Abstract
Maintenance of lasting synaptic efficacy changes requires protein synthesis. We report here a mechanism that might influence translation control at the level of the single synapse. Stimulation of metabotropic glutamate receptors in hippocampal slices induces a rapid protein kinase C-dependent translocation of multifunction kinase p90rsk to polyribosomes; concomitantly, there is enhanced phosphorylation of at least six polyribosome binding proteins. Among the polyribosome bound proteins are the p90rsk-activating kinase ERK-2 and a known p90rsk substrate, glycogen synthase kinase 3β, which regulates translation efficiency via eukaryotic initiation factor 2B. Thus metabotropic glutamate receptor stimulation could induce synaptic activity-dependent translation via translocation of p90rsk to ribosomes.
Synaptic regulation of local protein synthesis is attractive from the viewpoints of developmental plasticity and adult memory (1). The long-term maintenance of induced changes in synaptic efficacy requires protein synthesis; resulting localized alterations in protein composition could restrict modifications to activated synapses, without affecting inactive synapses of the same neuron. One mechanism proposed for this is “synaptic tagging,” in which locally activated kinases or phosphoproteins sequester relevant proteins synthesized in the cell body (2). A more direct mechanism is locally regulated protein synthesis at the synapse; in fact, protein synthesis has been observed in distal dendritic processes (3, 4) and in response to neurotransmitter administration in hippocampal slices (5). Local postsynaptic synthesis of proteins is supported by (i) the postsynaptic presence of polyribosomes (6–8), (ii) dendritically localized mRNAs (9–11), (iii) postsynaptic presence of components for translation (11); and (iv) the translation of transfected reporter-tagged mRNA in dendrites (12).
Protein translation is controlled by various translation factors (reviewed in refs. 13 and 14) and by other proteins that are tightly associated with polyribosomal aggregates. Many of them are phosphoproteins, and the state of their phosphorylation determines their effect on protein synthesis (15). The effects of phosphorylation of some translation factors have been described under in vitro conditions and in various non-neuronal cultured cells. Thus, the kinase p70S6K up-regulates translation of 5′ target of puromycin mRNAs upon mitogenic stimulation (16). Rapid stimulation of translation in PC12 and some other cell lines, in response to insulin or growth factors, is mediated by the phosphorylation state of eukaryotic initiation factor (eIF) 2B and the availability of free eIF4E, controlled by the phosphorylation degree of eIF4E-binding proteins (PHAS-1 and PHAS-2) (17–20). However, in neuronal tissue no receptor-mediated mechanism that up-regulates localized postsynaptic translation has been described. A receptor-mediated inhibition of protein synthesis in neurons has been described that involves N-methyl-d-aspartate-induced phosphorylation of eukaryotic elongation factor 2 (eEF-2) by activation of calcium/calmodulin-dependent protein kinase III (EF-2 kinase) (21, 22).
We previously have described polyribosomal formation and protein synthesis in response to group I metabotropic glutamate receptor (mGluR) agonist administration to a synaptoneurosome preparation and have shown this to require receptor-coupled phospholipase C activation of protein kinase C (PKC) (23–25). We report here specific mechanisms that appear to link PKC activation to polyribosomal formation and protein synthesis.
MATERIALS AND METHODS
In Situ Phosphorylation of Polyribosome-Binding Proteins (PRBPs).
Ten transverse hippocampal slices from single 12-day-old Long-Evans rats were prepared and incubated in an interface chamber in a medium containing 134 mM NaCl, 6.24 mM KCl, 1.3 mM MgSO4, 2 mM CaCl2, 16 mM NaHCO3, and 10 mM glucose (pH 7.4, 32°C). The medium was aerated with carbogen (95% O2, 5% CO2) throughout the experiment. After 60 min of preincubation, 200 μCi 33Pi (final concentration in the medium 100 μCi/ml, Amersham) was added, and the slices were labeled for 90 min. After removing the control slices, the remaining slices were stimulated for 10 min either with 0.1 mM 3,5-dihydroxyphenylglycine (DHPG) or 1 μM phorbol 12,13-dibutyrate (PDBu). Inhibitors were applied 10 min before stimulation. Immediately after incubation the slices were homogenized in ice-cold buffer A (125 mM NaCl/100 mM sucrose/50 mM Hepes/2 mM potassium acetate/2 mM DTT/1 μM okadaic acid, pH 7.5), treated with Triton X-100 (final concentration 1.2%), and centrifuged for 10 min at 10,000 g. The supernatants were centrifuged through 1 M sucrose at 400,000 g for 11 min, and the resulting pellet (polysomes) was washed in a buffer containing 50 mM Tris⋅HCl (pH 7.5), 1 mg/ml of heparin, 20 mM EDTA, 2 mM EGTA, 0.1 mM sodium orthovanadate, 20 μg/ml of aprotinin, 10 μg/ml of leupeptin, and 0.1 mg/ml of phenylmethylsulfonyl fluoride. The PRBPs then were released by a buffer containing 50 mM Tris⋅HCl (pH 7.5), 0.5 M potassium acetate, 100 mM NaF, 10 mM Na4P2O7, and the same protease inhibitors. The eluates were precipitated by 30% trichloroacetic acid, and the resulting pellet was washed with ethanol and dissolved in 60 μl of SDS-sample buffer (250 mM Tris/50 mM DTT/3 mM EDTA/4% SDS/20% glycerol, pH 8.0).
