Abstract
Myoglobin is a globular protein involved in oxygen storage and transport. No consensus yet exists on the atomic level mechanism by which oxygen and other small nonpolar ligands move between the myoglobin's buried heme, which is the ligand binding site, and surrounding solvent. This study uses room temperature molecular dynamics simulations to provide a complete atomic level picture of ligand migration in myoglobin. Multiple trajectories—providing a cumulative total of 7 μs of simulation—are analyzed. Our simulation results are consistent with and tie together previous experimental findings. Specifically, we characterize: (i) Explicit full trajectories in which the CO ligand shuttles between the internal binding site and the solvent and (ii) pattern and structural origins of transient voids available for ligand migration. The computations are performed both in sperm whale myoglobin wild-type and in sperm whale V68F myoglobin mutant, which is experimentally known to slow ligand-binding kinetics. On the basis of these independent, but mutually consistent ligand migration and transient void computations, we find that there are two discrete dynamical pathways for ligand migration in myoglobin. Trajectory hops between these pathways are limited to two bottleneck regions. Ligand enters and exits the protein matrix in common identifiable portals on the protein surface. The pathways are located in the “softer” regions of the protein matrix and go between its helices and in its loop regions. Localized structural fluctuations are the primary physical origin of the simulated CO migration pathways inside the protein.
Keywords: proteins, molecular dynamics, kinetics, structure-function relationships
Myoglobin is a small globular protein used by muscle cells for oxygen storage and transport. Historically, myoglobin has been an important starting point for structural and biophysical characterization of larger proteins. It has therefore been dubbed the “Hydrogen Atom” of molecular biology (1). However, despite nearly half a century of experimental and theoretical studies, the precise manner in which the oxygen ligand moves between heme iron and solvent in myoglobin remains unresolved. None of the over 250 static crystallographic structures of myoglobin in the Protein Data Bank show an obvious static path between the external solvent and the heme iron. It was recognized long ago that thermal fluctuations create a web of transient structural voids and, therefore, a dynamical pathway through which small nonpolar ligands such as O2, CO or NO can migrate inside the molecule. Numerous experimental and theoretical works have elucidated many aspects of the ligand migration process (2–17). Some of these studies have provided evidence for a single major pathway for ligand migration that would require movement of the distal histidine (“His gate”) (16, 18–23). However, other investigators have suggested the existence of multiple pathways for ligand diffusion in myoglobin (4, 5, 24–26).
Reconciling these two seemingly contradictory views requires the precise characterization of the dynamical pathways through which ligand moves. This is important for two reasons. First, myoglobin plays a vital role in cellular oxygen storage and transport. Second, myoglobin function offers the simplest example of a class of mechanisms by which protein binds ligand in solvent-inaccessible pockets. Molecular dynamics (MD) studies should have the potential to fill the gap in our understanding of ligand migration. However, trajectory studies until recently did not extend into the time range that would allow full simulation of ligand escape, inclusion, and migration between the heme-binding pocket and solvent at experimentally-realistic conditions.
Recent advances in computational power and methodology now permit this type of all-atom MD simulation to be performed at room temperature in a solvent bath of explicit water molecules. No further approximations are required, such as, for example, the use of high temperature ligand probes, which were employed to facilitate past studies (5).
Following previous computational and experimental work, we substitute CO for O2 in our simulations. This substitution is predicated on the greater availability of structural and other experimental data for the bound CO form of myoglobin. It is justified by the generally held view that the overall picture of CO diffusion in myoglobin should hold for other nonpolar ligands of similar size including molecular oxygen and nitric oxide (26–27).
In this study, we report (i) room-temperature MD simulations of explicit full escape and inclusion trajectories of CO beginning with photo-detached and solvated CO, respectively, in wild-type (WT) myoglobin and corresponding simulations in a V68F myoglobin mutant in which experimental studies have shown ligand binding kinetics to be retarded but binding affinity to be unchanged; and (ii) integration of free volume fluctuations in MD simulations of WT and mutant myoglobin with bound CO. The latter category of study utilizes a volumetric algorithm and associated PathFinder computer program to identify cavities sterically capable of accommodating CO in static structure snapshots [see Methods and supporting information (SI) Appendix].
