Abstract
RNA processing is altered during malignant transformation, and expression of the polypyrimidine tract-binding protein (PTB) is often increased in cancer cells. Although some data support that PTB promotes cancer, the functional contribution of PTB to the malignant phenotype remains to be clarified. Here we report that although PTB levels are generally increased in cancer cell lines from multiple origins and in endometrial adenocarcinoma tumors, there appears to be no correlation between PTB levels and disease severity or metastatic capacity. The three isoforms of PTB increase heterogeneously among different tumor cells. PTB knockdown in transformed cells by small interfering RNA decreases cellular growth in monolayer culture and to a greater extent in semi-solid media without inducing apoptosis. Down-regulation of PTB expression in a normal cell line reduces proliferation even more significantly. Reduction of PTB inhibits the invasive behavior of two cancer cell lines in Matrigel invasion assays but enhances the invasive behavior of another. At the molecular level, PTB in various cell lines differentially affects the alternative splicing pattern of the same substrates, such as caspase 2. Furthermore, overexpression of PTB does not enhance proliferation, anchorage-independent growth, or invasion in immortalized or normal cells. These data demonstrate that PTB is not oncogenic and can either promote or antagonize a malignant trait dependent upon the specific intra-cellular environment.
The polypyrimidine tract-binding protein (PTB),3 also termed heterogeneous nuclear ribonucleoprotein I, is a 57-kDa RNA-binding protein that binds preferentially to pyrimidinerich sequences (1–3). PTB contains four RNA recognition motifs (RRMs). RRM 1 and 2 at the N terminus of the protein are involved in the dimerization of PTB, whereas RRM 3 and 4 are responsible for high affinity interactions with RNA (4, 5). PTB has been shown to be involved in many aspects of pre-mRNA and mRNA metabolism. PTB participates in pre-mRNA splicing (6) and acts as a splicing repressor in alternative splicing of pre-mRNA (5, 7–12). PTB is also involved in 3′ end polyadenylation of pre-mRNA (13–15) and is important for translational regulation of certain RNA transcripts through internal ribosome entry sites (16–20). In addition, PTB shuttles between the nucleus and the cytoplasm (21), which is regulated through phosphorylation by 3′,5′-cAMP-dependent protein kinase (22).
Alternative splicing is a process that allows multiple different proteins to be made from the same pre-mRNA by either including or excluding particular exons during pre-mRNA splicing. PTB plays a key role in alternative site selection for many gene products by acting as a splicing repressor that prevents the inclusion of target exons (11, 23–25). Changes in alternative splicing sites have been previously correlated with malignant transformation (26–29), and the expression level of PTB has been found elevated in transformed cells. Such an elevation is responsible for the increases in fibroblast growth factor receptor-1α-exon skipping in glioblastoma multiforme tumors (26). Increases in PTB expression are also associated with changes in alternative splicing of multidrug resistance protein 1, which contributes to the drug-resistant phenotype associated with many cancers (28). In addition to PTB, changes in the expression levels of other factors involved in alternative splicing, such as SR proteins, have been found to impact the metastatic phenotype. A classic example demonstrated that a CD44 splice variant, CD44 v6, confers metastatic potential when expressed in nonmetastatic cells (30). Therefore, changes in alternative splicing dynamics in tumor cells likely modify gene expression, in which the expression of functionally altered proteins may directly contribute to the malignant phenotype (31). Being a splice repressor, PTB may influence the transformed phenotype through changing alternative splicing patterns.
PTB itself also undergoes alternative splicing and has three splicing isoforms. PTB1 is the smallest, whereas PTB2 and -4 have an additional 19 or 26 amino acids, respectively, between RRM 2 and 3 as a result of exon 9 inclusion (1, 2). These isoforms are differentially effective in the alternative splicing of α-tropomyosin. PTB4 has strongest influence and PTB1 the weakest on exon 3 skipping in vivo and in vitro (32). However, differential splicing efficiency of the individual PTB isoforms is not observed for all PTB substrates, as demonstrated by the equal efficiency of α-actinin exon skipping by all isoforms (32). Therefore, the differential expression of PTB isoforms may enormously influence gene expression because of the large number of PTB substrates present in the cell and thus differentially affect cellular behavior based on the amount of certain PTB substrates that are expressed in a given cell type.
