Abstract
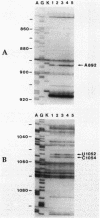
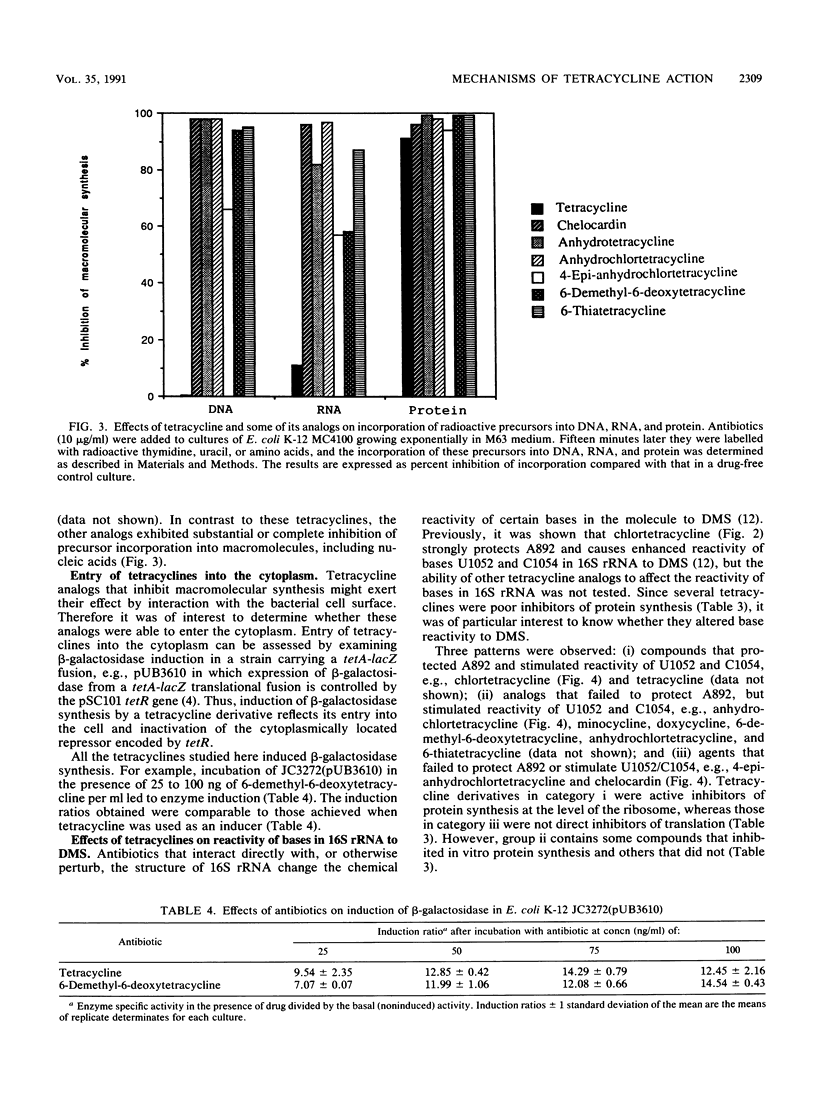
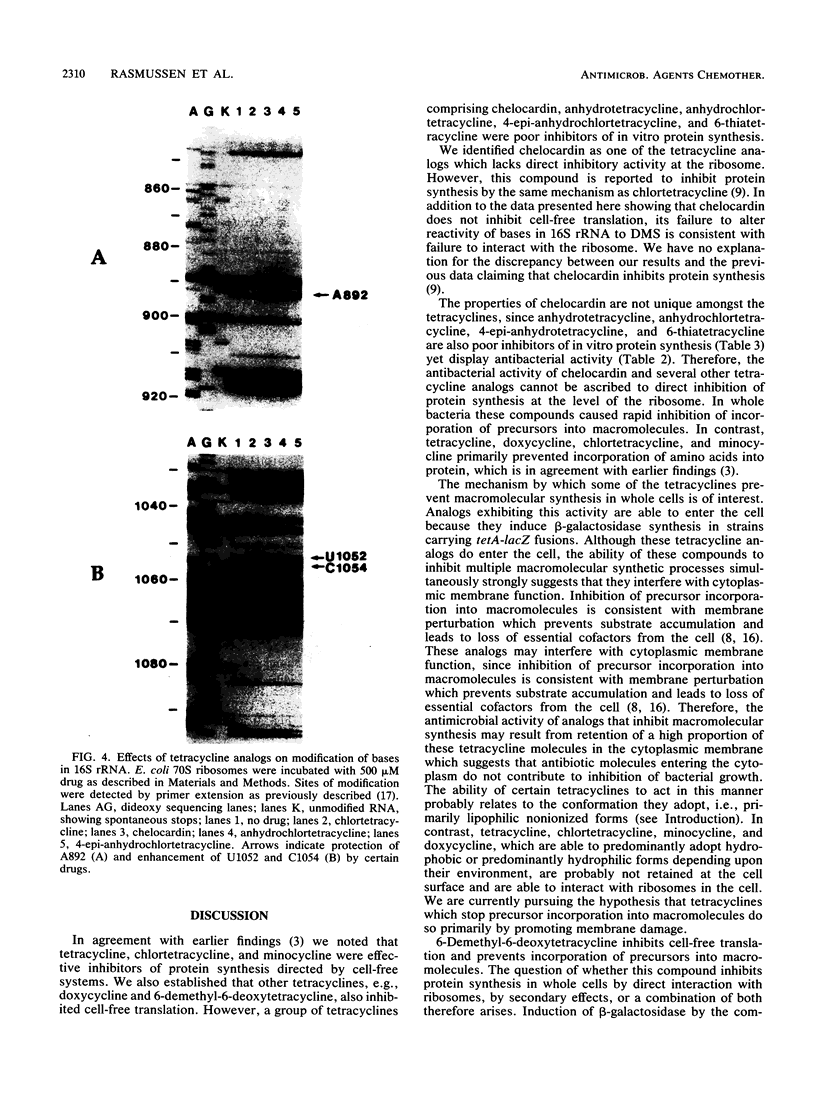
Tetracycline analogs fell into two classes on the basis of their mode of action. Tetracycline, chlortetracycline, minocycline, doxycycline, and 6-demethyl-6-deoxytetracycline inhibited cell-free translation directed by either Escherichia coli or Bacillus subtilis extracts. A second class of analogs tested, including chelocardin, anhydrotetracycline, 6-thiatetracycline, anhydrochlortetracycline, and 4-epi-anhydrochlortetracycline, failed to inhibit protein synthesis in vitro or were very poor inhibitors. Tetracyclines of the second class, however, rapidly inhibited the in vivo incorporation of precursors into DNA and RNA as well as protein. The class 2 compounds therefore have a mode of action that is entirely distinct from the class 1 compounds, such as tetracycline that are used clinically. Although tetracyclines of the second class entered the cytoplasm, the ability of these analogs to inhibit macromolecular synthesis suggests that the cytoplasmic membrane is their primary site of action. The interaction of class 1 and class 2 tetracyclines with ribosomes was studied by examining their effects on the chemical reactivity of bases in 16S rRNA to dimethyl sulfate. Class 1 analogs affected the reactivity of bases to dimethyl sulfate. The response with class 2 tetracyclines varied, with some analogs affecting reactivity and others (chelocardin and 4-epi-anhydrotetracycline) not.