] [PubMed] [Google Scholar]
- 22.Toller W, Rowan R, Knowlton N. Repopulation of zooxanthellae in the Caribbean corals Montastraea annularis and M. faveolata following experimental and disease-associated bleaching. Biol Bull. 2001;201:360–373. doi: 10.2307/1543614. [DOI] [PubMed] [Google Scholar]
- 23.Aronson R, Precht W. White-band disease and the changing face of Caribbean coral reefs. Hydrobiologia. 2001;460:25–38. [Google Scholar]
- 24.Gardner T, et al. Long-term region-wide declines in Caribbean corals. Science. 2003;301:958–960. doi: 10.1126/science.1086050. [DOI] [PubMed] [Google Scholar]
- 25.Garzon-Ferreira J, Gil-Agudelo D, Barrios L, Zea S. Stony coral diseases observed in southwestern Caribbean reefs. Hydrobiologia. 2001;460:65–69. [Google Scholar]
- 26.Porter J, et al. Patterns of spread of coral disease in the Florida Keys. Hydrobiologia. 2001;460:1–24. [Google Scholar]
- 27.LaJeunesse T. Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Marine Biol. 2002;141:387–400. [Google Scholar]
- 28.Thornhill D, et al. Multi-year, seasonal genotypic surveys of coral–algal symbioses reveal prevalent stability or post-bleaching reversion. Marine Biol. 2006;148:711–722. [Google Scholar]
- 29.Lewin R. Symbiosis and parasitism: Definitions and evaluations. BioScience. 1982;32:254–260. [Google Scholar]
- 30.Goff L. Symbiosis and Parasitism: Another viewpoint. BioScience. 1982;32:255–256. [Google Scholar]
- 31.Fine P. Vectors and vertical transmission: an epidemiological perspective. Ann NY Acad Sci. 1975;266:173–194. doi: 10.1111/j.1749-6632.1975.tb35099.x. [DOI] [PubMed] [Google Scholar]
- 32.Ewald P. Host parasite relations, vectors, and the evolution of disease severity. Annu Rev Ecol System. 1983;14:465–485. [Google Scholar]
- 33.Bull J. Perspective: Virulence. Evolution (Lawrence, Kans.) 1994;48:1423–1437. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 34.Frank S. Genetics of mutualism: The evolution of altruism between species. J Theor Biol. 1994;170:393–400. doi: 10.1006/jtbi.1994.1200. [DOI] [PubMed] [Google Scholar]
- 35.Frank S. Host control of symbiont transmission: The separation of symbionts into germ and soma. Am Nat. 1996;148:1113–1124. [Google Scholar]
- 36.Frank S. Host-symbiont conflict over mixing of symbiotic lineages. Proc R Soc London Ser B. 1996;263:339–344. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- 37.Wallace C. Reproduction, recruitment, and fragmentation in nine sympatric species of the coral genus Acropora. Marine Biol. 1985;8:217–233. [Google Scholar]
- 38.Baker A, Rowan R, Knowlton N. Symbiosis ecology of two Caribbean Acroporid corals. Proc 8th Int Coral Reef Symp; 1997. pp. 1295–1300. [Google Scholar]
- 39.Loh W, Carter D, Hoegh-Guldberg O. Brisbane, Australia: University of Queensland; 1998. Diversity of zooxanthellae from scleractinian corals of One Tree Island (The Great Barrier Reef) pp. 141–150. [Google Scholar]
- 40.Stat M, Gates R. Vectored introductions of marine endosymbiotic dinoflagellates into Hawaii. Biol Invasions. 2008;10:579–583. [Google Scholar]
- 41.Sachs J, Wilcox T. A shift to parasitism in the jellyfish symbiont Symbiodinium microadriaticum. Proc R Soc London Ser B. 2006;273:425–429. doi: 10.1098/rspb.2005.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, et al. Symbiont diversity in scleractinian corals form tropical reefs and subtropical nonreef communities in Taiwan. Coral Reefs. 2005;24:11–22. [Google Scholar]
- 43.Takabayashi M, Santos S, Cook C. Mitochondrial DNA phylogeny of the symbiotic dinoflagellates (Symbiodinium, Dinophyta) J Phycol. 2004;40:160–164. [Google Scholar]
- 44.Santos S, et al. Molecular phylogeny of symbiotic dinoflagellates inferred from partial chloroplast large subunit (23S)-rDNA sequences. Mol Phylogenet Evol. 2002;23:97–111. doi: 10.1016/S1055-7903(02)00010-6. [DOI] [PubMed] [Google Scholar]
- 45.Barbrook A, Visram S, Douglas A, Howe C. Molecular diversity of dinoflagellate symbionts of Cnidaria: The psbA minicircle of Symbiodinium. Protist. 2006;157:159–171. doi: 10.1016/j.protis.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Savage A, et al. Molecular diversity of symbiotic algae at the latitudinal margins of their distribution: Dinoflagellates of the genus Symbiodinium in corals and sea anemones. Marine Ecol Progr Ser. 2002;244:17–26. [Google Scholar]
- 47.Baillie B, Belda-Baillie C, Maruyama T. Conspecificity and Indo-Pacific distribution of Symbiodinium genotypes (Dinophyceae) from giant clams. J Phycol. 2000;36:1153–1161. [Google Scholar]
- 48.Yamamura N. Vertical transmission and evolution of mutualism from parasitism. Theor Popul Biol. 1993;44:95–109. [Google Scholar]