Although several studies have demonstrated altered PTB expression in cancer cells (31, 33), fundamental questions regarding the role of PTB in cancer cells remain unresolved. It is not clear whether increased PTB expression is a phenomenon common to cancers of all origins, or whether PTB isoform expression changes similarly among cancers from different origins, or whether increased PTB expression is important for the transformed phenotype. Recently, a study using siRNA knockdown technique demonstrated that PTB promotes the malignant phenotype in ovarian tumor cell lines (34). To further address these issues, we investigated the changes in PTB expression in cancer cells from multiple origins and compared the PTB isoform profiles in these cells. We examined the role of PTB in malignant transformation by knocking down PTB expression in cultured tumor and nontransformed cells. We found that PTB levels generally increase in cancer cells from a variety of tissues; however, the expression levels of the three individual PTB isoforms are heterogeneous among different cell types. PTB knockdown by siRNA significantly reduces the growth rate for both cancer and normal cell lines, and reduces anchorage-independent growth in tumor cells to a great extent than monolayer culture. In addition, PTB knockdown inhibits the invasive capacity of two cancer cell lines but increased invasion in another. However, overexpression of PTB in normal and immortalized cells does not increase proliferation or induce traits associated with transformation in vitro. Our findings suggest that PTB itself is not transforming but may support or interfere with malignancy depending on the specific cellular environment as it can promote transformed phenotypes in some cells while antagonizing them in others.
MATERIALS AND METHODS
Cell Culture and Tissue Specimens—HeLa (human cervical cancer), Wacar and Homa (normal human skin fibroblasts), HEK-293 (human embryonic kidney transformed with adenovirus 5 DNA), and NIH-3T3 (Mus musculus, fibroblasts) were maintained in Dulbecco's modified Eagle's medium. PC-3, PC-3M, PC-3M Pro4, and PC-3M LN4 prostate cancer cell lines were generous gifts from the laboratories of Dr. Zhou Wang and Dr. Chung Lee (Northwestern University) and cultured in RPMI 1640 medium. WI-38 normal lung fibroblasts were grown in minimal essential medium. All media were supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 100 units/ml penicillin and streptomycin unless otherwise noted. CG cells (human neuroblastoma), T84 cells (human colon carcinoma), and SAOS-2 (osteosarcoma) were cultured according to the protocols provided by ATCC Cultures™. Stably expressing NIH-3T3 cells were created by transfecting ∼2 × 106 cells with 2 μg of either PTB1-GFP vector (39) or constitutively active K-Ras vector (courtesy of Dr. William Hahn-Addgene plasmid 9051) or 2 μg of each at the same time and then selecting with 500 μg/ml G-418 (PTB-GFP) and/or 5 μg/ml puromycin (H-Ras) for 2 weeks. The resulting stable, nonclonal cell lines were utilized for assays within 1 month of creation. Human endometrial tissue samples were obtained by surgical resection, trypsinized, and seeded in culture (Robert H. Lurie Comprehensive Cancer Center of Northwestern University). Histopathological examination allowed the samples to be classified as benign, grade-1, grade-2, or grade-3 endometrial tumors. All cell culture products were obtained from Invitrogen, and all other reagents mentioned under “Materials and Methods” were obtained from Sigma unless otherwise noted.
