Abstract
Ribonucleotide reductase catalyzes the rate-limiting step in nucleotide biosynthesis and plays a central role in genome maintenance. Although a number of regulatory mechanisms govern RNR activity, the physiological impact of RNR deregulation had not previously been examined in an animal model. We demonstrate here that overexpression of the small RNR subunit potently and selectively induces lung neoplasms in transgenic mice and is mutagenic in cultured cells. Combining RNR deregulation with defects in DNA mismatch repair, the cellular mutation correction system, synergistically increased RNR-induced mutagenesis and carcinogenesis. Moreover, the proto-oncogene K-ras was identified as a frequent mutational target in RNR-induced lung neoplasms. Together, these results demonstrate that RNR deregulation promotes lung carcinogenesis through a mutagenic mechanism and establish a new oncogenic activity for a key regulator of nucleotide metabolism. Importantly, RNR-induced lung neoplasms histopathologically resemble human papillary adenocarcinomas and arise stochastically via a mutagenic mechanism, making RNR transgenic mice a valuable model for lung cancer.
Keywords: ribonucleotide reductase, lung cancer, mutagenesis, mismatch repair, K-ras
INTRODUCTION
An adequate and balanced supply of deoxyribonucleotide triphosphates (dNTPs) is essential for accurate DNA replication and repair. The rate limiting step in de novo dNTP biosynthesis is catalyzed by the enzyme ribonucleotide reductase (RNR). RNR reduces ribonucleoside diphosphate (NDP) to deoxyribonucleoside diphosphate (dNDP), phosphorylation of which yields dNTP. RNR is composed of two non-identical homodimeric subunits (1). The large R1 subunit harbors the catalytic site and is encoded by the Rrm1 gene in mammals. The small R2 subunit contains an oxygen-bridged dinuclear iron center that generates a tyrosyl free radical that is transferred to the R1 subunit for enzyme activity. Mammalian genomes contain two independent genes, Rrm2 and Rrm2b (p53R2), that encode closely related R2 proteins. A complex of Rrm2 and Rrm1 accounts for most RNR activity during S-phase. p53R2 was originally identified as a target gene for the p53 tumor suppressor protein and is transcriptionally induced following DNA damage (2, 3). In addition to its role in stress responses, p53R2 is expressed at low levels throughout the cell cycle and complexes with Rrm1 to produce dNTPs for mitochondrial DNA replication (4).
Because intracellular nucleotide concentrations have a major impact on DNA replication fidelity (5), RNR enzyme activity is tightly controlled by several regulatory mechanisms. During an unperturbed cell cycle, the transcription of Rrm1 and Rrm2 is undetectable in G0/G1 phase and reaches maximal levels in S-phase cells (6–8). However, owing to its long half-life, Rrm1 protein levels are nearly constant throughout the cell cycle and in excess relative to the R2 subunit. RNR enzyme activity is therefore determined in part by R2 protein levels. Rrm2 protein is absent during G0/G1-phase, peaks in S–phase, and then falls in mitosis following ubiquitination by the anaphase promoting complex (8–10). Consistent with a need for nucleotides during DNA repair, DNA damage and replication stress induce RNR expression in both yeast and mammalian cells, in a manner dependent on DNA damage checkpoint pathways (11, 12). While mammalian Rrm1 and Rrm2 proteins are cytoplasmic (13), p53R2 localizes to the nucleus in genotoxin-treated cells (2, 3), which may facilitate the localized production of nucleotides at DNA damage sites.
RNR enzyme activity also is controlled by two allosteric sites in the R1 subunit. A specificity site regulates the relative cellular concentration of each of the four dNTPs by influencing substrate choice, while an activity site regulates the total dNTP pool size by monitoring the ATP/dATP ratio. Analysis of the mutant Rrm1-D57N, which is insensitive to feedback inhibition by dATP due to a mutation in the activity site, indicates that loss of RNR allosteric control results in a mutator phenotype in both yeast and mammalian cells (14–16).
Although RNR is a major determinant of genomic integrity, the consequences of RNR deregulation in animals are unknown. We generated transgenic mice that overexpress Rrm1, Rrm2, or p53R2 and found that overexpression of either small RNR subunit induced spontaneous lung neoplasms and was mutagenic in cultured cells. Defects in DNA mismatch repair (MMR) synergistically increased RNR-induced mutagenesis and carcinogenesis, and activating mutations in the proto-oncogene K-ras were identified in lung neoplasms from Rrm2 and p53R2 transgenic mice. These results identify mutagenic and carcinogenic effects of RNR deregulation in vivo.
MATERIALS AND METHODS
Plasmids
Expression plasmids encoding mouse Rrm1, Rrm2, or p53R2 were constructed in the pCaggs expression vector as described in Supplementary Materials and Methods.
Transgenic mice
Transgenic mice were generated in the FVB/N strain background as described in Supplementary Materials and Methods. For analysis of tumor development, cohorts of mice were aged until moribund for 15 to 21 months. Msh6-null mice (17) were obtained from the Mouse Models of Human Cancers Consortium, bred with Rrm2Tg or p53R2Tg mice, and aged until moribund for 6 or 17 months. All mice were maintained identically, following guidelines approved by the Cornell University Institutional Laboratory Animal Use and Care Committee. Pathological assessments were performed as described in Supplementary Materials and Methods.
Northern blot analysis
Total RNA was isolated from mouse cells and tissues using RNA STAT-60 (Tel-Test Inc.), resolved on an agarose/formaldehyde gel, and hybridized with probes specific to mouse Rrm1, Rrm2, p53R2, or Gapdh.
Western blot analysis
Cell or tissue protein extracts were prepared as described in Supplementary Materials and Methods. The antibodies used were mouse anti-R1 (AD203, Bio Med Tek), goat anti-R2 (sc-115, Santa Cruz Biotechnology,), rabbit anti-p53R2 (2383, ProSci-inc,) and β-actin (A5441, Sigma).
Immunohistochemistry
Immunohistochemical detection of pro-SP-C and CC10 was performed on 5 μm paraffin sections using the Vectastain ABC kit (Vector Laboratories) as described in Supplementary Materials and Methods.
Generation of RNR overexpressing 3T3 cell pools
Pools of mouse NIH/3T3 fibroblasts that overexpress individual RNR genes were generated as described in Supplementary Materials and Methods.