SDS/PAGE Analysis.
From the resulting sample 30 μl was used for electrophoretic separation using a 5–20% gradient polyacrylamide SDS gel. The gel was dried, and the phosphoproteins were detected by phosphoimaging using the Bio-Imaging Analyzer BAS1000 (FUJIX, Tokyo). The phosphorylation degree of different proteins was quantified by scanning densitometry (FUJIX system software); stimulus-induced effects were calculated by measuring the appropriate peak areas and expressed as percent of the unstimulated sample.
In Vitro Phosphorylation Assays.
Synaptoneurosomes were prepared as described (24). PRBPs were prepared as above, and for each phosphorylation assay 10 μg of protein was used. The assay (65 μl) for determination of endogenous kinase activity included 24 mM Tris⋅HCl (pH 7.5), 12 mM MgCl2, and 8% glycerol (vol/vol). The protein kinase C assay (65 μl) included 24 mM Tris⋅HCl (pH 7.5), 12 mM MgCl2, 10 ng PKC (Upstate Biotechnology, Lake Placid, NY), alternatively 0.6 mM CaCl2 or 0.5 mM EGTA and 7.5 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)/Tris, pH 7.5 (1.2:1.2) or phosphatidylserine (100 μg/ml)/dioleoyl-sn-glycerol (20 μg/ml). The p90rsk-2 phosphorylation assay (65 μl) included 24 mM Tris⋅HCl (pH 7.5), 12 mM MgCl2, 8% glycerol (vol/vol), and 1 unit of p90rsk-2 (Upstate Biotechnology). The protein kinase A (PKA) assay (100 μl) included 50 mM Tris⋅HCl (pH 7.4), 10 mM MgSO4, 1 mM EGTA, 1 mM DTT, and 0.1 pmolar units of PKA (Sigma) and alternatively, 0.02 mM cAMP. The tubes were preincubated for 1 min at 30°C, and the reaction was started by addition of 33γ-ATP (50 μM, 1 μCi, Amersham). The reaction was terminated after 5 min by addition of 1 ml of ice-cold 30% trichloroacetic acid. After 60 min on ice the tubes were centrifuged for 15 min at 17,000 g, and the resulting pellet was washed twice with 500 μl of ethanol and dissolved in 60 μl of sample buffer. Samples of 25 μl were used for gel electrophoresis followed by autoradiography.
Western Blot Assay.
For the detection or determination of the amount of different kinases bound to polyribosomes, equal amounts of PRBPs (about 30 μg) from unstimulated and stimulated synaptoneurosomes were loaded on a gel (5–20% acrylamide). After separation proteins were electrotransferred onto nitrocellulose (0.45 μm pore size) in a transfer buffer (25 mM Tris/192 mM glycine/0.02% SDS/20% methanol) for 90 min at a constant current (200 mA) by using a tank blotting system. Additional protein binding sites on the nitrocellulose were saturated by incubation in 10 mM Tris-buffered saline with 0.1% Tween 20 (TBST; pH 7.4) containing 5% dry milk powder for 1 h at room temperature. After a short wash with TBST, an antibody specific for p90rsk-1 (rabbit polyclonal IgG, Santa Cruz Biotechnology), p90rsk-2 (goat polyclonal IgG (Santa Cruz Biotechnology), glycogen synthase kinase 3β (mAb, Transduction Laboratories, Lexington, KY), ERK-2 (mAb, Santa Cruz Biotechnology), or p70S6K (rabbit polyclonal IgG, Santa Cruz Biotechnology) was applied overnight at 4°C. The nitrocellulose was washed with TBST, and the immunoreactivity was revealed by using an appropriate peroxidase-conjugated anti-mouse IgG (Sigma, 1:10,000), anti-rabbit (Sigma, 1:7,500), or anti-goat antibody (Sigma 1:5,000) and the ECL system (Pierce).