Immunostaining—Cells were fixed with 4% paraformaldehyde in PBS for 10 min followed by 5 min of permeabilization with 0.5% w/v Triton X-100 in PBS at room temperature. Primary antibody was applied for 1 h, and cells were washed with PBS three times for 10 min. The primary antibody, SH54 (anti-PTB) (35), was used at a 1:300 dilution in PBS, and secondary anti-mouse antibodies conjugated to fluorescein isothiocyanate or Texas Red were used at a 1:200 dilution (Jackson ImmunoResearch Laboratories). Coverslips were analyzed with a Nikon Eclipse E800 microscope equipped with a SenSys cooled CCD camera (Photometrics). Images were captured using Metamorph image acquisition software (Universal Imaging).
Protein Electrophoresis and Immunoblotting—Protein extracts were prepared by sonicating tissue or cells in RIPA buffer containing 1% Nonidet P-40, 1% deoxycholic acid, sodium salt, 0.1% SDS, 10 mm Tris-HCl, pH 7.4, and 150 mm NaCl. Protein concentrations were determined with the BCA protein assay kit (Pierce). Equal amounts of each protein sample were separated on 10% SDS-polyacrylamide gel and transferred to nitrocellulose membrane. Antibodies used for Western blot analysis were rabbit or mouse anti-PTB (SH54) at a 1:800 dilution, rabbit anti-actin (Sigma) at a 1:2000 dilution, mouse anti-GFP (BD Biosciences) at a 1:1000 dilution, rabbit anti-K-Ras (Santa Cruz Biotechnology) at a 1:500 dilution, and horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibodies (Jackson ImmunoResearch) at a 1:10,000 dilution. SuperSignal West Pico Chemiluminescent Substrate (Pierce) detection reagents were used to detect immunoreactive bands.
Northern Blotting—To determine the RNA expression level of PTB in different cell lines by Northern analysis, total RNAs were extracted from different cell lines with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Total RNAs from different cell lines were loaded (5 μg/lane) and run on a 1% agarose gel and subsequently transferred onto GeneScreenPlus membranes (PerkinElmer Life Sciences) by capillary action with a high salt solution. Hybridization and washing conditions were standard as described previously (36). A 32P-labeled PTB probe was used to detect the expression level of RNA with labeled glyceraldehyde-3-phosphate dehydrogenase probe used as loading control. 32P was obtained from Amersham Biosciences.
RT-PCR—RNA was converted to cDNA, and the DNA was amplified by PCR with a forward primer (5′-ACCAGCCTCAACGTCAAGTA) and a reverse primer (5′-GGGTTGAGGTTGCTGACCAG) in a single reaction. These primers were designed to include the alternatively spliced region of PTB so ratios of isoforms could be directly compared among cell lines. Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) on total RNA obtained via TRIzol isolation from cell lines. PCR was carried out with 30 cycles at 95 °C for 30 s, 57 °C for 1 min, and 72 °C for 90 s. The PCR products were resolved on a 2% agarose gel. The intensity of the isoform bands were measured with Kodak MI software, which allowed for determination of isoform ratios.
Alternative Splicing Efficiency Assay—A caspase 2 minigene was used as described previously (37). HEK-293 cells were plated onto 6-well culture plates at 60–70% confluence. After 24 h, 4 μg of GFP-tagged PTB expression vectors and 1 μg of reporter minigene were introduced into cells by a standard calcium phosphate precipitation protocol. Total RNA was purified with RNeasy mini kit (Qiagen) from 6-well culture plates 36 h after transfection. Alternative splicing products of the caspase 2 minigene were detected using RT-PCR in the presence of [32P]dCTP (GE Healthcare) as described previously (37, 38). PCR products were fractionated with 6% polyacrylamide gel containing 1× TBE buffer and then detected and quantified using a PhosphorImager BAS-1800II (Fuji Film).
RNA Interference—Double-stranded RNA was chemically synthesized, deprotected, and purified by Dharmacon Research, Inc. One strand of the double-stranded RNA was homologous to the PTB mRNA sequence 5′-UGACAAGAGCCGUGACUAC(dTdT)-3′. The scramble control siRNA was from Ambion, Inc. (Silencer® negative control 2 siRNA). Transfection of siRNA duplexes into various cell lines was conducted as described previously according to the manufacturer's instructions (Oligofectamine reagent, Invitrogen) (39). Cells were utilized for subsequent experiments 72 h post-transfection.