Determination of mutation rates in mammalian and yeast cells
Mutation frequencies were determined in 3T3 cell pools and yeast strains by Hprt mutation and canavanine resistance assays, respectively, as described in Supplementary Materials and Methods.
Big Blue mutation rate assay
Big Blue C57Bl/6 mice, hemizygous for the λ shuttle vector, were obtained from Stratagene (La Jolla, CA) and bred with Rrm1Tg, Rrm2Tg, p53R2Tg, or Msh6−/−RNRTg mice. Mutation frequency in the resulting animals was determined as described in Supplementary Materials and Methods.
Sequencing of K-ras exons 1 and 2
DNA was prepared from tumor specimens isolated by laser microdissection, and K-ras exons 1 and 2 were PCR amplified and sequenced as described in Supplementary Materials and Methods.
RESULTS
Generation of RNR transgenic mice and analysis of transgene expression
Deregulation of RNR is mutagenic in yeast and cultured mammalian cells (14, 16). To test the consequences of RNR deregulation in an animal model, we set out to generate transgenic mice featuring broad, high level expression of the individual mouse RNR genes Rrm1, Rrm2, and p53R2, using pCaggs expression constructs that place the RNR genes under the control of chicken β-actin promoter and cytomegalovirus enhancer regulatory sequences. Six Rrm1, two Rrm2, and four p53R2 transgene-positive founders were generated and subsequently maintained on a pure FVB/N strain background. RNR transgenic mice appeared grossly normal and were fertile. When bred with wild-type FVB mice, p53R2 hemizygotes produced fewer than the expected number of transgene positive offspring (205 p53R2 transgene positive and 349 transgene negative mice were identified among 554 mice genotyped at weaning).
Endogenous and transgenic Rrm1, Rrm2 and p53R2 mRNA expression was tested in a variety of organs by Northern blot analysis. The endogenous Rrm1 and Rrm2 genes were coordinately expressed, with highest expression in proliferative tissues such as testis and thymus (Fig. 1A, left panels). Expression of the endogenous p53R2 gene was undetectable in all tested wild-type FVB tissues. Importantly, Rrm2Tg and p53R2Tg mice showed high-level transgene expression in all tissues, with overexpression being highest in muscle (Fig. 1A, right panels). Rrm1 overexpression was only observed in muscle and testis of Rrm1Tg mice. For technical reasons, the Rrm1 transgene included additional non-coding cDNA sequences and was microinjected as a linearized construct without removal of plasmid backbone sequences, which may contribute to the relatively poor transgene expression.
Figure 1. Widespread overexpression of ribonucleotide reductase genes in transgenic mice. (A).

Northern blot analysis of RNR expression in wild-type and RNR transgenic mice. Total RNA was extracted from the indicated tissues from wild-type FVB mice (left panels), or RNR transgenic mice (right panels) and subjected to Northern blot hybridization with the indicated probes specific for Rrm1, Rrm2 or p53R2. Positions of endogenous and transgene-derived RNR transcripts are indicated. (B) Western blot analysis of RNR protein expression in the indicated tissues from wild type (WT) and RNR transgenic (Tg) mice, as well as lung neoplasms from the corresponding transgenic strains (Tumor 1, 2). Total protein from the indicated tissues was subjected to immunoblotting with antibodies specific to Rrm1, Rrm2 or p53R2. Duplicate membranes were immunoblotted for β-actin as a loading control.
Consistent with results from the Northern blot analyses, immunoblotting revealed that the Rrm2 and p53R2 proteins were highly overexpressed in all tested tissues from Rrm2Tg and p53R2Tg mice (Fig. 1B). Although Northern blotting failed to identify p53R2 expression in wild-type tissues (Fig. 1A), low levels of p53R2 protein were apparent in most wild-type FVB tissues. Rrm1 protein overexpression was limited to muscle and to a lesser extent lung in Rrm1Tg mice as compared to wild-type littermates. Together, these results establish the restricted overexpression of the large RNR subunit Rrm1 and the widespread, high level overexpression of the small RNR subunits Rrm2 and p53R2 in transgenic mice.
Overexpression of the small RNR subunit promotes lung carcinogenesis
In order to identify spontaneous neoplasms and other abnormalities in RNR transgenic mice, we established a cohort consisting of 52 Rrm1Tg, 75 Rrm2Tg, and 81 p53R2Tg mice, as well as 49 transgene-negative control mice, and aged them until they exhibited clinical illness. Notably, a significantly increased frequency of lung neoplasms was observed in Rrm2Tg and p53R2Tg mice (Table 1). 72% of Rrm2Tg and 74% of p53R2Tg animals developed spontaneous lung neoplasms. By contrast, 31% of transgene-negative controls developed lung neoplasms, a frequency consistent with the reported incidence for aged wild-type FVB mice (18). The lung neoplasm incidence in Rrm1Tg mice was 31%, identical to that of the control animals and significantly less than that of Rrm2Tg or p53R2Tg mice (Chi-square analysis, p<0.05). Lung neoplasms were observed in multiple independent Rrm2Tg and p53R2Tg lines, indicating that transgene integration site effects did not account for the neoplastic phenotype. Signs of clinical illness arose following a latency of 16–18 months for all genotypes. No differences in lung neoplasm incidence between sexes was noted for any of the transgenic lines. The frequency of epithelial hyperplasia of alveoli also was increased in p53R2Tg and especially Rrm2Tg mice. Other neoplasms, including papilloma, histiocytic sarcoma, mammary carcinoma, and lymphoblastic lymphoma, were observed in 13% of Rrm1Tg, 12% of Rrm2Tg, and 12% of p53R2Tg mice, but only 2% of transgene-negative mice.
Table 1.
Lung neoplasm characteristics in RNR overexpressing mice
| Mouse genotype | # of animals | % of mice with lung neoplasms | % of mice with hyperplasia§ | Average lung neoplasm size (mm) ± SD | % of mice with multiple lung neoplasms | % of mice with lung adenocarcinoma |
|---|---|---|---|---|---|---|
| WT FVB† | 49 | 31% | 12% | 4.04 ± 3.98 | 8% | 6% |
| Rrm1Tg | 52 | 31% | 15% | 3.96 ± 3.59 | 8% | 10% |
| Rrm2Tg | 75 | 72%* | 44%* | 6.68 ± 4.22 | 53%* | 40%* |
| p53R2Tg | 81 | 74%* | 20% | 4.26 ± 3.44 | 47%* | 21% |
NOTE: Mice were aged until moribund for up to 21 months, euthanized by asphyxiation using carbon dioxide, and subjected to pathological examination as described in Materials and Methods.