RESULTS
Rat hippocampal slices were used to study changes in protein phosphorylation in brain tissue, under conditions that are closely related to normal in vivo situations. This use of slice tissue means that changes detected in protein phosphorylation represent the involvement of a whole phosphorylation system, including a given kinase, possible other downstream-activated kinases, and related phosphatases. To examine links between PKC activation and protein translation we focused on phosphoproteins tightly bound to polyribosomes, which are resistant to removal by heparin-containing buffers but are eluted by 0.5 M potassium acetate.
In untreated hippocampal slices from 12-day-old rats we detected a basal endogenous phosphorylation of at least six heparin-resistant PRBPs (19, 22, 33, 48, 85, and 90 kDa), pointing to an ongoing kinase activity affecting these substrates. This basal substrate phosphorylation was not changed if PKC inhibitors (calphostin C or GF109203X) were present in the incubation medium, indicating that some other kinases must be responsible. Stimulation with 0.1 mM DHPG (a mGluR1 agonist) for 10 min increased the phosphorylation of these proteins (Fig. 1, Table 1). Because DHPG can activate both PKC and PKA, we separated the effects of these two kinases by activating either PKC alone (with phorbol ester, PDBu), or PKA (by increasing the intracellular cAMP concentration with forskolin, an adenylate cyclase activator). Although forskolin had no effect, addition of 1 μM PDBu for 10 min increased endogenous PRBP phosphorylation in a manner similar to that after DHPG stimulation (Fig. 1). Furthermore, inhibition of PKC activity during DHPG stimulation by 10 μM calphostin C or 10 μM GF109203X blocked the increase in PRBP phosphorylation, indicating that PKC plays a crucial role in the increased phosphorylation of PRBP after mGluR stimulation, in contrast to the basal level that does not require PKC. Thus, phosphorylation experiments performed on hippocampal slices point to a certain basal level of kinase activity, maintaining a steady phosphorylation of at least six PRBP. After stimulation of mGluR by DHPG, phosphorylation is increased, either by PKC itself or by PKC-dependent activation of other kinases.
Figure 1.
PhosphorImager record of in situ phosphorylation of PRBPs isolated from hippocampal slices of 12-day-old rats, labeled with 33P. (A) Stimulation with 0.1 mM DHPG for 10 min increased endogenous phosphorylation of proteins with molecular masses of 90, 85, 48, 33, 22, and 19 kDa (arrows). A comparable effect could be observed after addition of a PKC activator, 1 μM PDBu, but not after stimulation with 50 μM forskolin. This enhanced phosphorylation elicited by DHPG was not detectable if 10 μM calphostin C, an inhibitor of PKC, or 10 μM GF109203X, an inhibitor of both PKC and p90rsk, was present during stimulation. (B) Scanning profiles obtained from the shown autoradiographs. Black line: unstimulated samples; gray line: stimulated samples (DHPG, PDBu, and forskolin). Arrows indicate peaks corresponding to the indicated bands in the autoradiography.
Table 1.
Changes in the phosphorylation of different PRBPs isolated from hippocampal slices of 12-day-old rats after 10-min stimulation with 0.1 mM DHPG
| Protein | % control | SEM | N |
|---|---|---|---|
| 90 kDa | 163.9** | 22.9 | 8 |
| 85 kDa | 136.2 | 8.7 | 5 |
| 48 kDa | 166.2** | 15.1 | 8 |
| 33 kDa | 174.1* | 11.3 | 6 |
| 22 kDa | 144.0* | 11.2 | 8 |
| 19 kDa | 146.4* | 10.6 | 8 |
Control = 100%, **P < 0.01; *P < 0.05, according to Wilcoxon pair test.
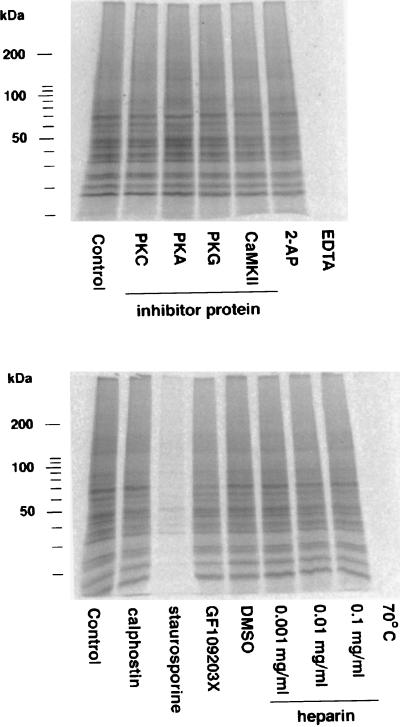
We then performed an in vitro phosphorylation assay to study the phosphorylation of the PRBP by PKC. To enrich for postsynaptic polyribosomes, we prepared synaptoneurosomes, synaptically joined intact presynaptic and postsynaptic entities (24, 26), from 12-day-old rat cerebral cortex, and eluted the PRBP from these polyribosomes with 0.5 M K+ after a heparin wash. Surprisingly, in a control sample without exogenous added PKC, we detected a strong kinase among the eluted proteins. This kinase activity was not influenced by the addition of specific inhibitor peptides against PKA, PKC, protein kinase G, or CaMKII, by 2-aminopurine (nonspecific PKR inhibitor), or heparin (casein kinase II inhibitor). Therefore none of these kinases is responsible for the observed phosphorylation activity in the PRBP fraction. However, 10 μM staurosporine (a nonspecific inhibitor of several Ser/Thr kinases), 50 mM EDTA, or heating the protein (70°C, 5 min) blocked the kinase activity (Fig. 2).