Results and Discussion
We begin by providing the reader with a panorama of the migration pathways, connecting bottlenecks, and surface portal features that emerges from and is common to all our simulations. Descriptions of results specific to each of the two categories of computer simulations then follows. Technical details of the computations are provided in SI Appendix.
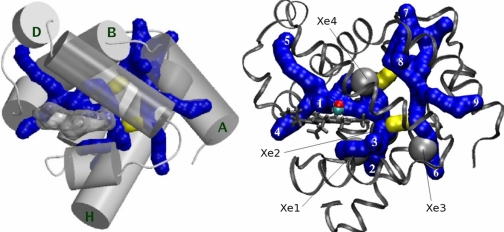
Our studies consistently show that CO ligand migrates along two discrete dynamic pathways in the protein matrix. The position of the pathways and connecting bottlenecks relative to the protein helix scaffold is shown in Fig. 1Left. The pathways in this figure are illustrated by a series of 1.4 Å radius fused spheres. The positions of the fused sphere centers have been sampled every 10 ps from CO atom coordinates in a representative set of trajectories. The representative set of trajectories is itself drawn only from those simulations in which the CO ligand makes a full transit between binding site and solvent.
Fig. 1.
Pathways of CO migration in myoglobin. (Left) Ligand migration pathways and bottlenecks relative to α-helices in myoglobin. Pathways are shown in blue. Helices are rendered as 4 Å radius cylinders. Helices A, B, D, and H are labeled; helices A, E, and F and the molecular surface of the heme group are rendered transparent. The major pathway is on the heme side of the protein; the minor pathway is on the opposite side, touching helix A. Two bottlenecks shown in yellow connect the two pathways. A full 360° 3D view of the pathways is available in Movie S1, in SI Appendix. (Right) Pathways, bottlenecks, and portals relative to xenon-binding sites. Portals numbered in white correspond to the description in Table 2. The CO is shown in the binding site.
In our explicit trajectory simulations, ligand migration occurs more frequently along the major pathway on the “heme side” of the protein and less frequently along the minor pathway on the “opposite” side near helix A, as will be detailed below. We also find trajectories that occasionally hop between these discrete pathways. These hops occur through the two bottleneck regions shown in yellow that connect the major and minor pathway. The binding site lies on the major pathway; thus, every trajectory that connects the binding site with the solvent must pass through the major pathway. Lastly, we find that the ligand enters and exits the two discrete pathways through nine distinct regions or portals on the protein surface. The pathways and portals are shown relative to experimentally determined xenon-binding sites (4) in Fig. 1 Right.
In what follows, we describe our room-temperature MD simulations of ligand migration between the binding site and solvent first in WT carbon monoxide myoglobin (MbCO) and secondly in the V68F MbCO mutant. Lastly, we describe our analysis of free volume fluctuations in MD simulations of both WT and mutant MbCO.
Wild-type Myoglobin.
We analyzed a total of 68 individual CO migration trajectories in WT myoglobin. These 90-ns-long trajectories began with two types of initial positions of the ligand: (i) Photo-detached CO in the heme binding pocket and (ii) CO placed in the water bulk at least 15 Å away from the protein surface.
Table 1 summarizes important statistics on all trajectories broken down by initial starting conditions. In the 20 CO trajectories beginning in the heme pocket after simulated photo-detachment, the CO molecule escapes the protein in 11 instances. None of these 11 trajectories re-enters the protein. The remaining nine trajectories diffuse in the protein but do not reach the solvent. Of the 48 CO trajectories beginning in the solvent, in 16 instances the CO molecule enters the protein and reaches the distal heme pocket; the remaining 32 trajectories diffuse into the protein but do not reach the binding site. Of these 16 instances in which CO reaches the binding site, 5 involve entries without subsequent exit, and 11 involve entry, visitation of the heme pocket, and subsequent escape into the solvent.
Table 1.
Trajectory count statistics of ligand entries and escapes in WT myoglobin
| Trajectory start | Number | Entries | Escapes |
|---|---|---|---|
| Photo-detached CO | 20 | 0 | 11 |
| Solvated CO | 48 | 16 | 11 |
The subset of trajectories that make at least one complete transit between heme-binding site and solvent sample the full natural movement of the ligand through the protein matrix and are thus collectively referred to as “complete transit” trajectories. In these “complete transit” trajectories, we observe a total of 38 entry/escape events: Their detailed breakdown is given in Table 2. Of these 38 entry/escape events, 31 occur along the major pathway and 7 along the minor pathway. The average travel time of CO between solvent and the binding site is 11.9 ns if the trajectory stays exclusively within the major pathway. The average travel time of CO between solvent and the binding site is 28.9 ns if entry/exit occurs through the minor pathway. Thus, in this partial kinetic window on the ligand migration events, transit through the major pathway alone occurs two to three times more rapidly, on average, than if the ligand trajectory also passes through the minor pathway.
