Abstract
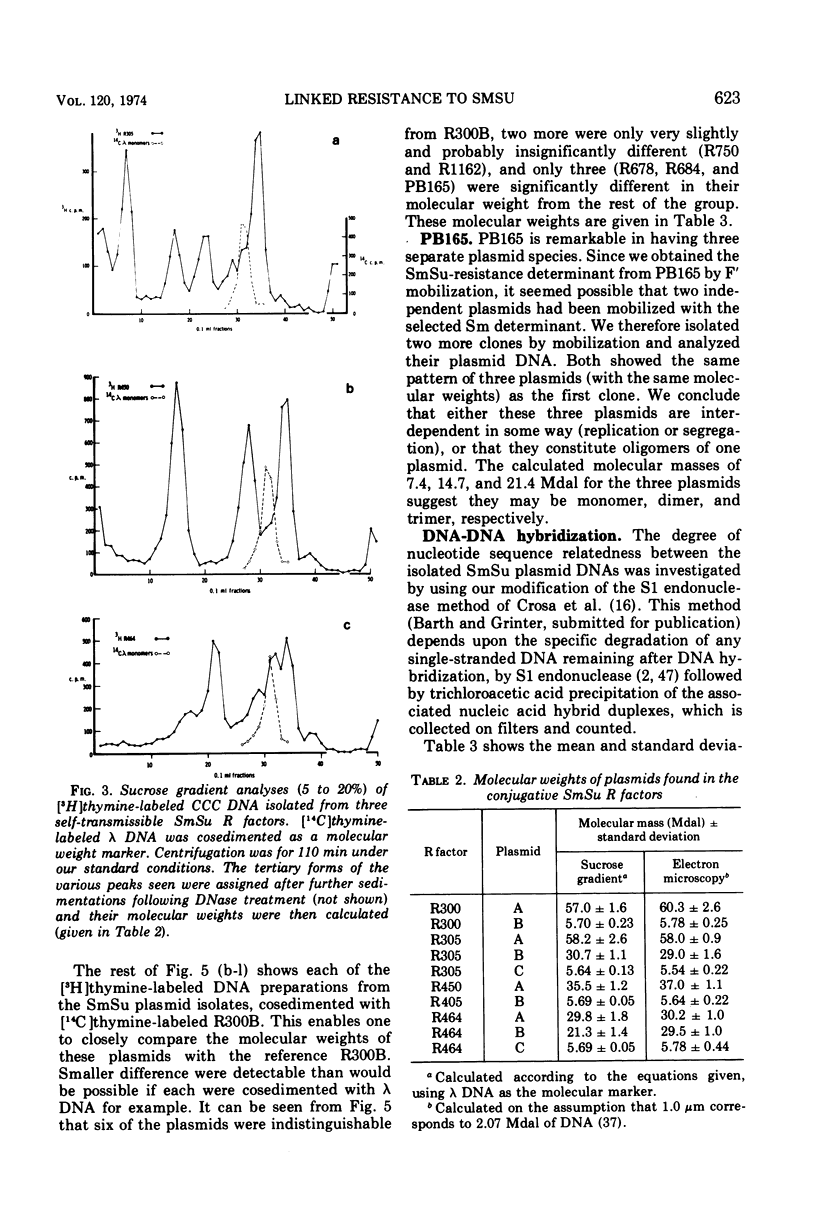
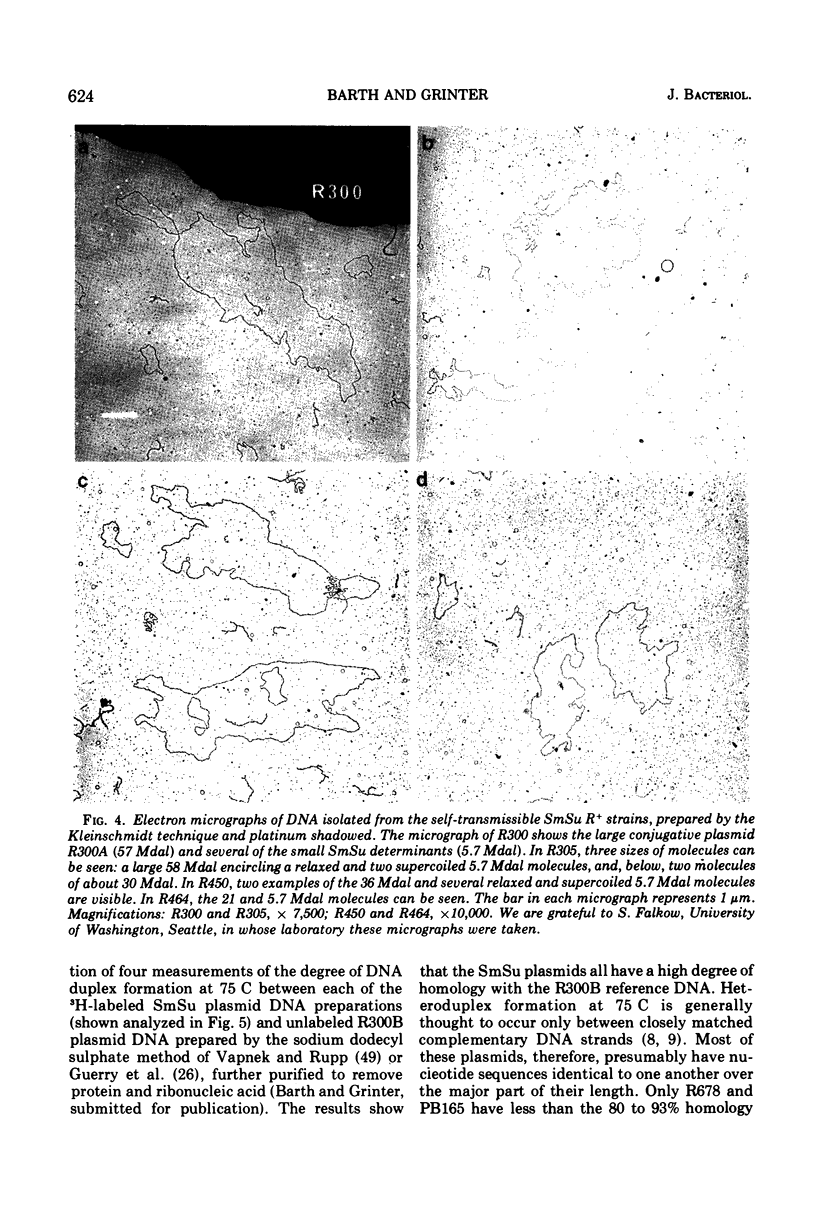
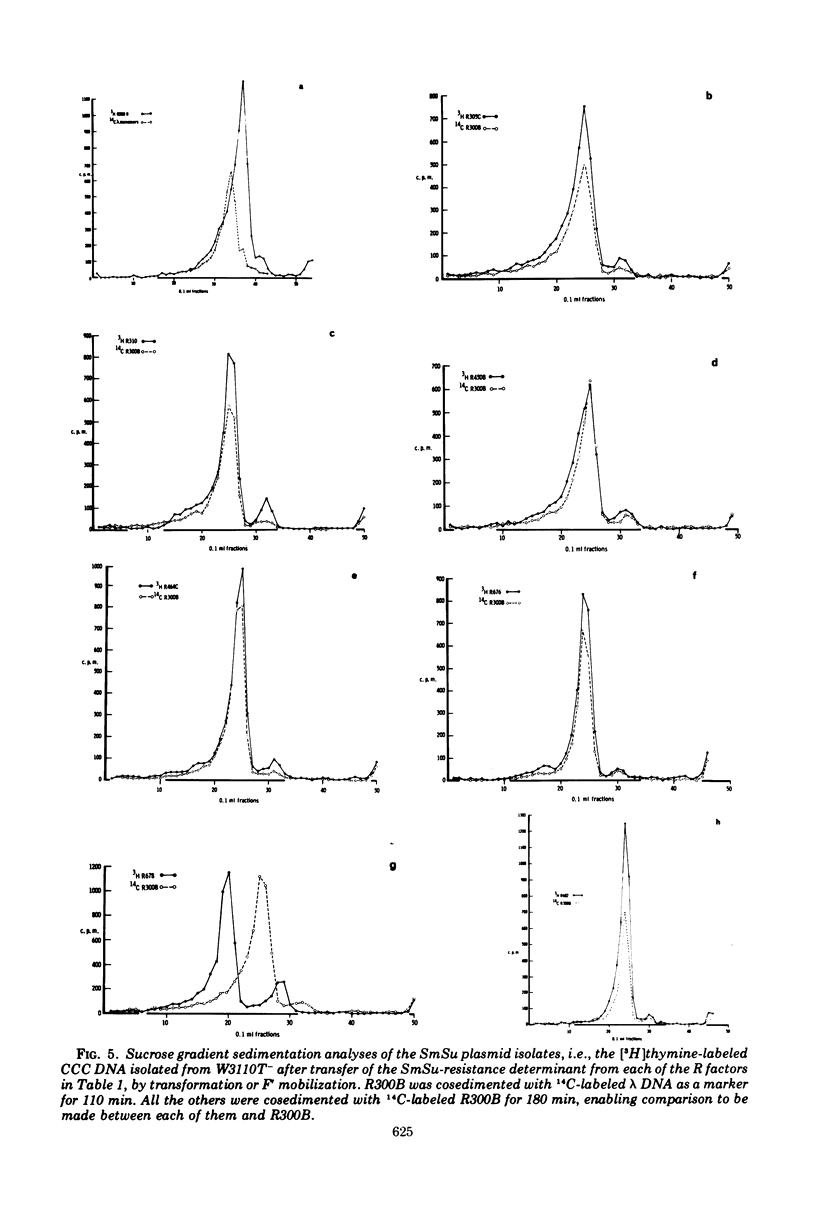
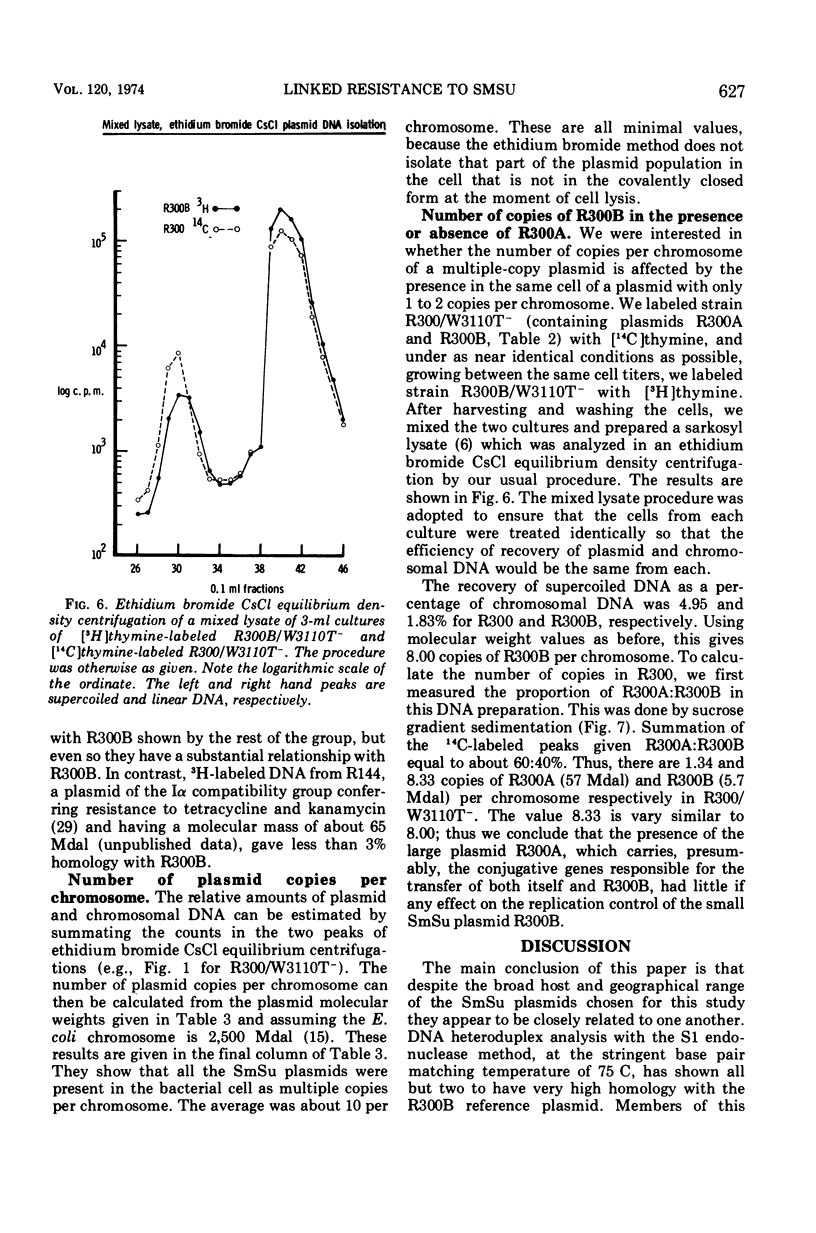
Bacterial strains showing linked resistance to streptomycin (Sm) and sulfonamides (Su) were chosen representing a wide taxonomic and geographical range. Their SmSu resistances were transferred to Escherichia coli K-12 and then plasmid deoxyribonucleic acid (DNA) was isolated by ethidium bromide CsCl centrifugation. The plasmid DNA was examined by electron microscopy and analyzed by sedimentation through 5 to 20% neutral sucrose gradients. Plasmid DNA from strains having transmissible SmSu resistance consisted of two or three molecular species, one of which had a molecular mass of about 5.7 Mdal (106 daltons), the others varying between 20 to 60 Mdal. By using transformation or F′ mobilization, we isolated the SmSu-resistance determinant from any fellow resident plasmids in each strain and again isolated the plasmid DNA. Cosedimentation of each of these with a differently labeled reference plasmid DNA (R300B) showed 9 out of 12 of the plasmids to have a molecular mass not significantly different from the reference (5.7 Mdal); two others were 6.3 and 9.2 Mdal, but PB165 consisted of three plasmids of 7.4, 14.7, and 21.4 Mdal. Three separate isolations of the SmSu determinant from PB165 gave the same three plasmids, which we conclude may be monomer, dimer, and trimer, respectively. DNA-DNA hybridizations at 75 C demonstrated 80 to 93% homology between reference R300B DNA and each isolated SmSu plasmid DNA, except for the 9.2-Mdal plasmid which had 45% homology and PB165 which had 35%. All the SmSu plasmids were present as multiple copies (about 10) per chromosome. The conjugative plasmid of R300 (present as 1.3 copies per chromosome) has been shown to have negligible effect on the number of copies of its accompanying SmSu plasmid R300B. We conclude that the SmSu plasmids are closely related and probably have a common evolutionary origin.