Figure 2.
Detection of polyribosome-bound kinase activity. In vitro phosphorylation assay of PRBP (10 μg per lane) prepared from cerebral cortical synaptoneurosomes from 12-day-old rats (24). This fraction includes ribosome-bound kinases as well as substrates. Incorporated 33P was detected by PhosphorImaging. The activity of the polyribosome-bound kinases was not blocked by addition of specific inhibitor peptides against PKC (fragment 19–36, Sigma), PKA (PKI, Sigma), PKG (Arg-Lys-Arg-Ala-Arg-Lys-Glu, Sigma), or CaMKII (fragment 290–309, Sigma), each 2 μg per assay, or inhibitors against protein kinase R (100 mM 2-aminopurine = 2-AP), casein kinase II (increasing concentrations of heparin), PKC (10 μM calphostin C). GF109203X (10 μM), an inhibitor of PKC and p90rsk, had a small inhibitory effect. The kinase activities could be blocked completely by capturing divalent cations (Mg2+, Ca2+) with 50 mM EDTA or heating the fraction to 70°C for 5 min and inhibited almost completely by 10 μM staurosporine, a nonspecific inhibitor of Ser/Thr kinases.
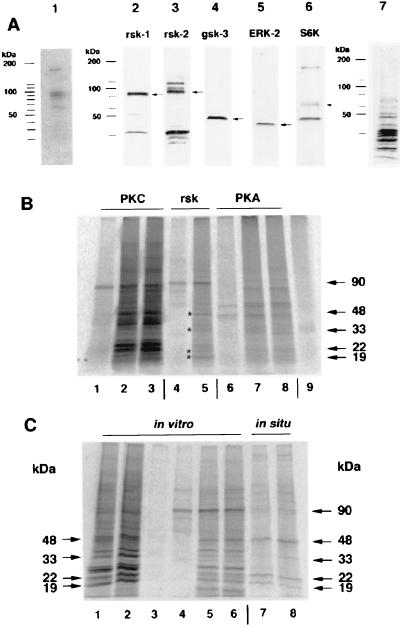
To identify the unknown polyribosome-bound kinase(s) we transferred the electrophoretically separated PRBPs to a poly(vinylidene difluoride) membrane, renatured the proteins in a renaturation buffer (10 mM Tris⋅HCl/140 mM NaCl/2 mM EDTA/2 mM DTT/1% BSA/0.1% Nonidet P-40) and allowed them to autophosphorylate in the presence of added Mg2+ and [γ-33P]ATP (27). This assay revealed five autophosphorylating kinases among PRBPs, with molecular masses of 180, 95, 90, 85, and 65 kDa (Fig. 3A). In addition to these kinases, three other kinases with molecular masses of 130, 40, and 42 kDa could be detected if this kinase assay was performed with a peptide in the gel matrix corresponding to the phosphorylation site of eIF2B (28, 29). The 85-kDa kinase was identified as p90rsk-1 by two different antisera to p90rsk-1. The related kinase p90rsk-2 with a molecular mass of 90 kDa is also present in this fraction. Furthermore, p70S6K (65 kDa), glycogen synthase kinase 3β (gsk-3β, 49 kDa), and ERK-2 (42 kDa) could be identified by Western blot assay (Fig. 3A).
Figure 3.