Table 2.
Ligand entry/escape event properties for ″complete transit″ trajectories broken down by pathway and portal
Care must be taken when comparing these trajectory averages with experimental observables: Trajectories that did not show complete transit between the binding site and the solvent on the 90-ns time scale were not included in the calculation of the above average travel times. In SI Appendix, we describe how the macroscopic CO rate constants for both bi-molecular recombination and exit-to-solvent can be estimated from our entire trajectory statistics and simulation parameters; the main results of these estimates and comparison with experiment are summarized in Conclusions.
In both pathways, the distribution of the associated transit times is predicted to be quite broad. While a noticeable fraction of ligands are able to shuttle between the binding site and the solvent within only a few nanoseconds (see SI Appendix), more than half of the simulated photo-detached trajectories did not escape the protein within 90 ns. Remarkably, even in those trajectories that did not lead to a complete transit of CO between the binding site and the solvent, the ligands that diffused into the protein matrix were still found to traverse portions of the same pathways shown in Fig. 1 Right. The ligand movements within the pathways consist of a series of relatively long dwellings in selected “transient docking” sites and short hops between them. In agreement with the consensus emerging from time-resolved experiments (8, 28, 29), these docking sites involve Xe(4) and Xe(1) cavities on the major pathway. As seen in Fig. 1 Right, the major pathway also overlaps with Xe(2) cavity, in agreement with a recent suggestion (30). A detailed analysis of the ligand trajectories predicts additional transient docking sites located at the branching points of the pathways, that is, regions where separate “branches” of the same pathway meet or where the connectors (bottlenecks) attach to the pathways. As seen in Fig. 1 Right, some of these regions are in direct contact with the Xe sites, but some are not related to the Xe sites, in particular those on the minor pathway. The overall shape of the major pathway is consistent with the “horseshoe” schematic pathway around the heme group proposed on the basis of transient grating spectroscopy measurement of CO escape kinetics (25).
There are nine distinct surface portals on the protein surface through which ligand enters and exits: Five are associated with the major pathway, and four are associated with the minor pathway. Every portal associated with the major pathway contains at least one residue that Huang and Boxer (24) identified as significantly affecting ligand binding kinetics in their mutagenesis studies. Two of the portals in the minor pathway share this property; we speculate that residues near the other two portals may not have been identified by Huang and Boxer because CO rarely enters or exits through them (see Table 2).
The existence of the minor pathway is supported by site-directed mutagenesis and is also consistent with the existence of a transient docking site formed by the A–B and G–H helix corners (31). In our simulations of photo-detached CO, a docking site in this location was visited by the trajectory that eventually exited through portal 7 of the minor pathway.
The detailed atom-level picture of ligand pathways and portals presented in Fig. 1 Right and Table 2 may help reconcile the single vs. multiple pathway dispute mentioned in the Introduction. Ligand entry/escape through portals in the major pathway occurs four to five times more frequently than through the minor pathway; within the major pathway itself, portal 1 dominates. This agrees with experiments that emphasize the major role of the His gate pathway that links the binding site to the exterior (through portal 1, using our terminology). On the other hand, this is not the only way for the ligand to travel between the solvent and its binding site. We find that other portals to solvent exist—both on the major and minor pathways—in agreement with experiments that suggest multiple routes to solvent.
Site-directed mutagenesis (23) in combination with kinetic measurements (24, 25, 32–34) may be used to further investigate and refine the relative roles that different parts of the predicted pathways play in determining the overall ligand migration kinetics. The precise, atomic-resolution coordinates of the complete pathways for CO migration in myoglobin made available by this work, along with predicted relative significance of various parts of the pathways (see the SI Appendix and Table 2), should greatly facilitate the selection of specific site mutations for such studies and help rationalize the outcomes of the experiments. For example, the effect of a mutation that blocks a part of the minor pathway should be relatively small (assuming that the rest of the protein is unaffected by the mutation) compared with an equivalent mutation that disrupts the major pathway. At the same time, the existence of multiple entry portals and routes connecting the binding cavity with the solvent ensures that blocking any single one of them cannot completely prevent the ligand from reaching the binding site from the solvent, or ligand escape from the binding site.