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ando T. A nuclease specific for heat-denatured DNA in isolated from a product of Aspergillus oryzae. Biochim Biophys Acta. 1966 Jan 18;114(1):158–168. doi: 10.1016/0005-2787(66)90263-2. [DOI] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Beacham I. R., Ahmad S. I., Pritchard R. H. The inducer of the deoxynucleoside phosphorylases and deoxyriboaldolase in Escherichia coli. Biochim Biophys Acta. 1968 Jul 23;161(2):554–557. doi: 10.1016/0005-2787(68)90132-9. [DOI] [PubMed] [Google Scholar]
- Baxter-Gabbard K. L. A simple method for the large-scale preparation of sucrose gradients. FEBS Lett. 1972 Jan 15;20(1):117–119. doi: 10.1016/0014-5793(72)80031-0. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Characterization of multiple circular DNA forms of colicinogenic factor E-1 from Proteus mirabilis. Biochemistry. 1968 Oct;7(10):3513–3520. doi: 10.1021/bi00850a028. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Falkow S. Genetics of the Enterobacteriaceae. C. Molecular relationships among members of the Enterobacteriaceae. Adv Genet. 1971;16:81–118. doi: 10.1016/s0065-2660(08)60355-7. [DOI] [PubMed] [Google Scholar]
- Bryan L. E., Van Den Elzen H. M., Tseng J. T. Transferable drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1972 Jan;1(1):22–29. doi: 10.1128/aac.1.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee J. N., Datta N., Hedges R. W. R factors from Proteus rettgeri. J Gen Microbiol. 1972 Oct;72(3):543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- Cooper S., Helmstetter C. E. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968 Feb 14;31(3):519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- Crosa J. H., Brenner D. J., Falkow S. Use of a single-strand specific nuclease for analysis of bacterial and plasmid deoxyribonucleic acid homo- and heteroduplexes. J Bacteriol. 1973 Sep;115(3):904–911. doi: 10.1128/jb.115.3.904-911.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzin F., Vogt M., Dieckmann M., Berg P. Induction of virus multiplication in 3T3 cells transformed by a thermosensitive mutant of polyoma virus. II. Formation of oligometric polyoma DNA molecules. J Mol Biol. 1970 Feb 14;47(3):317–333. doi: 10.1016/0022-2836(70)90305-0. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Compatibility groups among fi - R factors. Nature. 1971 Nov 26;234(5326):222–223. doi: 10.1038/234222a0. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E., Maas W. K. Inhibition of replication of an F'lac episome in Hfr cells of Escherichia coli. J Bacteriol. 1968 Feb;95(2):531–539. doi: 10.1128/jb.95.2.531-539.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D. Molecular weights of coliphages and coliphage DNA. IV. Molecular weights of DNA from bacteriophages T4, T5 and T7 and the general problem of determination of M. J Mol Biol. 1970 Dec 28;54(3):567–577. doi: 10.1016/0022-2836(70)90127-0. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Humphreys G. O., Anderson E. S. Molecular studies of R factor compatibility groups. J Bacteriol. 1973 Jul;115(1):387–398. doi: 10.1128/jb.115.1.387-398.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., Falkow S. Polynucleotide sequence relationships among some bacterial plasmids. J Bacteriol. 1971 Jul;107(1):372–374. doi: 10.1128/jb.107.1.372-374.