WT FVB refers to transgene-negative control mice.
Includes mice that had both epithelial hyperplasia of alveoli and lung neoplasms.
Statistically significant difference (p<0.05) relative to WT FVB mice. Incidences were compared by Chi-square analysis. Neoplasm sizes were compared by t-test analysis.
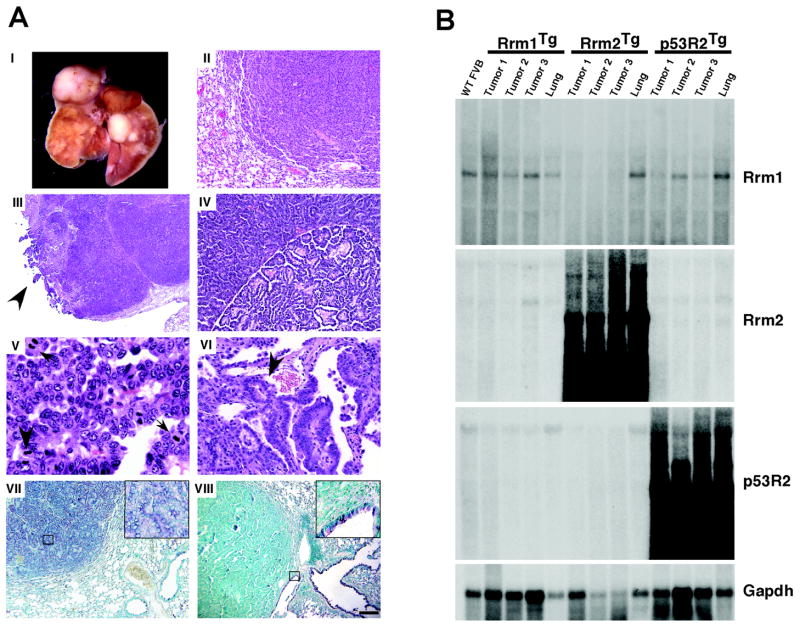
The lung neoplasms in Rrm2Tg and p53R2Tg mice displayed several features consistent with a substantial lung cancer predisposition. A significantly greater lung neoplasm multiplicity was observed for Rrm2Tg and p53R2Tg mice, and the lung neoplasms in Rrm2Tg mice were also considerably larger than those from control animals (Table 1). The lung neoplasms from Rrm2Tg and p53R2Tg mice ranged from adenoma to advanced adenocarcinoma (Fig. 2A I–VI), and resembled human glandular pulmonary neoplasms, particularly adenocarcinomas. RNR-induced lung adenocarcinomas were primarily of the papillary subtype and exhibited pleural invasion, heterogeneous growth pattern, nuclear atypia, high mitotic index, and blood vessel invasion (Fig. 2A III–VI). A greater frequency of adenocarcinoma was observed in Rrm2Tg and p53R2Tg mice as compared to Rrm1Tg or transgene-negative mice (Table 1), with Rrm2 overexpression in particular eliciting pathologically advanced neoplasms. Together, these data indicate that overexpression of either small RNR subunit in mice promotes lung neoplasm formation, with Rrm2 being more potent than p53R2 with respect to tumor size, multiplicity, and malignancy.
Figure 2. Histopathological and molecular analysis of lung neoplasms from RNR transgenic mice. (A).
(I) Lungs from a Rrm2Tg mouse with multiple independent neoplasms affecting several lobes. (II–VI) H&E-stained sections of lung neoplasms. (II) Solid adenoma from a p53R2Tg mouse. (III–VI) Papillary adenocarcinomas from Rrm2Tg or p53R2Tg mice showing pleural invasion (arrow) (III), regional variation in growth pattern (IV), multiple mitotic figures (arrows) (V), and blood vessel invasion (arrow) (VI). (VII, VIII) Immunohistochemical staining of RNR-induced lung neoplasms for Pro-SP-C (VII) or CC10 (VIII) by the ABC method, with methyl green counterstain. Inserts show higher magnification views of the boxed regions. Calibration bar: II, IV: 50 μm; III: 241 μm; V: 10 μm; VI: 25 μm; VII, VIII: 100 μm. (B) Northern blot analysis of lung neoplasms from RNR transgenic mice. Total RNA was prepared from lung neoplasms (Tumor 1, Tumor 2, Tumor 3) or normal lung tissue (Lung) from RNR transgenic mice, as well as from wild-type FVB lung tissue (WT FVB). Northern blotting was performed with the indicated radiolabeled probes.
To investigate the possible cell type of origin for RNR-induced lung neoplasms, we performed immunohistochemistry using antibodies against Clara cell antigen (CC10) and surfactant apoprotein-C (SP-C), markers that distinguish Clara and alveolar type II cells, respectively. Eight of eight lung neoplasms from Rrm2Tg and p53R2Tg mice were positive for SP-C (Fig. 2A VII), while none was positive for CC10 (Fig. 2A VIII). Adjacent bronchioles, on the other hand, were positive for CC10 and negative for SP-C as expected. These results suggest that RNR-induced lung neoplasms arose from alveolar type II cells or their progenitors.
To confirm a causative role for RNR overexpression in lung carcinogenesis, we analyzed the expression of Rrm1, Rrm2, and p53R2 in lung neoplasms by Northern (Fig. 2B) and Western (Fig. 1B) blotting. Lung neoplasms from Rrm2Tg and p53R2Tg animals showed prominent RNR overexpression, consistent with a causative role for RNR in the genesis of these lung lesions. By contrast, lung neoplasms from Rrm1Tg mice did not display high level transgene expression, providing further evidence that carcinogenesis in Rrm2Tg and p53R2Tg mice is highly specific. Overall, these data identify a novel oncogenic activity for the small RNR subunit.
Increased mutation frequency following RNR overexpression in cultured 3T3 cells
We hypothesized that RNR overexpression induced lung neoplasms through a mutagenic mechanism because defects in RNR allosteric control result in increased mutation frequencies in yeast and mammalian cells (14, 16). To determine if RNR overexpression was similarly mutagenic, we generated Rrm1, Rrm2, or p53R2 overexpressing NIH/3T3 cell pools using the same expression constructs as used to generate the transgenic mice. Overexpression of individual RNR genes in these cell pools was confirmed by Northern and Western blotting (Fig. 3A and B).