In Vivo Br-UTP Incorporation—Seventy two hours after transfection with PTB siRNA, cells were rinsed once in Glycerol Buffer (20 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 25% glycerol, 0.5 mm EGTA, 0.5 mm phenylmethylsulfonyl fluoride) and permeabilized in the Glycerol Buffer with 50 μg/ml digitonin for 3 min. Cells were then incubated in transcription mixture (100 mm KCl, 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm EGTA, 25% glycerol, 2 mm ATP, 0.5 mm CTP, 0.5 mm GTP, 0.2 mm Br-UTP) at 37 °C for 5 min and then fixed in 4% (w/v) paraformaldehyde in PBS for 10 min. Fixed cells were double-labeled with anti-PTB at a 1:300 dilution and anti-BrdUrd that also recognizes Br-UTP (Sigma) at a 1:50 dilution and subsequently prepared as described above.
Anchorage-independent Growth Assay—Seventy two hours after transfection, cells from the PTB siRNA and the control siRNA-treated dishes were trypsinized and counted using a hemocytometer. The same number of cells (about 5 × 104) from all experimental conditions were added into 2 ml of 1.5% (w/v) methylcellulose media and seeded onto 1% agarosecoated 35-mm Petri dishes and allowed to grow for 10 days. At this point, pictures were taken using phase microscopy to show the colony formation. Then the media containing the cells were removed from the dish, put in a 15-ml tube, vigorously pipetted and vortexed to break up the colonies, allowed to sit for 5 min, and then gently mixed as to suspend the cells homogeneously while avoiding air bubbles. The cell number relative to control was determined by measuring the scattering at 650 nm using a spectrophotometer (Beckman DU-64).
Invasion Assay—Invasive activity was determined via the transwell Matrigel invasion (Boyden chamber) assay. Transwell inserts (0.8 μm; BD Biosciences) were coated with Matrigel (100 μg in 100 μl, for 1 h at room temperature), and coated inserts were then washed with PBS and used immediately. Seventy two hours after siRNA transfection, 2 × 105 cells from each experimental condition were added to the upper chamber in 500 μl of serum-free medium. Twenty four hours after incubation at 37 °C, the noninvading cells were removed from the upper chamber with a cotton swab, and invading cells adherent to the bottom of membrane were fixed and stained using a Diff-Quick staining kit (DADE AG). Invading cells were counted by tallying the number of cells in 10 random fields under a ×20 objective using an ocular micrometer. Data were expressed as average relative (compared with control) number of migrating cells in 10 fields from six experiments (40).
Plasminogen Activator Assay—Net plasminogen activator activity in conditioned media was quantified using a coupled assay to monitor plasminogen activation and the resulting plasmin hydrolysis of a colorimetric substrate (d-Val-Leu-Lys-p-nitroanilide; Sigma) as described previously (41, 42).