A group of kinases is bound to polyribosomal complexes prepared from synaptoneurosomes. (A) Detection of autophosphorylation activity in kinases from the 0.5 M K+ fraction. Lane 1, PRBPs (25 μg) were prepared as described (15, 16) and after electrophoretic separation blotted to a poly(vinylidene difluoride) membrane. The proteins were renatured, and the presence of kinases was detected by autophosphorylation in the presence of [γ-33P]-ATP according to the method of Ferrell (27). This procedure revealed five kinases with molecular masses of about 65, 85, 90, 95, and 180 kDa. Identification of different kinases among PRBP by Western blot assay. Lane 2, p90rsk-1. Lane 3, p90rsk-2, the antiserum to p90rsk-2 crossreacts with two unknown additional proteins with molecular masses of about 100 and 115 kDa. Lane 4, gsk-3β. Lane 5, ribosomal S6 kinase p70S6K. Lane 6, ERK-2 (p42MAPK). Lane 7, an amido black protein staining of PRBPs on nitrocellulose. (B) In vitro phosphorylation of PRBPs by added kinases after blocking the activity of all endogenous kinases by heating the fraction to 70°C for 5 min. Lane 1, autophosphorylated form of PKC alone; lane 2, PRBP and PKC without activators; lane 3, PRBP and PKC with activators; lane 4, autophosphorylated p90rsk-2; lane 5, PRBP and p90rsk-2 (* indicates the position of proteins that are also phosphorylated in situ); lane 6, autophosphorylated PKA; lane 7, PRBP and PKA; lane 8, PRBP, PKA, and cAMP; lane 9, PRBP. Note that the specific activities of the kinases are not equal, because p90rsk-2 is only partially purified. (C) Phosphorylation pattern of PRBP under different conditions. Lanes 1–2, all polyribosome-bound kinases (lane 1 with 10 μM GF109203X and lane 2 without inhibitor). Lanes 3–6, effect of p90rsk-2 (lane 3, heated PRBP; lane 4, p90rsk-2 alone, lane 5 and 6 p90rsk-2 with heated PRBP). Lanes 7–8, phosphorylation pattern of PRBP under in situ conditions. All endogenous phosphorylated proteins (arrows on right) are in vitro substrates for p90rsk-2. The 85- and 90-kDa protein phosphorylated under in situ conditions might be p90rsk-1 and p90rsk-2 itself.
To characterize possible substrates of p90rsk-2, PKC, and PKA, we inhibited endogenous kinase activity in the PRBP fraction by heating (5 min, 70°C), then added back each kinase in commercially available partially purified form. The 19- and 22-kDa proteins were phosphorylated by all three kinases although to different degrees. The 48-kDa protein was affected by both PKC and p90rsk-2, and the 33-kDa protein was a good substrate for p90rsk-2, but a poor substrate for PKC (Fig. 3B). The addition of exogenous p90rsk-2 alone produced the same phosphorylation pattern as the native set of proteins (minus the 85-kDa protein) (Fig. 3C).
We next measured the endogenous phosphorylation of ribosome-bound proteins in DHPG-treated hippocampal slices to verify that the in vitro phosphorylation corresponds to an effect in hippocampal slices. Five of the six phosphoproteins that showed increased phosphorylation after mGluR stimulation of slices were identical in size to PRBP substrates phosphorylated in vitro by p90rsk-2; the remaining 85-kDa band was possibly the p90rsk-1 kinase itself (Fig. 3 A and C).
Thus the in vitro phosphorylation assays revealed the existence of kinases that are bound tightly to polyribosomes. PKC, which is not tightly associated to polyribosomes, can phosphorylate some of the phosphoproteins involved, but did not affect the 85- and 90-kDa proteins and affected the 33-kDa protein only weakly. Surprisingly, ribosome-bound p90rsk can phosphorylate all of the substrates that correspond to the PRBPs prepared from hippocampal slices. Thus, p90rsk may represent a likely pathway mediating the effects of mGluR stimulation.
Because PKC activation in hippocampal slices resulted in increased phosphorylation of all polysome-associated p90rsk substrates, we asked whether PKC might activate the p90rsk already bound to ribosomes, or whether it might mediate a translocation of the kinase to ribosomes. We found no evidence that PKC itself phosphorylates the p90rsk (Fig. 3B, lane 3), and there is no evidence that the specific kinase activity of p90rsk was changed. However, antibody staining of p90-rsk-1 showed that DHPG stimulation results in increased p90rsk-1 binding to polyribosomes in hippocampal slices (Fig. 4A, Left) indicating translocation of additional p90rsk-1 to polyribosomes. To prove that this effect did not merely reflect the increased polyribosomal complex formation observed previously after mGluR stimulation (25), we also used a Western blot of a synaptoneurosomal preparation to determine the relative amount of p90rsk in the whole PRBP fraction by measuring the density of bands stained by anti-p90rsk-1. A cortical synaptoneurosome preparation was divided into identical aliquots; stimulation of synaptoneurosomes by 0.1 mM DHPG (for 1.5 min) increased the relative amount of polyribosome-associated p90rsk-1 by 73.5% (n = 9, P < 0.05, Fig. 4A, Center). Furthermore, DHPG stimulation in the presence of 1 μM calphostin C, a specific PKC inhibitor, did not cause this effect (Fig. 4A), confirming that PKC mediates translocation of p90rsk-1 to polyribosomes. We also found both the p90-rsk-activating kinase ERK-2 and gsk-3β, a known substrate for p90rsk, among the proteins bound tightly to polyribosomes; however, the amount of both kinases did not change after receptor stimulation (Figs. 3A and 4B).