Figure 3. Increased mutation frequency in RNR overexpressing NIH/3T3 cell pools. (A).

Northern blot analysis of RNR expression in stable 3T3 cell pools transfected with either pCaggs empty vector or pCaggs RNR genes. Total RNA was extracted from the indicated cell lines and subjected to Northern blot hybridization with probes specific for Rrm1, Rrm2, p53R2, or Gapdh. (B) Western blot analysis of RNR protein expression in RNR overexpressing 3T3 cells. Total protein was extracted from the indicated cell lines and subjected to immunoblotting with antibodies specific to Rrm1, Rrm2, or p53R2. Duplicate membranes were immunoblotted for β-actin as a loading control. Samples in (A) and (B) were run on single blots, which were then cropped to remove extraneous lanes. (C) Mutation frequency at the Hprt locus in Rrm1, Rrm2 and p53R2 overexpressing 3T3 cells. Mutation frequency was determined by Hprt assay.
We then measured mutation frequency using the Hprt mutation detection assay, which identifies cells harboring Hprt mutations by virtue of their resistance to 6-thioguanine (6-TG) (19). In a representative experiment (Fig. 3C), a significantly increased mutation frequency was observed in a Rrm2 overexpressing cell pool (9.0 × 10−6) as compared to Rrm1 overexpressing or empty plasmid vector cell pools (less than 0.7 × 10−6 and 0.8 × 10−6, respectively). Three independent Rrm2 overexpressing cell pools showed a consistently increased mutation frequency that was 9.9- to 16.0-fold greater than that observed for vector control cells. A p53R2 overexpressing cell pool showed a more modestly but nevertheless significantly increased mutation frequency of 3.0 × 10−6 (Fig. 3C). However, mutation frequency in p53R2 overexpressing cells varied, with three p53R2 overexpressing cell pools showing an elevated mutation frequency that was 4.2- to 11.2-fold greater than that for vector control cells while two other p53R2 overexpressing cell pools displayed no increase in mutation frequency. Whether this variability is due to differences in expression levels between individual cell pools, or to the fact that the mutation frequencies measured were near the lower end of sensitivity for this assay, has not been determined. However, three independent Rrm1 overexpressing cell pools and another five empty plasmid vector cell pools showed no detectable increase in mutation frequency.
To determine the nature of the mutations conferring 6-TG resistance, we sequenced the Hprt gene from individual colonies (20). Interestingly, four of seven Hprt mutations from the Rrm2 overexpressing cell pool shown in Fig. 3C were G→T substitutions (Supplementary Table 1), which are relatively rare among reported spontaneous Hprt mutations (21). Similar results were obtained in a separate experiment with an independent Rrm2 overexpressing cell pool (Supplementary Table 1). One of six mutations from the p53R2 overexpressing cell pool shown in Fig. 3C also was a G→T mutation, but no G→T mutations were observed among six 6-TG resistant clones from a second independent experiment (Supplementary Table 1). Collectively, the results indicate that overexpression of the small RNR subunit causes a mutator phenotype.
Combined defects in RNR regulation and MMR result in synergistic increases in mutagenesis and carcinogenesis
To further evaluate a role for mutagenesis in RNR-induced lung carcinogenesis, we investigated whether combining RNR deregulation with a defect in MMR, the repair system that suppresses mutation accumulation, would cause a synergistic increase in mutagenesis and carcinogenesis. In eukaryotes, a complex of Msh2-Msh6 is responsible for recognizing base-base mispairs and single base insertion/deletions, while a Msh2-Msh3 complex detects larger insertion/deletion loops (22). We first tested this hypothesis in S. cerevisiae by measuring the mutation rate by canavanine resistance assay in strains with deregulated RNR activity and mutations in MMR genes. To deregulate budding yeast RNR, we utilized the rnr1-D57N mutant in which a single amino acid change in the R1 activity site makes the enzyme insensitive to feedback inhibition by dATP (16). Consistent with published reports (14), rnr1-D57N yeast exhibited a 3.4-fold increase in mutation rate relative to the wild-type strain, which had a mutation rate of 1.5 × 10−7 (Fig. 4A). MMR defective strains also displayed elevated mutation rates (msh2Δ: 28.4-fold; msh3Δ: 2.9-fold; msh6Δ: 9.8-fold), similar to previous reports (23). Notably, rnr1-D57N msh2Δ and rnr1-D57N msh6Δ double mutants displayed approximately multiplicative increases in mutation rate relative to the single mutants (61.4-fold and 23.8-fold, respectively). Multiplicative increases in mutagenesis are seen for mutations that affect factors acting in series in a common pathway (24), suggesting that the Msh2-Msh6 complex corrects DNA mismatches induced by RNR deregulation. By contrast, combining rnr1-D57N with msh3Δ resulted in only an additive increase in mutation rate (rnr1-D57N msh3Δ: 3.9-fold). The spectrum of mutations arising in WT, rnr1-D57N, msh2Δ, and msh6Δ strains was consistent with previous publications (14, 23, 25) and included primarily base substitutions, as well as frameshift mutations for msh2Δ (Supplementary Table 2). The frequency of frameshift mutations involving single nucleotide insertions or deletions was substantially increased in rnr1-D57N msh2Δ and rnr1-D57N msh6Δ strains relative to the single mutants.
Figure 4. Genetic interactions between RNR and mismatch repair. (A).

Canavanine mutation rate assay for RNR1(WT) and rnr1-D57N strains on MMR-deficient backgrounds (msh3Δ, msh2Δ, msh6Δ, or WT) of S. cerevisiae. The forward mutation rate (per generation) to canavanine resistance was measured for the indicated single and double mutant combinations. Error bars show the 95% confidence interval. (B) Survival curves for Msh6−/−RNRTg (Rrm2Tg or p53R2Tg) mice. Mice were aged until moribund for up to 17 months. Survival curves were generated using SPSS software. The following number of animals was analyzed for each genotype: Msh+/+ (11), Msh6+/− (23), Msh6+/−Rrm2Tg (22), Msh6+/−p53R2Tg (11), Msh6−/− (34), Msh6−/−Rrm2Tg (20), Msh6−/− p53R2Tg (17). (C, D) Mutation frequency at the λ cII locus in lung (C) or spleen (D) tissues from RNR overexpressing and control mice. Genomic DNA was isolated from 3-month old mice of the indicated genotypes and packaged into infectious phage. Mutation frequency was determined based on the ratio of the number of mutant phage obtained to the total number of phage analyzed.