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cammack K. A., Wade H. E. The sedimentation behaviour of ribonuclease-active and -inactive ribosomes from bacteria. Biochem J. 1965 Sep;96(3):671–680. doi: 10.1042/bj0960671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chopra I., Hacker K., Misulovin Z., Rothstein D. M. Sensitive biological detection method for tetracyclines using a tetA-lacZ fusion system. Antimicrob Agents Chemother. 1990 Jan;34(1):111–116. doi: 10.1128/aac.34.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. Cell-free synthesis of proteins coding for mobilisation functions of ColE1 and transposition functions of Tn3. Gene. 1979 May;6(1):29–42. doi: 10.1016/0378-1119(79)90083-0. [DOI] [PubMed] [Google Scholar]
- Konisky J. Colicins and other bacteriocins with established modes of action. Annu Rev Microbiol. 1982;36:125–144. doi: 10.1146/annurev.mi.36.100182.001013. [DOI] [PubMed] [Google Scholar]
- Mao J. C., Robishaw E. E. Mode of action of beta-chelocardin. Biochim Biophys Acta. 1971 Apr 29;238(1):157–160. doi: 10.1016/0005-2787(71)90019-0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed D., Van Stolk B. J., Douthwaite S., Noller H. F. Interconversion of active and inactive 30 S ribosomal subunits is accompanied by a conformational change in the decoding region of 16 S rRNA. J Mol Biol. 1986 Oct 5;191(3):483–493. doi: 10.1016/0022-2836(86)90143-9. [DOI] [PubMed] [Google Scholar]
- Sahl H. G. Influence of the staphylococcinlike peptide Pep 5 on membrane potential of bacterial cells and cytoplasmic membrane vesicles. J Bacteriol. 1985 May;162(2):833–836. doi: 10.1128/jb.162.2.833-836.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Zubay G. In vitro synthesis of protein in microbial systems. Annu Rev Genet. 1973;7:267–287. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]