V68F Mutant Myoglobin.
Additional support for our picture of ligand migration in WT myoglobin comes from analysis of the V68F mutant. Ligand binding kinetics have been studied in dozens of myoglobin mutants. The V68F mutant was chosen for study for two reasons: First, valine is highly conserved in this position across many species variants of WT myoglobin, and second, the V68F mutation significantly alters ligand binding kinetics without changing binding affinities (32).
For the mutant structure, we report nine separate 90-ns simulations of photo-detached CO initially in the distal heme pocket. CO did not escape to solvent in five of these nine trajectories. The non-escaping CO in these trajectories remained in the distal pocket close to their starting coordinates. This was in strong contrast to the behavior of non-escaping CO in the WT studies, which were significantly displaced from their starting coordinates. In the remaining four trajectories, CO escaped the distal pocket and reached solvent, notably through the same pathways as identified for the WT structure in Fig. 1 Right. The average escape time after photo-detachment is four times longer than in the corresponding WT myoglobin trajectories. The observation is consistent with the suggested steric constriction of the pathways leading out of the distal pocket. This was originally proposed to explain the experimental observation that the rates of ligand uptake and escape are both significantly reduced in this mutant (32).
PathFinder Analysis of Free Volume Fluctuations.
Our explicit ligand trajectory simulations, though long enough to exhibit multiple full ligand migration events, only capture the fastest of them. We use the following geometric analysis—PathFinder—to complement the necessarily limited sampling of the phase space by explicit trajectories. In this approach, we analyze static snapshots of the protein structure generated by MD simulations to determine the spaces in the protein that do not exclude ligand solely on the basis of a steric potential model of the protein interior. This scaled particle (35, 36) viewpoint on migration pathways provides a picture of where a small nonpolar ligand can and cannot go in the protein matrix whose fluctuations are completely decoupled from the ligand motion.
Because the snapshots for this analysis come from separate, 15-ns long MD simulations in which the ligand is not allowed to move (see SI Appendix for details), these results are independent from the previously described analysis of actual ligand motion. Insofar as these simulations extend over time scales long enough to establish simulated protein compressibility in close agreement with experiment, we argue that our analysis captures the bulk of free volume fluctuations in the protein and thus possible dynamical pathways. The PathFinder algorithm, associated trajectories, and computation of protein compressibility are more fully described in SI Appendix.
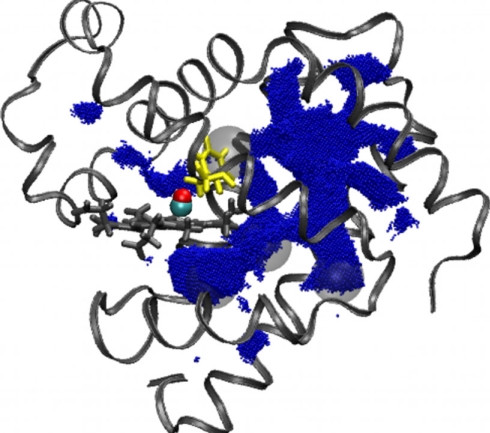
We first applied PathFinder to MD snapshots representing WT myoglobin. In each snapshot, we generally observe a pattern of only a few disconnected cavities large enough to hold the ligand; the pattern can vary significantly from snapshot to snapshot (1 ps apart), suggesting that the underlying structural fluctuations occur on time scales much shorter than that of ligand migration between the binding site and the solvent. The time integrated union of transient cavities from multiple snapshots delineates the pathways formed by free volume fluctuations. Comparison of the pathways formed by free volume fluctuations with the visitation map of explicit ligand migration trajectories is shown in Fig. 2. These side-by-side figures show remarkable qualitative agreement between the free volume analysis and the ligand migration trajectories. Moreover, detailed features such as the bottlenecks (shown in yellow in Fig. 1) that connect major and minor dynamical pathways are also observed in the free volume analysis. It is the existence of two distinct regions of conformational space available to ligand migration that rationalizes the proposed classification scheme based on major and minor pathways.