The synergistic effects of RNR deregulation and MMR deficiency on mutation rates in yeast prompted us to further test genetic interactions between RNR and MMR in mice, by crossing RNR transgenic mice with Msh6-null mice (17). If RNR overexpression induces lung carcinogenesis through a mutagenic mechanism, Msh6 deficiency would be predicted to accelerate lung carcinogenesis in RNR transgenic mice. A cohort of Msh6−/−, Msh6+/−, or Msh6+/+ mice that also carried either the Rrm2 or p53R2 transgene was established and examined for survival and cancer susceptibility. Interestingly, the median lifespan for Msh6−/−p53R2Tg mice (136 days) was significantly reduced compared to that of transgene-negative Msh6−/− mice (258 days; p<0.05; logrank test) (Fig. 4B). The reduced survival of Msh6−/−p53R2Tg mice was associated with early onset lymphomagenesis (Supplementary Table 3). Because these Msh6−/−p53R2Tg mice died at a young age, we could not evaluate whether Msh6 deficiency cooperated with p53R2 overexpression in inducing lung neoplasms.
The survival rate for Msh6−/−Rrm2Tg and transgene negative Msh6−/− mice was not significantly different (p=.975; logrank test), suggesting that Rrm2 overexpression, unlike p53R2 overexpression, did not enhance lymphomagenesis (Fig. 4B). However, that 90% of Msh6−/−Rrm2Tg mice had developed lung neoplasms despite their shortened lifespan of approximately 10 months was suggestive of a synergistic genetic interaction (Supplementary Table 3). To directly test whether lung carcinogenesis was accelerated in Msh6−/−Rrm2Tg mice, we sacrificed a cohort of Msh6−/−Rrm2Tg mice and littermate controls at 6 months of age. 3 of 18 Msh6+/+Rrm2Tg mice and 3 of 17 Msh6+/−Rrm2Tg mice had developed lung neoplasms by 6 months, while no lung neoplasms were observed in transgene-negative Msh6+/+ or Msh6+/− littermates (Table 2). Lung neoplasms were also observed in 2 of 13 Msh6−/− mice. Msh6 deficiency strongly accelerated Rrm2-induced lung carcinogenesis, as 13 of 13 Msh6−/−Rrm2Tg mice developed lung neoplasms by 6 months of age, with 9 of these mice carrying multiple lung neoplasms.
Table 2.
Combining RNR overexpression with mismatch repair deficiency results in a synergistic increase in lung carcinogenesis
| Mouse genotype | # of animals | % of mice with lung neoplasms | % of mice with multiple lung neoplasms | # of lung neoplasms per mouse† ± SD | Average lung neoplasm size (mm) ± SD | % of mice with lymphoma |
|---|---|---|---|---|---|---|
| Msh6−/−Rrm2Tg | 13 | 100%* | 69%* | 2.9 ± 1.99 | 1.30 ± 0.54 | 31% |
| Msh6−/− | 13 | 15% | 0% | 1.0 ± 0 | 1.25 ± 1.06 | 8% |
| Msh6+/−Rrm2Tg | 17 | 18% | 0% | 1.0 ± 0 | 0.73 ± 0.68 | 12% |
| Msh6+/− | 10 | 0% | 0% | N/A | N/A | 0% |
| Msh6+/+Rrm2Tg | 18 | 17% | 6% | 1.33 ± 0.58 | 1.23 ± 0.25 | 0% |
| Msh6+/+ | 14 | 0% | 0% | N/A | N/A | 0% |
NOTE: Mice were aged for 6 months, euthanized by asphyxiation using carbon dioxide, and subjected to pathological examination as described in Materials and Methods. Only mice that lived to 6 months were included. Four Msh6−/−Rrm2Tg mice died before 6 months due to lymphoma, one of which also had a lung neoplasm. Four Msh6−/− mice died before 6 months due to lymphoma.
Values refer to the average number of lung neoplasms per mouse among tumor-bearing animals only.
Statistically significant difference (p<0.01) relative to Msh6−/−, Msh6+/−Rrm2Tg, or Msh6+/+Rrm2Tg mice as determined by Fisher’s Exact test.
To determine whether combining RNR overexpression with MMR deficiency would increase mutation frequency in vivo, we analyzed the mutation frequency at the λ phage cII locus in lung tissue from 3 month old RNRTg mice, with or without Msh6 deficiency, using the Big Blue transgene system (26). There was no difference in mutation frequency in RNR transgenic mice as compared to wild-type mice (Fig. 4C and Supplementary Table 4), possibly because the Big Blue system is relatively insensitive due to a high background mutation frequency. However, the mutation frequency in Msh6−/−Rrm2Tg (55.2 ± 20.9 × 10−5) and Msh6−/−p53R2Tg (54.0 ± 15.4 × 10−5) lung tissues was consistently higher than that in Msh6−/− lung tissue (31.1 ± 10.4 × 10−5), although these differences were not statistically significant. By contrast, the mutation frequency was similar in spleen tissue from Msh6−/−Rrm2Tg (52.0 ± 20.1 × 10−5) and Msh6−/− (50.5 ± 16.3 × 10−5) mice, but slightly elevated in Msh6−/−p53R2Tg (63.7 ± 10.7 × 10−5) animals (Fig. 4D and Supplementary Table 5). Together, these results indicate that MMR deficiency synergizes with RNR overexpression in a tissue specific manner to increase mutagenesis and carcinogenesis.
RNR-induced lung neoplasms display a unique signature of K-ras activating mutations
A mutagenic mechanism implies that RNR overexpression triggers additional genetic alterations while promoting tumor development. Because mutations in codons 12 and 61 of the K-ras proto-oncogene are often observed in human and mouse lung cancers (27, 28), we examined the frequency of K-ras mutations in microdissected lung neoplasms from the RNR cohort. 100% of Rrm2-induced lung neoplasms and 79% of p53R2-induced lung neoplasms carried K-ras activating mutations (Supplementary Table 6), indicating that RNR-induced lung carcinogenesis frequently involves K-ras activating mutations. 56% and 100% of the rare lung neoplasms from transgene-negative control and Rrm1Tg mice, respectively, also had K-ras mutations.