Figure 4.
mGluR induced translocation of p90rsk-1 to polyribosomes. (A, Left) Stimulation of hippocampal slices with 0.1 mM DHPG for 10 min induced an increase in the amount of polyribosome-bound p90rsk-1. This increase is mediated by an enhanced binding of p90rsk-1 to polyribosomes as revealed by Western blot assay. (Center) PRBPs were prepared from cerebral cortical synaptoneurosomes that were untreated (C), stimulated with 0.1 mM DHPG for 1.5 min (DHPG), or stimulated with 0.1 mM DHPG in presence of 10 μM calphostin C (DHPG/calph.). Identical amounts of proteins were loaded on each lane. P90rsk-1 is indicated by an arrow. (Right) Quantification of nine independent experiments; C = control 100%; *P < 0.05%, Wilcoxon pair test. (B) Blots were washed and restained with antibody to gsk-3β; no significant changes in the amount of polyribosome-bound gsk-3β were found (n = 4).
DISCUSSION
We demonstrate that p90rsk-1, p90rsk-2, gsk-3β, ERK-2, and p70S6K kinases are components of the protein translation complex. The only kinases previously described from the ribosomal fraction are a heparin-sensitive, casein kinase II-related kinase (30, 31), and protein kinase R (PKR or double-stranded RNA-activated protein kinase; ref. 32). We could not detect a heparin-sensitive kinase in our fraction, even with a high concentration of this inhibitor; possibly it was removed or inhibited by pretreatment of the polyribosomal pellet with the heparin/EDTA buffer. PKR binds primarily to 40 and 60 S ribosome subunits; it is thought to be activated by double-stranded RNA during viral infection and to inhibit protein translation by phosphorylation of eIF2α (33). There is no evidence for a basal activity of ribosome-bound PKR in our assay because the addition of 2-aminopurine, an inhibitor of this kinase, had no effect.
The multifunction kinase p90rsk is a member of a family of at least three different serine-threonine kinases (rsk 1–3) with an unusual structure consisting of two nonidentical kinase domains. The N-terminal domain is chiefly responsible for substrate phosphorylation; both domains are responsible for intramolecular autophosphorylation (34–36). P90rsk was described originally as a kinase that phosphorylates ribosomal S6 protein in vitro (37), but it subsequently was shown that the unrelated kinase p70S6K is responsible for in vivo phosphorylation of S6 (38–40). No direct association of p90rsk with ribosomal complexes has been described so far. A broad range of different stimuli activate this kinase, including growth factors, insulin, heat shock, and phorbol ester or oncogenic transformations (41–43); activation requires Ser/Thr phosphorylation. The mitogen-activated protein kinases (MAP) ERK-1 (p44MAPK) and ERK-2 (p42MAPK) have been implicated as upstream activators and a MAP kinase-dependent translocation of p90rsk into the nucleus has been observed (36). Interestingly, ERK-2 is also tightly associated to the translational complex; however, the amount does not change upon receptor stimulation, which might indicate that activation of p90rsk occurs after translocation to polyribosomes. A number of known substrates for the p90rsk are localized in the nucleus, such as c-fos, serum response factor, Nur 77, and the cyclic AMP response element-binding protein CBP (44), whereas others are cytosolic, e.g., IκBα, SOS, and gsk-3β (45–48).
In addition to the p90rsk upstream-activating kinase, ERK-2, the p90rsk-regulated kinase, gsk-3β, also is associated tightly to polyribosomes. The changed ratio between the two kinases, p90rsk and gsk-3β, might control the general efficiency of translation. Stimulation by mGluR induced a translocation of p90rsk but not gsk-3β to the ribosomes. This proximity might be important, because gsk-3β is a constitutively active kinase and phosphorylation by p90rsk inhibits its kinase activity (47). One of the known gsk-3β substrates is the translation initiation factor eIF2B. Phosphorylation of this translation initiation factor decreases translation (49, 50). Thus, inhibition of gsk-3β by p90rsk phosphorylation would result in a rapid increase in translation.