RESULTS
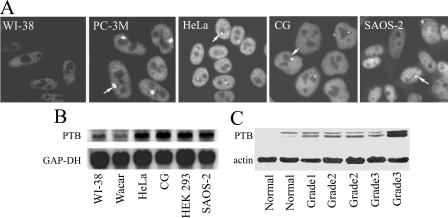
Transformed Cell Lines and Cancer Tissues Express Increased Levels of PTB—To evaluate the expression level of PTB in tumor cell lines and human tissue samples, we used both immunofluorescent staining and Western blotting. For the immunofluorescent staining, tumor and normal cells were immunolabeled in parallel, and the images were captured under the same image acquisition settings. The results show a significant increase in nuclear labeling intensity in tumor cells over that in normal cells as exemplified in PC-3M (a human prostate cancer cell line), HeLa (human cervical cancer), CG cells (human neuroblastoma), SAOS-2 (osteosarcoma), versus WI-38 (a normal human lung fibroblast cell line) (Fig. 1A). Many malignant cells also contain the perinucleolar compartment (Fig. 1A, arrowheads), a nuclear structure that is highly enriched with PTB (43). To quantify the expression of total PTB protein in cancer cell lines from various tissue origins, we performed Western blotting using the anti-PTB antibody SH54 (35). The panel of human cancer cells examined (HeLa, CG, T84, SAOS-2, HEK-293, PC-3, PC-3M, PC-3M LN4, and PC-3M Pro4) was derived from a broad spectrum of tissue types and represents cells of varying degrees of malignancy. The normal cell lines evaluated were WI-38 and Homa, which are human fibroblasts. Western blotting demonstrates that the level of PTB protein is generally increased in transformed cell lines examined when compared with the normal cell lines (Fig. 2B). The increases in protein expression is consistent with the increased level of steady state PTB mRNA in tumor cells as measured by Northern blotting (Fig. 1B). Densitometry quantification of the PTB protein levels shows that most tumor cell lines express PTB at a level 2-fold, or greater, than normal cells (data not shown). To evaluate whether the increases in PTB levels are correlated with the degree of malignancy, we compared PTB expression in three prostate cancer cell lines of varying levels of malignancy with the parental PC-3 line. PC-3 cells were originally isolated from a human prostate cancer, and PC-3M was created by implanting a PC-3 xenograft tumor into a nude mouse, allowing distant metastases to form, and subsequently removing a metastatic lesion to culture (44). PC-3M LN4 is enriched with highly metastatic cells through four iterations of inoculating PC-3M cells into the mouse prostate and isolating the metastatic tumor cells. In contrast, PC-3M Pro4 is highly concentrated with nonmetastatic tumor cells through four iterations of inoculating PC-3M cells into mouse prostate and isolating tumor cells localized to the prostate (45). If PTB level is directly related to the metastatic capacity, we would expect PTB expression in the PC-3 panel of cell lines to correlate with their metastatic capacity; however, PTB is expressed at a higher level in the PC-3M cells compared with PC-3 cells, and there is little difference among the three PC-3M derivatives (Fig. 2B).
FIGURE 1.
PTB expression is generally elevated in transformed cell lines and endometrial carcinomas. A, immunofluorescent staining against PTB demonstrates higher nuclear labeling intensity in tumor cells over that in normal cells as exemplified in PC-3M (human prostate cancer cell line) HeLa (human cervical cancer), CG (human neuroblastoma), and SAOS-2 cells (human osteosarcoma) versus WI-38 (normal human lung fibroblast). B, Northern blotting analysis shows the increased level of steady state PTB mRNA in tumor cells versus normal cell lines. C, Western blotting of lysates from five endometrial carcinoma and two normal endometrial tissue samples shows that PTB expression increases in tumor tissues but does not correlate with the histological tumor grading. GAP-DH, glyceraldehyde-3-phosphate dehydrogenase.
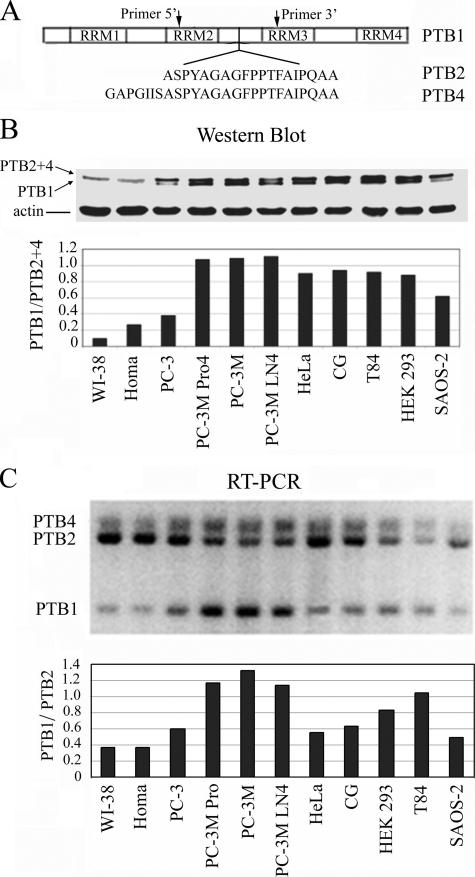
FIGURE 2.