Sequence analysis revealed that the lung neoplasms from Rrm2Tg and p53R2Tg mice exhibited distinct mutation spectra relative to those from transgene-negative and Rrm1Tg mice (Supplementary Table 7). In particular, 50% of the K-ras codon 12 mutations from Rrm2-induced lung neoplasms were G→T transversions (GGT→GTT, G12V), as were 30% of those from p53R2-induced lung neoplasms. By contrast, lung neoplasms from transgene-negative and Rrm1Tg mice showed exclusively G→A transitions (GGT→GAT, G12D) in K-ras codon 12. We conclude that K-ras activating mutations, common events in lung carcinogenesis, are central to Rrm2- and p53R2-induced lung carcinogenesis and arise through a mechanism that appears distinct from that underlying spontaneous lung tumor development in wild-type animals.
DISCUSSION
RNR enzyme activity has long been positively correlated with cancer cell division (29), and RNR inhibition is an effective strategy for suppressing tumor proliferation and survival (30). Yet, investigation of the effects of RNR deregulation in animal models has been incomplete. We report that overexpression of Rrm2 or p53R2 specifically induces lung but not other neoplasms at high frequency in transgenic mice. Previous studies indicated that human RRM2 has transforming activity in cultured cells (31), while p53R2 has been suggested to have tumor suppressor activity based on its regulation by p53 and its role in the DNA damage response (2). RNR may be an example of a growth regulator that has dual roles both as a tumor suppressor and oncogene. While impaired RNR function can trigger genomic instability by limiting nucleotide availability for DNA replication and repair purposes, RNR hyperactivity may be equally detrimental due to its mutagenic effects. Interestingly, the genomic regions containing human RRM2 (2p25-2p24) and p53R2 (8q23.1) are commonly amplified in human lung cancers (32–35), raising the possibility that RNR deregulation might have a causative role in human lung carcinogenesis. Because RNR is a DNA damage-inducible enzyme, our results also suggest that increased RNR levels due to chronic DNA damage in the lungs of smokers may contribute to tumor development.
In contrast to Rrm2Tg and p53R2Tg mice, Rrm1Tg mice did not show increased lung carcinogenesis. This might be due to the relatively limited overexpression of the Rrm1 transgene, or the fact that the R2 subunit is the limiting component of the enzyme (7, 36). However, Rrm1 demonstrates tumor suppressor activity both in cultured cells and human lung cancer patients (37–39). Consistent with our findings, Rrm1 overexpression in another mouse model also did not result in any overt spontaneous phenotypes and instead was reported to suppress chemical carcinogenesis in the lung (40). Thus, lung tumor induction might be specific to the small RNR subunit and independent of RNR enzyme activity.
We determined that RNR-induced lung tumorigenesis proceeded through a mutagenic mechanism. Overexpression of Rrm2 or p53R2, but not Rrm1, in 3T3 cells resulted in a significant increase in mutation frequency. Additional experiments in budding yeast indicated that MMR normally corrects base mispairs that arise due to RNR deregulation, as multiplicative increases in mutation rate were observed when the allosteric site mutant rnr1-D57N was combined with MMR gene mutations. A similar genetic interaction between RNR and MMR was observed in mice. Msh6-null mice develop primarily lymphoma (17), and p53R2 overexpression cooperated with Msh6-deficiency to cause an earlier onset of lymphomagenesis and shortened lifespan in Msh6−/−p53R2Tg mice as compared to Msh6−/− controls. We also observed that Msh6 deficiency strongly accelerated Rrm2-induced lung carcinogenesis, with 100% of Msh6−/−Rrm2Tg mice developing lung neoplasms by 6 months of age. The accelerated lung carcinogenesis in Msh6−/−Rrm2Tg mice was associated with increased mutation frequency in lung tissue, while the accelerated lymphomagenesis in Msh6−/−p53R2Tg mice correlated with a modestly elevated mutation frequency in spleen tissue. The synergy observed between these pathways raises the possibility that aberrant RNR expression may be selected for in MMR-deficient cancers.
A key question arising from this study is the molecular basis for mutagenesis and lung tumor induction by Rrm2 and p53R2 overexpression. One possibility is that increased RNR expression leads to dNTP level alterations that impair replication fidelity and trigger mutations in growth regulatory genes. Abnormal nucleotide levels result in increased base misinsertion during DNA replication as well as decreased proof-reading due to enhanced polymerization rates (5). Consistent with the notion that regulators of nucleotide biosynthesis can influence cell transformation, overexpression of another enzyme involved in dNTP biosynthesis, thymidylate synthase, transforms cultured cells (41) and promotes tumor formation in transgenic mice (42).
Alternatively, carcinogenesis due to R2 subunit overexpression could be independent of nucleotide metabolism. One possibility is that free radical production by Rrm2 and p53R2 contributes to cell transformation. During each catalytic cycle the small RNR subunit generates a tyrosyl radical that normally is transferred to the active site in Rrm1 for use in NDP reduction (1). R2 protein overexpression might lead to increased radical generation and the formation of reactive oxygen species (ROS), which cause oxidative DNA damage and are mutagenic. ROS also have mitogenic effects and can play a direct role in neoplastic transformation (43). Notably, human RRM2 protein generates ROS in vitro, although recombinant p53R2 was reported in the same study to have antioxidant activity, despite the fact that both RRM2 and p53R2 generate tyrosyl free radicals (44). G→T transversions, a signature of oxidative DNA damage, were detected at K-Ras codon 12 in lung neoplasms from Rrm2Tg and p53R2Tg mice, and also at the Hprt locus in Rrm2 and p53R2 overexpressing 3T3 cells. Because MMR corrects mismatches arising from both replication errors (22) and oxidative DNA damage (45), the multiplicative increases in mutagenesis and carcinogenesis observed when combining RNR overexpression with MMR deficiency are compatible with both dNTP level alterations and increased ROS production as possible mechanisms of action.
The possibility that R2 subunit overexpression induces mutagenesis and tumorigenesis through excessive free radical production may account for the observation that RNR transgenic mice, despite broad RNR overexpression, develop lung but not other neoplasms at high frequency. The lung is an oxygen-rich environment with a high basal level of ROS (46) and thus may be more susceptible to increased free radical production. Alternatively, it could be that the mutational targets of RNR dictate the tissue specificity. Indeed, activated K-ras preferentially induces lung neoplasms in mice (47). Other more trivial explanations for the lung specific carcinogenesis, such as subtle transgene expression level differences or varying DNA repair efficiencies among tissues, also cannot be ruled out.