Fig. 2.
Comparison of ligand migration pathways determined by free volume analysis of transient cavities and mapping of explicit CO visitations. (Left) The results of the PathFinder analysis in the WT myoglobin, represented by an ensemble of 15,000 individual snapshots, 1 ps apart. Pathway volumes are represented by blue points where ligand molecules might be positioned in transient cavities large enough to hold the ligand. The CO is shown in the binding site. (Right) The positions occupied by CO traveling through the protein matrix in the MD trajectories in WT myoglobin. Light-blue points are positions of CO that exited/entered the protein via portals in the major pathway. Dark-blue points are positions of CO that entered/escaped via portals in the minor pathway. The coordinates are sampled every 10 ps. A movie showing a time series of a representative sample of the trajectories is available in Movie S2, in SI Appendix.
Synthesis of the explicit MD ligand trajectory data and free-volume fluctuation analysis allows us to better understand the atomic-level mechanism of ligand migration at various conceptual levels. In particular, it can be seen that the migration pathways are located between the α-helices of the myoglobin scaffold, as displayed in Fig. 1 Left. This is consistent with the relative structural incompressibility within α-helices: Large structural fluctuations that facilitate ligand migration occur in the “softer” space between the helices and in the flexible loop regions. The overall agreement between the ligand migration pathways inferred from explicit MD trajectories and the pattern of free space available to the ligand movement through free-volume fluctuations suggest that small nonpolar ligands such as CO do not play a major role in determining the location of these pathways. Rather, these pathways are, by and large, determined by the fluctuations of the protein matrix itself.
PathFinder analysis of the V68F mutant reveals that a bulky phenylalanine side-chain blocks the route between the distal pocket and the rest of the protein interior, as shown in Fig. 3. Thus, when the F68 side chain is in the position found in the x-ray structure, as it is in Fig. 3, ligand escape from the distal pocket can occur only through portals 1 (the “His gate”), 4, or 5. All other escape routes available in WT Mb are blocked in the V68F mutant. Indeed, two of four escape events in the photo-detached CO simulations discussed above occurred through portals 1 and 5 of the major pathway. The other two escapes from the distal pocket were coincident with a transient ≈90° rotation of the F68 side chain that opened the rest of the pathway: These two trajectories left the protein through portal 3 of the major pathway and through the minor pathway, respectively. We note that these two events could not be predicted based solely on our analysis of free-volume fluctuations for the time scales studied here. We speculate that time-resolved studies might be able to test our predictions of the effect of V68F mutation on the ligand migration pathways.
Fig. 3.
The results of PathFinder analysis in the V68F mutant of myoglobin. F68 is yellow. The F68 residue is shown in the position found in the x-ray structure. Each blue point represents a cavity large enough to hold a ligand molecule. The CO is shown in the binding site.
Conclusions
We conclude that CO migration in myoglobin occurs along two discrete pathways in the protein matrix: The major pathway is more frequently visited by ligand in both escape and inclusion trajectories than the minor pathway. Trajectories can hop between these pathways via two bottleneck regions. We also conclude that ligand enters and exits the protein matrix through nine distinct surface portals.
Two different types of computational studies independently and consistently support our identification of pathway, bottleneck, and portal features. In the first type of study, we explicitly simulated the room-temperature escape and inclusion dynamics of photo-detached and solvated CO, in both WT and mutant myoglobin. The time scales of these simulations extend into the leading edge of kinetically-relevant experimental times for CO diffusion in the protein matrix. Thus, our simulated trajectories feature multiple events of complete transit of ligand between its binding site on the heme group and the solvent, in both directions. In the second type of study, we identified sterically allowed pathways by integrating the free-volume fluctuations in simulations of WT and mutant myoglobin. The free-volume cavities accessible to ligand were determined by geometric analysis of structural snapshots using the PathFinder algorithm developed as part of this study. The map of ligand visitation sites explicitly observed in the simulated trajectories closely follows the independent mapping of integrated free-volume fluctuations. This remarkable consistency points to localized transient thermal fluctuations that occur in the protein structure as the likely primary physical origin of the migration pathways for small nonpolar ligands inside the protein. We suggest that the predicted location of the pathways relative to the protein scaffold—in between the helices and in the loop regions—is therefore determined by the natural propensity of larger fluctuations to occur in these “softer” domains of the protein matrix.