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., van Embden J., Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974 Feb;117(2):619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Coetzee J. N., Dennison S. R factors from Proteus morganii. J Gen Microbiol. 1973 Aug;77(2):249–259. doi: 10.1099/00221287-77-2-249. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Datta N. Plasmids determining I pili constitute a compatibility complex. J Gen Microbiol. 1973 Jul;77(1):19–25. doi: 10.1099/00221287-77-1-19. [DOI] [PubMed] [Google Scholar]
- Hedges R. W. R factors from Providence. J Gen Microbiol. 1974 Mar;81(1):171–181. doi: 10.1099/00221287-81-1-171. [DOI] [PubMed] [Google Scholar]
- Hershey A. D., Burgi E., Ingraham L. COHESION OF DNA MOLECULES ISOLATED FROM PHAGE LAMBDA. Proc Natl Acad Sci U S A. 1963 May;49(5):748–755. doi: 10.1073/pnas.49.5.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V., LeBlanc D. J., Falkow S. Membrane attachment of R-factor deoxyribonucleic acid in compatible and incompatible cell pairs following conjugation. J Bacteriol. 1973 Sep;115(3):1208–1211. doi: 10.1128/jb.115.3.1208-1211.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y. THE EFFECT OF ACRIDINE DYES ON MATING TYPE FACTORS IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1960 Jan;46(1):57–64. doi: 10.1073/pnas.46.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER A. D., HOGNESS D. S. The transformation of Escherichia coli with deoxyribonucleic acid isolated from bacteriophage lambda-dg. J Mol Biol. 1960 Dec;2:392–415. doi: 10.1016/s0022-2836(60)80050-2. [DOI] [PubMed] [Google Scholar]
- KAISER A. D. The production of phage chromosome fragments and their capacity for genetic transfer. J Mol Biol. 1962 Apr;4:275–287. doi: 10.1016/s0022-2836(62)80005-9. [DOI] [PubMed] [Google Scholar]