- 49.Ewald P. Transmission modes and evolution of the parasitism-mutualism continuum. Ann NY Acad Sci. 1987;503:295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x. [DOI] [PubMed] [Google Scholar]
- 50.Yamamura N. Evolution of mutualistic symbiosis: A differential equation model. Res Population Ecol. 1996;38:211–218. [Google Scholar]
- 51.Holte A, Houck M, Collie N. Potential role of parasitism in the evolution of mutualism in astigmatid mites: Hemisarcoptes cooremani as a model. Exp Appl Acarol. 2001;25:97–107. doi: 10.1023/a:1010655610575. [DOI] [PubMed] [Google Scholar]
- 52.Gou W, et al. Phylogenetic analysis of a free-living strain of Symbiodinium isolated from Jiaozhou Bay, P.R. China. J Exp Marine Biol Ecol. 2003;296:135–144. [Google Scholar]
- 53.Coffroth M, Lewis C, Santos S, Weaver J. Environmental populations of symbiotic dinoflagellates in the genus Symbiodinium can initiate symbioses with reef cnidarians. Curr Biol. 2006;16:R985–R988. doi: 10.1016/j.cub.2006.10.049. [DOI] [PubMed] [Google Scholar]
- 54.Herre E, Knowlton N, Mueller U, Rehner S. The evolution of mutualisms: Exploring the paths between conflict and cooperation. Trends Ecol Evol. 1999;14:49–53. doi: 10.1016/s0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- 55.Moran N. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lutzoni F, Pagel M. Accelerated evolution as a consequence of transitions to mutualism. Proc Natl Acad Sci USA. 1997;94:11422–11427. doi: 10.1073/pnas.94.21.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pochon X, Montoya-Burgos J, Stadelmann B, Pawlowski J. Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Mol Phylogenet Evol. 2006;38:20–30. doi: 10.1016/j.ympev.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 58.Shaked Y, de Vargas C. Pelagic photosymbiosis: rDNA assessment of diversity and evolution of dinoflagellate symbionts and planktonic foraminiferal hosts. Marine Ecol Progr Ser. 2006;325:59–71. [Google Scholar]
- 59.Herre E, et al. Molecular phylogenies of figs and their pollinator wasps. J Biogeogr. 1996;23:521–530. [Google Scholar]
- 60.Pellmyr O, Leebens-Mack J, Huth C. Nonmutualistic yucca moths and their evolutionary consequences. Nature. 1996;380:155–156. doi: 10.1038/380155a0. [DOI] [PubMed] [Google Scholar]
- 61.Pellmyr O, Thompson J, Brown J, Harrison R. Evolution of pollination and mutualism in the yucca moth lineage. Am Nat. 1996;148:827–847. [Google Scholar]
- 62.Sachs J, Simms E. Pathways to mutualism breakdown. Trends Ecol Evol. 2006;21:585–592. doi: 10.1016/j.tree.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 63.Reddell P, Bowen G. Frankia source affects growth, nodulation, and nitrogen fixation in Casuarina species. New Phytol. 1985;100:115–122. doi: 10.1111/j.1469-8137.1985.tb02850.x. [DOI] [PubMed] [Google Scholar]
- 64.Francis R, Read D. Mutualism and antagonism in the mycorrhizal symbiosis, with special reference to impacts on plant community structure. Can J Bot. 1995;1(Supp 1):S1301–S1309. [Google Scholar]
- 65.Nishiguchi M, Nair V. Evolution of symbiosis in the Vibrionaceae: A combined approach using molecules and physiology. Int J System Evol Microbiol. 2003;53:2019–2026. doi: 10.1099/ijs.0.02792-0. [DOI] [PubMed] [Google Scholar]
- 66.Loram J, Trapido-Rosenthal H, Douglas A. Functional significance of genetically different symbiotic algae Symbiodinium in a coral reef symbiosis. Mol Ecol. 2007;16:4849–4857. doi: 10.1111/j.1365-294X.2007.03491.x. [DOI] [PubMed] [Google Scholar]
- 67.Kinziee R, III, Takayama M, Santos S, Coffroth M. The adaptive bleaching hypothesis: Experimental tests of critical assumptions. Biol Bull. 2001;200:51–58. doi: 10.2307/1543084. [DOI] [PubMed] [Google Scholar]
- 68.Seutin G, White B, Boag P. Preservation of avian blood and tissue samples for DNA analyses. Can J Zool. 1991;69:82–90. [Google Scholar]
- 69.Dempster E, et al. Rapid DNA extraction from ferns for PCR-based analyses. BioTechniques. 1999;27:62–64. doi: 10.2144/99271bm13. [DOI] [PubMed] [Google Scholar]
- 70.Rowan R, Powers D. Molecular genetic identification of symbiotic dinoflagellates (zooxanthellae) Marine Ecol Progr Ser. 1991;71:65–73. [Google Scholar]
- 71.Posada D, Crandall K. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 72.Swofford D. PAUP*, Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer; 1999. version 4.0b10. [Google Scholar]
- 73.Kimura M. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 74.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution (Lawrence, Kans.) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 75.Gates R, et al. Free amino acids exhibit anthozoan “host factor” activity: they induce the release of photosynthate from symbiotic dinoflagellates in vitro. Proc Natl Acad Sci USA. 1995;92:7430–7434. doi: 10.1073/pnas.92.16.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.