Although Rrm2 and p53R2 encode related R2 proteins, they did not give identical results in our experiments. While overexpression of either was capable of inducing lung neoplasms, Rrm2 overexpression elicited larger and more malignant tumors. p53R2 overexpression, on the other hand, significantly accelerated lymphomagenesis in Msh6-null mice, suggesting a broad effect of p53R2 overexpression. Rrm2 also was more mutagenic than p53R2 in cultured cells, and induced a greater proportion of G→T transversions in both the Hprt and K-ras genes. One possible explanation for the partially distinct phenotypes associated with Rrm2 and p53R2 is that both dNTP alterations and ROS production can contribute to neoplastic transformation, and that these activities differ between Rrm2 and p53R2. The distinct subcellular localizations of Rrm2 and p53R2 (2, 3, 13) could contribute to such differing effects on dNTP biosynthesis or ROS production.
Mouse models hold great promise for facilitating the development of diagnostic tools, prognostic markers, and therapeutics for lung cancer, the leading cause of cancer death world-wide. Like human lung adenocarcinomas (48), the RNR-induced lung neoplasms expressed SP-C, a marker of type II alveolar cells. Furthermore, RNR-induced lung neoplasms arose with moderate latency in a stochastic process associated with an elevated mutation rate, suggesting that this may be a particularly authentic model for lung cancer. A mutagenic mechanism for RNR-induced lung carcinogenesis implies that several genetic alterations are required for lung carcinogenesis. Consistent with this model, we observed activating K-ras mutations at very high frequency in RNR-induced lung neoplasms. K-ras has been reported to be mutated in 90% of mouse lung neoplasms and as many as 25% of human lung adenocarcinomas (27, 28). That G→T transversions in K-ras codon 12 were detected in RNR-induced lung neoplasms further validates this lung cancer model, as G→T transversions are the most common mutations at K-ras codon 12 in human lung cancers and correlate with a poorer prognosis (49, 50). Continued use of the RNR lung cancer model has great potential for revealing additional genetic alterations that contribute to lung tumor initiation and progression.
Supplementary Material
Table S1. Mutational spectrum at the Hprt locus in RNR overexpressing cell pools
Table S2. Mutational spectrum at the CAN1 locus in wild-type and rnr1-D57N yeast strains that vary in mismatch repair status
Table S3. Combining RNR overexpression with mismatch repair deficiency results in a synergistic increase in tumorigenesis
Table S4. Analysis of mutation frequencies at the cII locus of a bacteriophage λ transgene in lung tissue from Msh6−/− RNRTg and control mice
Table S5. Analysis of mutation frequencies at the cII locus of a bacteriophage λ transgene in spleen tissue from Msh6−/− RNRTg and control mice
Table S6. Mutations in K-ras codons 12 and 61 in lung tumors from RNR transgenic mice
Table S7. Mutational spectrum at K-ras codons 12 and 61 in RNR-induced and control lung neoplasms
Acknowledgments
We thank Dr. Phil Leder for his support of this project, Dr. Lars Thelander for Rrm1 and Rrm2 cDNA clones, Dr. Andrei Chabes for yeast strains 4069-4C and 4069-8C, Charlene Manning and Dr. Jianrong Lu for assistance with plasmid cloning, Nick Fuda for contributions to the yeast mutation rate analyses, Anne Harrington for performing transgene microinjections, and Dr. Ruth Collins and Dr. Sylvia Lee for helpful discussions.
Financial Support: This work was supported in part by NIH grants GM53085 (EA), CA96823 (AYN), and RR017595 (AYN). XX was supported by a Genomics Scholar Award from the Cornell University Center for Vertebrate Genomics, JLP was supported by NIH training grant T32GM07617, and JAS was a Research Fellow of the National Cancer Institute of Canada supported with funds from the Terry Fox Run.
References
- 1.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka H, Arakawa H, Yamaguchi T, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–9. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 3.Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19:4283–9. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 4.Thelander L. Ribonucleotide reductase and mitochondrial DNA synthesis. Nat Genet. 2007;39:703–4. doi: 10.1038/ng0607-703. [DOI] [PubMed] [Google Scholar]
- 5.Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–14. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 6.Bjorklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29:5452–8. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- 7.Mann GJ, Musgrove EA, Fox RM, Thelander L. Ribonucleotide reductase M1 subunit in cellular proliferation, quiescence, and differentiation. Cancer Res. 1988;48:5151–6. [PubMed] [Google Scholar]
- 8.Eriksson S, Graslund A, Skog S, Thelander L, Tribukait B. Cell cycle-dependent regulation of mammalian ribonucleotide reductase. The S phase-correlated increase in subunit M2 is regulated by de novo protein synthesis. J Biol Chem. 1984;259:11695–700. [PubMed] [Google Scholar]
- 9.Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci U S A. 2003;100:3925–9. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem. 2000;275:17747–53. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 11.Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–9. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 12.Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–41. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 13.Engstrom Y, Rozell B. Immunocytochemical evidence for the cytoplasmic localization and differential expression during the cell cycle of the M1 and M2 subunits of mammalian ribonucleotide reductase. EMBO J. 1988;7:1615–20. doi: 10.1002/j.1460-2075.1988.tb02987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 15.Reichard P, Eliasson R, Ingemarson R, Thelander L. Cross-talk between the allosteric effector-binding sites in mouse ribonucleotide reductase. J Biol Chem. 2000;275:33021–6. doi: 10.1074/jbc.M005337200. [DOI] [PubMed] [Google Scholar]
- 16.Caras IW, Martin DW., Jr Molecular cloning of the cDNA for a mutant mouse ribonucleotide reductase M1 that produces a dominant mutator phenotype in mammalian cells. Mol Cell Biol. 1988;8:2698–704. doi: 10.1128/mcb.8.7.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edelmann W, Yang K, Umar A, et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–77. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 18.Mahler JF, Stokes W, Mann PC, Takaoka M, Maronpot RR. Spontaneous lesions in aging FVB/N mice. Toxicol Pathol. 1996;24:710–6. doi: 10.1177/019262339602400606. [DOI] [PubMed] [Google Scholar]