- KORN D., WEISSBACH A. Thymineless induction in Escherichia coli K12 (lambda). Biochim Biophys Acta. 1962 Nov 26;61:775–790. doi: 10.1016/0926-6550(62)90060-9. [DOI] [PubMed] [Google Scholar]
- Lawn A. M., Meynell G. G., Meynell E., Datta N. Sex pili and the classification of sex factors in the enterobacteriaceae. Nature. 1967 Oct 28;216(5113):343–346. doi: 10.1038/216343a0. [DOI] [PubMed] [Google Scholar]
- LeBlanc D. J., Falkow S. Studies on superinfection immunity among transmissible plasmids in Escherichia coli. J Mol Biol. 1973 Mar 15;74(4):689–701. doi: 10.1016/0022-2836(73)90057-0. [DOI] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milliken C. E., Clowes R. C. Molecular structure of an R factor, its component drug-resistance determinants and transfer factor. J Bacteriol. 1973 Feb;113(2):1026–1033. doi: 10.1128/jb.113.2.1026-1033.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard R. H., Zaritsky A. Effect of thymine concentration on the replication velocity of DNA in a thymineless mutant of Escherichia coli. Nature. 1970 Apr 11;226(5241):126–131. doi: 10.1038/226126a0. [DOI] [PubMed] [Google Scholar]
- Skovgaard N., Nielsen B. B. Salmonella in pigs and animal feeding stuffs in England and Wales and in Denmark. J Hyg (Lond) 1972 Mar;70(1):127–140. doi: 10.1017/s0022172400022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. R., Humphreys G. O., Anderson E. S. Genetic and molecular characterisation of some non-transferring plasmids. Mol Gen Genet. 1974 Mar 27;129(3):229–242. doi: 10.1007/BF00267915. [DOI] [PubMed] [Google Scholar]
- Vapnek D., Rupp W. D. Identification of individual sex-factor DNA strands and their replication during conjugation in thermosensitive DNA mutants of Escherichia coli. J Mol Biol. 1971 Sep 28;60(3):413–424. doi: 10.1016/0022-2836(71)90178-1. [DOI] [PubMed] [Google Scholar]