Significantly, the atomic-level characterization of ligand migration and other properties of myoglobin determined in this computational work are consistent with a number of independent experimental studies. These include Laue crystallographic studies, transient grating spectroscopy of ligand escape kinetics, and kinetic studies of ligand uptake in mutant variants. The MD simulations themselves are consistent with experimental measurements of overall myoglobin compressibility and structural identification of internal cavities through xenon inclusion studies. The CO bimolecular rebinding rate of ≈(0.3 μs M)−1 estimated from our simulations (see SI Appendix) is remarkably close to the experimental value of (0.5 μs M)−1 reported for CO by Olson and Phillips in their 1996 review (37). The corresponding estimated CO exit-to-solvent rate of (113 ns)−1 is consistent with earlier IR studies (37–39). Additionally, we point out that though the above results were obtained for CO ligand, experimental work suggests that the overall picture of migration channels should also hold for other nonpolar ligands of similar size including molecular oxygen (27).
The techniques featured here in the investigation of ligand diffusion in myoglobin—explicit full-ligand migration-trajectory analysis and geometric analysis of integrated free volume over shorter time spans—are conceptually simple, yet powerful enough to be able to make kinetic predictions from atomic structure of a medium-sized protein. These features should make the methodology particularly attractive for investigating structure–kinetics relationships in other biologically-relevant systems that have not been studied nearly as extensively as myoglobin.
Methods
MD Simulations.
All MD simulations are all-atom explicit solvent and are performed at 300 K. Each trajectory simulates 90 ns of ligand migration. Details of the MD protocols, timings, choice of force fields, and ligand and heme models are described in SI Appendix.
Structures.
We used two different structures for WT MbCO from the sperm whale: PDB ID 1A6G solved at 1.15-Å resolution by x-ray diffraction (40); and PDB ID 2MB5 solved by neutron diffraction (41). The V68F MbCO mutant was represented by PDB ID 1MLJ solved at 2.0-Å resolution by x-ray diffraction. Preparation of structures and insensitivity of the results presented here to the choice of structure are detailed in SI Appendix.
Analysis of Volume Fluctuations.
The steric potential model of the protein is built by assigning a hard-sphere van der Waals radius to each atom in the simulation; the ligand is modeled as a single hard sphere of appropriate radius. To compute the pattern of voids in the protein matrix available for ligand migration, we have developed the volumetric PathFinder algorithm, which finds the points in a static snapshot of protein (plus a solvent shell) structure on which we can place a single atom sphere without colliding with other atoms in the structure. By integrating free volumes over time, PathFinder provides an alternate picture of possible ligand migration pathways. Details of the algorithm, its implementation, and the input parameters are in SI Appendix.
Supplementary Material
Acknowledgments.
We thank N. Saha and J. Shu for work at the early stages of the project; N. Polys for graphics suggestions; C. Dahn and J. Ruscio for optimization of PathFinder; J. Olson for stimulating discussions; P. H. Mishra for the help with the computational analysis of myoglobin compressibility; and the Advanced Research Computing Facility (System X) at Virginia Tech for compute hours. We also thank reviewer “one” for sketching out how macroscopic kinetic rates could be estimated from these simulation results. This work was partially supported by National Institutes of Health Grant GM076121 and A Support Program for Innovative Research Strategies (ASPIRES) seed grant from Virginia Tech. M.G.P. and A.V.O. acknowledge National Science Foundation Grant CBA 9601399 for supporting earlier studies at Duke University that were a prelude to the present work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710825105/DCSupplemental.