- 19.Fenwick RG. The HGPRT system. In: Gottesman M, editor. Molecular Cell Genetics. 1. New York: Wiley; 1985. pp. 333–73. [Google Scholar]
- 20.Wijnhoven SW, Kool HJ, van Oostrom CT, et al. The relationship between benzo[a]pyrene-induced mutagenesis and carcinogenesis in repair-deficient Cockayne syndrome group B mice. Cancer Res. 2000;60:5681–7. [PubMed] [Google Scholar]
- 21.Zhang LH, Vrieling H, van Zeeland AA, Jenssen D. Spectrum of spontaneously occurring mutations in the hprt gene of V79 Chinese hamster cells. J Mol Biol. 1992;223:627–35. doi: 10.1016/0022-2836(92)90979-t. [DOI] [PubMed] [Google Scholar]
- 22.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–9. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau PJ, Flores-Rozas H, Kolodner RD. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol Cell Biol. 2002;22:6669–80. doi: 10.1128/MCB.22.19.6669-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–73. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–20. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 26.Jakubczak JL, Merlino G, French JE, et al. Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc Natl Acad Sci U S A. 1996;93:9073–8. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You M, Candrian U, Maronpot RR, Stoner GD, Anderson MW. Activation of the Ki-ras protooncogene in spontaneously occurring and chemically induced lung tumors of the strain A mouse. Proc Natl Acad Sci U S A. 1989;86:3070–4. doi: 10.1073/pnas.86.9.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills NE, Fishman CL, Rom WN, Dubin N, Jacobson DR. Increased prevalence of K-ras oncogene mutations in lung adenocarcinoma. Cancer Res. 1995;55:1444–7. [PubMed] [Google Scholar]
- 29.Elford HL, Freese M, Passamani E, Morris HP. Ribonucleotide reductase and cell proliferation. I. Variations of ribonucleotide reductase activity with tumor growth rate in a series of rat hepatomas. J Biol Chem. 1970;245:5228–33. [PubMed] [Google Scholar]
- 30.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6:409–31. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- 31.Fan H, Villegas C, Wright JA. Ribonucleotide reductase R2 component is a novel malignancy determinant that cooperates with activated oncogenes to determine transformation and malignant potential. Proc Natl Acad Sci U S A. 1996;93:14036–40. doi: 10.1073/pnas.93.24.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong MP, Fung LF, Wang E, et al. Chromosomal aberrations of primary lung adenocarcinomas in nonsmokers. Cancer. 2003;97:1263–70. doi: 10.1002/cncr.11183. [DOI] [PubMed] [Google Scholar]
- 33.Goeze A, Schluns K, Wolf G, Thasler Z, Petersen S, Petersen I. Chromosomal imbalances of primary and metastatic lung adenocarcinomas. J Pathol. 2002;196:8–16. doi: 10.1002/path.1009. [DOI] [PubMed] [Google Scholar]
- 34.Pei J, Balsara BR, Li W, et al. Genomic imbalances in human lung adenocarcinomas and squamous cell carcinomas. Genes Chromosomes Cancer. 2001;31:282–7. doi: 10.1002/gcc.1145. [DOI] [PubMed] [Google Scholar]
- 35.Lui WO, Tanenbaum DM, Larsson C. High level amplification of 1p32-33 and 2p22-24 in small cell lung carcinomas. Int J Oncol. 2001;19:451–7. doi: 10.3892/ijo.19.3.451. [DOI] [PubMed] [Google Scholar]
- 36.Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260:9114–6. [PubMed] [Google Scholar]
- 37.Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22:2135–42. doi: 10.1038/sj.onc.1206232. [DOI] [PubMed] [Google Scholar]
- 38.Fan H, Huang A, Villegas C, Wright JA. The R1 component of mammalian ribonucleotide reductase has malignancy-suppressing activity as demonstrated by gene transfer experiments. Proc Natl Acad Sci U S A. 1997;94:13181–6. doi: 10.1073/pnas.94.24.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–8. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 40.Gautam A, Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66:6497–502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- 41.Rahman L, Voeller D, Rahman M, et al. Thymidylate synthase as an oncogene: a novel role for an essential DNA synthesis enzyme. Cancer Cell. 2004;5:341–51. doi: 10.1016/s1535-6108(04)00080-7. [DOI] [PubMed] [Google Scholar]
- 42.Chen M, Rahman L, Voeller D, et al. Transgenic expression of human thymidylate synthase accelerates the development of hyperplasia and tumors in the endocrine pancreas. Oncogene. 2007;26:4817–24. doi: 10.1038/sj.onc.1210273. [DOI] [PubMed] [Google Scholar]
- 43.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 44.Xue L, Zhou B, Liu X, et al. Structurally dependent redox property of ribonucleotide reductase subunit p53R2. Cancer Res. 2006;66:1900–5. doi: 10.1158/0008-5472.CAN-05-2656. [DOI] [PubMed] [Google Scholar]
- 45.Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–51. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Rahman I. Oxidative stress, chromatin remodeling and gene transcription in inflammation and chronic lung diseases. J Biochem Mol Biol. 2003;36:95–109. doi: 10.5483/bmbrep.2003.36.1.095. [DOI] [PubMed] [Google Scholar]
- 47.Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 48.Linnoila RI, Mulshine JL, Steinberg SM, Gazdar AF. Expression of surfactant-associated protein in non-small-cell lung cancer: a discriminant between biologic subsets. J Natl Cancer Inst Monogr. 1992:61–6. [PubMed] [Google Scholar]
- 49.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2:411–8. [PubMed] [Google Scholar]
- 50.Rodenhuis S, Slebos RJ, Boot AJ, et al. Incidence and possible clinical significance of K-ras oncogene activation in adenocarcinoma of the human lung. Cancer Res. 1988;48:5738–41. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Mutational spectrum at the Hprt locus in RNR overexpressing cell pools
Table S2. Mutational spectrum at the CAN1 locus in wild-type and rnr1-D57N yeast strains that vary in mismatch repair status
Table S3. Combining RNR overexpression with mismatch repair deficiency results in a synergistic increase in tumorigenesis
Table S4. Analysis of mutation frequencies at the cII locus of a bacteriophage λ transgene in lung tissue from Msh6−/− RNRTg and control mice
Table S5. Analysis of mutation frequencies at the cII locus of a bacteriophage λ transgene in spleen tissue from Msh6−/− RNRTg and control mice
Table S6. Mutations in K-ras codons 12 and 61 in lung tumors from RNR transgenic mice
Table S7. Mutational spectrum at K-ras codons 12 and 61 in RNR-induced and control lung neoplasms