Because PKC does not directly phosphorylate p90rsk, it might act by phosphorylation of an anchoring protein. Further studies are necessary to identify the binding partner that fixes the kinase to the polyribosomal complex. However, ERK-2 or gsk-3β are possible binding partners there.
Increased phosphorylation of a set of PRBPs in response to mGluR stimulation suggests a possible mechanism for a synaptically driven control of protein translation in neurons. Among these tightly bound proteins are kinases that have not previously been found to be associated with polyribosomes. We identified p90rsk-1, p90rsk-2, ERK-2, and gsk-3β as polysome-associated proteins. Stimulation of mGluR induced a PKC-dependent translocation of p90rsk to polyribosomes within 90 sec. Possibly, the already polyribosome-bound ERK-2 then might completely activate p90rsk, which, in turn, could up-regulate translation by phosphorylating and inhibiting gsk-3β. We therefore propose that mGluR activation may prime protein synthesis via p90rsk translocation to polyribosomes and subsequent phosphorylation of gsk-3β.
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (An200/2–1) to F.A., the FRAXA Research Foundation, and National Institutes of Health Grants MH 35321 and R03HD35565-01.
ABBREVIATIONS
- mGluR
metabotropic glutamate receptor
- PKC
protein kinase C
- DHPG
3,5-dihydroxyphenylglycine
- PDBu
phorbol 12,13-dibutyrate
- PRBP
polyribosome-binding protein
- PKA
protein kinase A
- eIF
eukaryotic initiation factor
- gsk-3β
glycogen synthase kinase 3β
References
- 1.Bailey C H, Bartsch D, Kandel E R. Proc Natl Acad Sci USA. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey U, Morris R G M. Nature (London) 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 3.Leski M L, Steward O. Neurochem Res. 1996;21:681–690. doi: 10.1007/BF02527725. [DOI] [PubMed] [Google Scholar]
- 4.Torre E R, Steward O. J Neurosci. 1996;16:5967–5978. doi: 10.1523/JNEUROSCI.16-19-05967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feig S, Lipton P. J Neurosci. 1993;13:1010–1021. doi: 10.1523/JNEUROSCI.13-03-01010.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martone M E, Pollock J A, Jones Y Z, Ellisman M H. J Neurosci. 1996;16:7437–7446. doi: 10.1523/JNEUROSCI.16-23-07437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenough W T, Hwang G, Gorman C. Proc Natl Acad Sci USA. 1985;82:4549–4552. doi: 10.1073/pnas.82.13.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steward O, Levy W. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.St. Johnston S D. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 10.Steward O, Kleiman R, Banker G. In: Basic and Clinical Aspects of Neuroscience. Harrison P J, editor. Vol. 6. Berlin: Springer; 1994. pp. 17–29. [Google Scholar]
- 11.Tiedge H, Brosius J. J Neurosci. 1997;16:7171–7181. doi: 10.1523/JNEUROSCI.16-22-07171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crino P B, Eberwine J. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 13.Hershey J W B, Mathews M B, Sonenberg N. Translation Control. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. [Google Scholar]
- 14.Sachs A B, Sarnow P, Hentze M W. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 15.Morley S J, Traugh J A. Biochimie. 1993;75:985–989. doi: 10.1016/0300-9084(93)90149-m. [DOI] [PubMed] [Google Scholar]
- 16.Jefferies H B J, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G, Marshall C A, Lin T-A, Kwon G, Munivenkatappa R B, Hill J R, Lawrence J C L, Jr, McDaniel M L. J Biol Chem. 1998;273:4485–4491. doi: 10.1074/jbc.273.8.4485. [DOI] [PubMed] [Google Scholar]
- 18.Fadden P, Haystead T A J, Lawrence J C., Jr J Biol Chem. 1997;272:10240–10247. doi: 10.1074/jbc.272.15.10240. [DOI] [PubMed] [Google Scholar]
- 19.Whalen S G, Gingras A-C, Amankwa L, Mader S, Branton P E, Aebersold R, Sonenberg N. J Biol Chem. 1996;271:11831–11837. doi: 10.1074/jbc.271.20.11831. [DOI] [PubMed] [Google Scholar]
- 20.Kleijn M, Welsh G I, Scheper G C, Voorma H O, Proud C G, Thomas A A M. J Biol Chem. 1998;273:5536–5541. doi: 10.1074/jbc.273.10.5536. [DOI] [PubMed] [Google Scholar]