References
- 1.Frauenfelder H, McMahon BH, Fenimore PW. Myoglobin: The hydrogen atom of biology and a paradigm of complexity. Proc Natl Acad Sci USA. 2003;100:8615–8617. doi: 10.1073/pnas.1633688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin RH, et al. Dynamics of carbon monoxide binding by heme proteins. Science. 1973;181:541–543. doi: 10.1126/science.181.4099.541. [DOI] [PubMed] [Google Scholar]
- 3.Henry ER, Sommer JH, Hofrichter J, Eaton WA. Geminate recombination of carbon monoxide to myoglobin. J Mol Biol. 1983;166:443–451. doi: 10.1016/s0022-2836(83)80094-1. [DOI] [PubMed] [Google Scholar]
- 4.Tilton RF, Jr, Kuntz ID, Jr, Petsko GA. Cavities in proteins: Structure of a metmyoglobin–xenon complex solved to 1.9 Å. Biochemistry. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 5.Elber R, Karplus M. Enhanced sampling in molecular dynamics: Use of the time-dependent hartree approximation for a simulation of carbon monoxide diffusion through myoglobin. J Am Chem Soc. 1990;112:9161–9175. [Google Scholar]
- 6.Schlichting I, Berendzen J, Phillips GN, Sweet RM. Crystal structure of photolysed carbonmonoxy-myoglobin. Nature. 1994;371:808–812. doi: 10.1038/371808a0. [DOI] [PubMed] [Google Scholar]
- 7.Srajer V, et al. Photolysis of the carbon monoxide complex of myoglobin: Nanosecond time-resolved crystallography. Science. 1996;274:1726–1729. doi: 10.1126/science.274.5293.1726. [DOI] [PubMed] [Google Scholar]
- 8.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Ligand binding and conformational motions in myoglobin. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 9.Chu K, et al. Structure of a ligand-binding intermediate in WT carbonmonoxy myoglobin. Nature. 2000;403:921–923. doi: 10.1038/35002641. [DOI] [PubMed] [Google Scholar]
- 10.Schotte F, et al. Watching a protein as it functions with 150-ps time-resolved x-ray crystallography. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 11.Lim M, Jackson TA, Anfinrud PA. Ultrafast rotation and trapping of carbon monoxide dissociated from myoglobin. Nat Struct Biol. 1997;4:209–214. doi: 10.1038/nsb0397-209. [DOI] [PubMed] [Google Scholar]
- 12.Dadusc G, Ogilvie JP, Schulenberg P, Marvet U, Miller RJ. Diffractive optics-based heterodyne-detected four-wave mixing signals of protein motion: from “protein quakes” to ligand escape for myoglobin. Proc Natl Acad Sci USA. 2001;98:6110–6115. doi: 10.1073/pnas.101130298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bossa C, et al. Extended molecular dynamics simulation of the carbon monoxide migration in sperm whale myoglobin. Biophys J. 2004;86:3855–3862. doi: 10.1529/biophysj.103.037432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossa C, et al. Molecular dynamics simulation of sperm whale myoglobin: effects of mutations and trapped CO on the structure and dynamics of cavities. Biophys J. 2005;89:465–474. doi: 10.1529/biophysj.104.055020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hummer G, Schotte F, Anfinrud P. Unveiling functional protein motions with picosecond x-ray crystallography and molecular dynamics simulations. Proc Natl Acad Sci USA. 2004;101:15330–15334. doi: 10.1073/pnas.0405295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perutz MF, Mathews FS. An x-ray study of azide methaemoglobin. J Mol Biol. 1966;21:199–202. doi: 10.1016/0022-2836(66)90088-x. [DOI] [PubMed] [Google Scholar]
- 17.Case DA, Karplus M. Dynamics of ligand binding to heme proteins. J Mol Biol. 1979;132:343–368. doi: 10.1016/0022-2836(79)90265-1. [DOI] [PubMed] [Google Scholar]
- 18.Ringe D, Petsko GA, Kerr DE, Ortiz de Montellano PR. Reaction of myoglobin with phenylhydrazine: A molecular doorstop. Biochemistry. 1984;23:2–4. doi: 10.1021/bi00296a001. [DOI] [PubMed] [Google Scholar]
- 19.Johnson KA, Olson JS, Phillips GN., Jr Structure of myoglobin-ethyl isocyanide. Histidine as a swinging door for ligand entry. J Mol Biol. 1989;207:459–463. doi: 10.1016/0022-2836(89)90269-6. [DOI] [PubMed] [Google Scholar]
- 20.Olson JS, et al. The role of the distal histidine in myoglobin and haemoglobin. Nature. 1988;336:265–266. doi: 10.1038/336265a0. [DOI] [PubMed] [Google Scholar]