Differential expression patterns of PTB isoforms. A, schematic illustration of PTB isoforms and PCR primers used to detect all PTB splicing isoforms. B, Western blotting shows PTB levels are generally higher in transformed cell lines when compared with normal cell lines. The shortest isoform, PTB1, shows significant increases in all cancer cell lines tested compared with normal cells, in which PTB1 is often below the level of detection. The normal cell lines used were WI-38 (lung fibroblasts) and Homa (human skin fibroblasts). C, quantitative RT-PCR shows that the ratio of PTB1:PTB2 is substantially increased in some tumor cell lines but not in all.
To evaluate the expression of PTB in human tumor tissues, we examined the levels of PTB expression in freshly isolated normal endometrial and endometrial adenocarcinoma cells. PTB expression in cells isolated from five adenocarcinoma tissues of varying grades was compared with two normal endometrial tissue cells by Western blotting, and the results show the total level of PTB is increased in tumor tissues, which is consistent with the observations in cancer cell lines (Fig. 1C). However, there is no obvious correlation between the expression level of PTB and tumor grades as exemplified by the grade 3 tumor samples, in which one shows a substantial increase in PTB expression although the other is comparable with or slightly less than the grade 1 tumor (Fig. 1C). The lack of correlation between the levels of PTB and the severity of the disease are consistent with the findings from the prostate cancer PC-3 cell line derivatives (Fig. 2B). Together, these data demonstrate that PTB levels generally increase in cancer cells independent of the tissue origin or degree of malignancy, as characterized by metastatic capacity or histological grading.
Differential PTB Isoform Expression in Cancer Cells—Although PTB levels are generally elevated in cancer cells, the ratios of the three PTB isoforms are heterogeneous among the cancer cell lines (Fig. 2B). The shortest isoform, PTB1, shows significant increases in all cancer cell lines tested compared with normal cells, in which PTB1 is often below the level of detection (Fig. 2B). Because PTB2 and PTB4 cannot be resolved on SDS gels, we evaluated the isoform expression by RT-PCR, which also ensures that the different bands observed on the Western blot are indeed because of alternative splicing rather than post-translational modifications. A set of primers was designed to include the alternatively spliced region that amplifies all three variants (Fig. 2A). The amplification of all variants in a single reaction provides an internal control for quantification of the proportion of each RNA isoform from a given cell line and thus allows comparisons between unrelated cell lines. Because the ratio of PTB1:PTB4 is relatively unchanged in all tested samples, we focused on the ratio of PTB1:PTB2 (Fig. 2C). In normal cells (WI-38 and Homa), PTB2 and -4 are the predominant isoforms so that the ratio of PTB1: PTB2 in these cells is less than 0.4 (Fig. 2C). This is consistent with the findings by Western blot (Fig. 2B) (the top PTB band represents PTB2 and -4), in which PTB1 is not detected. In comparison, cancer cell lines have heterogeneous ratios of PTB isoforms. Some of the cell lines, including PC-3M, PC-3M Pro4, PC-3M LN4, HEK-293, and T84 cell lines (Fig. 2B) and endometrial adenocarcinoma cells (Fig. 1C), show increased expression of the small isoform PTB1 over the other isoforms (Fig. 2B and Fig. 1C). The changes at the protein level are consistent with the findings at the RNA level as detected by RT-PCR (Fig. 2C), in which the ratio of PTB1:PTB2 significantly increases in the corresponding cancer cells, reaching as high as 1.32 for PC-3M cells. In contrast, other cancer cell lines, PC-3, HeLa, GC, and SAOS-2 maintain an isoform ratio of PTB1:PTB2 that is closer to the normal cells (Fig. 2, B and C). The heterogeneity of PTB isoform expression profiles in tumor cells from various origins and of varying levels of malignancy suggests that isoform switching toward PTB1 is not directly correlated with the malignant transformation. Our findings are consistent with another report where two prostate cancer cell lines predominantly increase the expression of PTB1, although HeLa expressed predominantly PTB2/4 (46).