- 21.Marin P, Nastiuk K L, Daniel N, Girault J-A, Czernik A J, Glowinski J, Nairn A C, Premont J. J Neurosci. 1997;17:3445–3454. doi: 10.1523/JNEUROSCI.17-10-03445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheetz A J, Nairn A C, Constantine-Paton M. Proc Natl Acad Sci USA. 1997;94:14770–14775. doi: 10.1073/pnas.94.26.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiler I J, Greenough W T. Mol Cell Neurosci. 1991;2:305–314. doi: 10.1016/1044-7431(91)90060-2. [DOI] [PubMed] [Google Scholar]
- 24.Weiler I J, Irwin S A, Klintsova A Y, Spencer C M, Brazelton A D, Miyashiro K, Comery T A, Patel B, Eberwine J, Greenough W T. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiler I J, Greenough W T. Proc Natl Acad Sci USA. 1993;90:7168–7171. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollingsworth E B, McNeal E T, Burton J L, Williams R, Daly J, Creveling D. J Neurosci. 1985;5:2240–2253. doi: 10.1523/JNEUROSCI.05-08-02240.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrell J E J, Martin G S. In: Methods in Enzymology. Hunter T, Sefton B M, editors. Vol. 200. New York: Academic; 1991. pp. 430–435. [DOI] [PubMed] [Google Scholar]
- 28.Kameshita I, Fujisawa H. Anal Biochem. 1989;183:139–143. doi: 10.1016/0003-2697(89)90181-4. [DOI] [PubMed] [Google Scholar]
- 29.Welsh G I, Patel J C, Proud C G. Anal Biochem. 1997;244:16–21. doi: 10.1006/abio.1996.9838. [DOI] [PubMed] [Google Scholar]
- 30.Thoen C, Slegers H. Biochim Biophys Acta. 1985;825:268–279. doi: 10.1016/0167-4781(85)90014-4. [DOI] [PubMed] [Google Scholar]
- 31.Issinger O G. Biochem J. 1977;165:511–518. doi: 10.1042/bj1650511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu S, Romano P R, Wek R C. J Biol Chem. 1997;272:14434–14441. doi: 10.1074/jbc.272.22.14434. [DOI] [PubMed] [Google Scholar]
- 33.Galabru J, Hovanessian A G. J Biol Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 34.Fisher T L, Blenis J. Mol Cell Biol. 1996;16:1212–1219. doi: 10.1128/mcb.16.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vik T A, Ryder J W. Biochem Biophys Res Commun. 1997;235:398–402. doi: 10.1006/bbrc.1997.6794. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Bjorbaek C, Moller D E. J Biol Chem. 1996;271:29773–29779. doi: 10.1074/jbc.271.47.29773. [DOI] [PubMed] [Google Scholar]
- 37.Erikson E, Maller J L. Second Messenger Phosphoproteins. 1988;2:135–143. [PubMed] [Google Scholar]
- 38.Thomas G. Biochem Soc Trans. 1993;21:901–904. doi: 10.1042/bst0210901. [DOI] [PubMed] [Google Scholar]
- 39.Price D J, Grove J R, Calvo V, Avruch J, Bierer B E. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 40.Chung J, Kuo C J, Crabtree G R, Blenis J. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 41.Jurivich D A, Chung J, Blenis J. J Cell Physiol. 1991;148:252–259. doi: 10.1002/jcp.1041480210. [DOI] [PubMed] [Google Scholar]
- 42.Chen R H, Chung J, Blenis J. Mol Cell Biol. 1991;11:1861–1867. doi: 10.1128/mcb.11.4.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erikson E, Stefanovic D, Blenis J, Erikson R L, Maller J L. Mol Cell Biol. 1987;7:3147–3155. doi: 10.1128/mcb.7.9.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. Cell. 1997;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 45.Douville E, Downward J. Oncogene. 1997;15:373–383. doi: 10.1038/sj.onc.1201214. [DOI] [PubMed] [Google Scholar]
- 46.Ghoda L, Lin X, Greene W C. J Biol Chem. 1997;272:21281–21288. doi: 10.1074/jbc.272.34.21281. [DOI] [PubMed] [Google Scholar]
- 47.Sutherland C, Leighton I A, Cohen P. Biochem J. 1993;296:15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schouten G J, Vertegaal A C O, Whiteside S, T, Israel A, Toebes M, Dorsman J C, van der Eb A J, Zantema A. EMBO J. 1997;16:3133–3144. doi: 10.1093/emboj/16.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welsh G I, Miyamoto S, Price N T, Safer B, Proud C G. J Biol Chem. 1996;271:11410–11413. doi: 10.1074/jbc.271.19.11410. [DOI] [PubMed] [Google Scholar]
- 50.Welsh G I, Proud C G. Biochem J. 1993;294:625–629. doi: 10.1042/bj2940625. [DOI] [PMC free article] [PubMed] [Google Scholar]