- 21.Springer BA, et al. Discrimination between oxygen and carbon monoxide and inhibition of autooxidation by myoglobin. Site-directed mutagenesis of the distal histidine. J Biol Chem. 1989;264:3057–3060. [PubMed] [Google Scholar]
- 22.Bellelli A, Blackmore RS, Gibson QH. Ligand binding to a hemoprotein lacking the distal histidine. The myoglobin from aplysia limacina (Val(E7)) J Biol Chem. 1990;265:13595–13600. [PubMed] [Google Scholar]
- 23.Scott EE, Gibson QH, Olson JS. Mapping the pathways for O2 entry into and exit from myoglobin. J Biol Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 24.Huang X, Boxer SG. Discovery of new ligand binding pathways in myoglobin by random mutagenesis. Nat Struct Biol. 1994;1:226–229. doi: 10.1038/nsb0494-226. [DOI] [PubMed] [Google Scholar]
- 25.Nishihara Y, Sakakura M, Kimura Y, Terazima M. The escape process of carbon monoxide from myoglobin to solution at physiological temperature. J Am Chem Soc I. 2004;126:11877–11888. doi: 10.1021/ja038877w. [DOI] [PubMed] [Google Scholar]
- 26.Cohen J, Arkhipov A, Braun R, Schulten K. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Biophys J. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tetreau C, Blouquit Y, Novikov E, Quiniou E, Lavalette D. Competition with xenon elicits ligand migration and escape pathways in myoglobin. Biophys J. 2004;86:435–447. doi: 10.1016/S0006-3495(04)74120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunori M, Gibson QH. Cavities and packing defects in the structural dynamics of myoglobin. EMBO Rep. 2001;2:674–679. doi: 10.1093/embo-reports/kve159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourgeois D, et al. Complex landscape of protein structural dynamics unveiled by nanosecond Laue crystallography. Proc Natl Acad Sci USA. 2003;100:8704–8709. doi: 10.1073/pnas.1430900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teeter MM. Myoglobin cavities provide interior ligand pathway. Protein Sci. 2004;13:313–318. doi: 10.1110/ps.03334304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoenborn BP. Structure of alkaline metmyoglobin–xenon complex. J Mol Biol. 1969;45:297–303. doi: 10.1016/0022-2836(69)90106-5. [DOI] [PubMed] [Google Scholar]
- 32.Egeberg KD, et al. The role of Val68(E11) in ligand binding to sperm whale myoglobin. Site-directed mutagenesis of a synthetic gene. J Biol Chem. 1990;265:11788–11795. [PubMed] [Google Scholar]
- 33.Quillin ML, et al. Structural and functional effects of apolar mutations of the distal valine in myoglobin. J Mol Biol. 1995;245:416–436. doi: 10.1006/jmbi.1994.0034. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Baskin J, Xia T, Zewail A. Human myoglobin recognition of oxygen: Dynamics of the energy landscape. Proc Natl Acad Sci USA. 2004;101:18000–18005. doi: 10.1073/pnas.0408379102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiss H. Scaled particle methods in the statistical thermodynamics of fluids. Adv Chem Phys. 1966;9:1–84. [Google Scholar]
- 36.Rashin A, Iofin M, Honig B. Internal cavities and buried waters in globular proteins. Biochemistry. 1986;25:3619–3625. doi: 10.1021/bi00360a021. [DOI] [PubMed] [Google Scholar]
- 37.Olson JS, Phillips GN., Jr Kinetic pathways and barriers for ligand binding to myoglobin. J Biol Chem. 1996;271:17593–17596. doi: 10.1074/jbc.271.30.17593. [DOI] [PubMed] [Google Scholar]
- 38.Lim M, Jackson TA, Anfinrud PA. Binding of CO to myoglobin from a heme pocket docking site to form nearly linear Fe-C-O. Science. 1995;269:962–966. doi: 10.1126/science.7638619. [DOI] [PubMed] [Google Scholar]
- 39.Lim M, Jackson TA, Anfinrud PA. Mid-infrared vibrational spectrum of CO after photodissociation from heme: Evidence for a ligand docking site in the heme pocket of hemoglobin and myoglobin. J Chem Phys. 1995;102:4355–4366. [Google Scholar]
- 40.Vojtchovský J, Chu K, Berendzen J, Sweet RM, Schlichting I. Crystal structures of myoglobin-ligand complexes at near-atomic resolution. Biophys J. 1999;77:2153–2174. doi: 10.1016/S0006-3495(99)77056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng XD, Schoenborn BP. Neutron diffraction study of carbonmonoxymyoglobin. J Mol Biol. 1991;220:381–399. doi: 10.1016/0022-2836(91)90020-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.