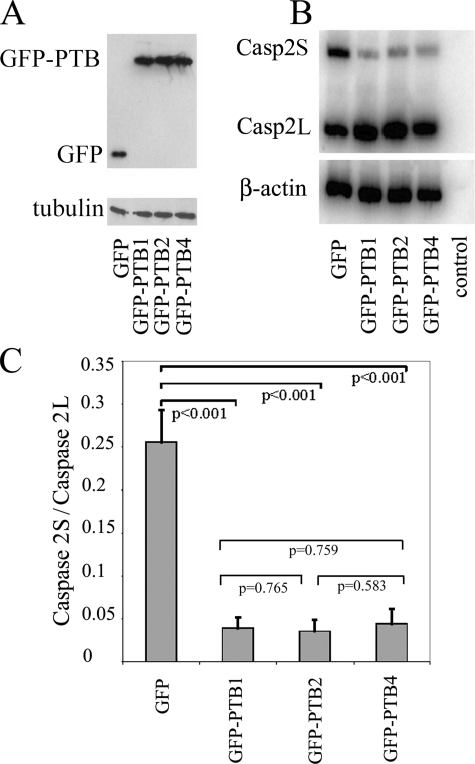
A previous study (32) showed that PTB isoforms have different effects on exon 3 exclusion of α-tropomyosin but have the same efficiency in excluding both the NW and SM exons of α-actinin pre-mRNA. These findings indicate substrate-dependent splicing activity for different PTB isoforms. To further evaluate the impact of each isoform on alternative splicing, we examined the effects of PTB isoforms on the inclusion of caspase 2 exon 9 (47). Although the full-length caspase 2 (caspase 2L) functionally promotes apoptosis, an inclusion of exon 9 generates a truncated protein product (caspase 2S) by frameshift, which inhibits programmed cell death (48). HEK-293 cells were transfected either with a construct expressing GFP alone or with a construct expressing GFP-tagged PTB isoforms. The transfection efficiency and expression levels of these proteins were very similar among the three isoforms as measured by Western blot (Fig. 3A). The expression of all three GFP-PTB variants significantly shifts the caspase 2 minigene from the short (caspase 2S) to the long (caspase 2L) form (Fig. 3B). Although GFP-PTB4 appears to be slightly less efficient than the other isoforms, the differences are not significant (Fig. 3C). Therefore, three PTB isoforms have similar influence in the splice site selection for caspase 2 and promote the formation of caspase 2L. Together with a previous study (32), these results suggest that PTB isoforms may have distinct or similar splicing regulatory efficacy, depending on the splice substrate. The similar or differential influence of PTB isoforms on a large number of PTB substrates could generate a very complex expression pattern of different protein products in different cell populations.
FIGURE 3.
Effects of PTB isoforms on caspase 2 alternative splicing. A, expression of PTB isoforms in transfected cells was confirmed by Western blot. B, overexpression of PTB isoforms resulted in decreases in caspase 2S (Casp2S) and increases in caspase 2L (Casp2L) as detected by RT-PCR. C, ratio of caspase 2S/caspase 2L was measured by densitometry from the RT-PCR experiments. The ratio of caspase 2S to 2L was significantly different from the control (GFP), but there was no difference among different PTB isoform groups.
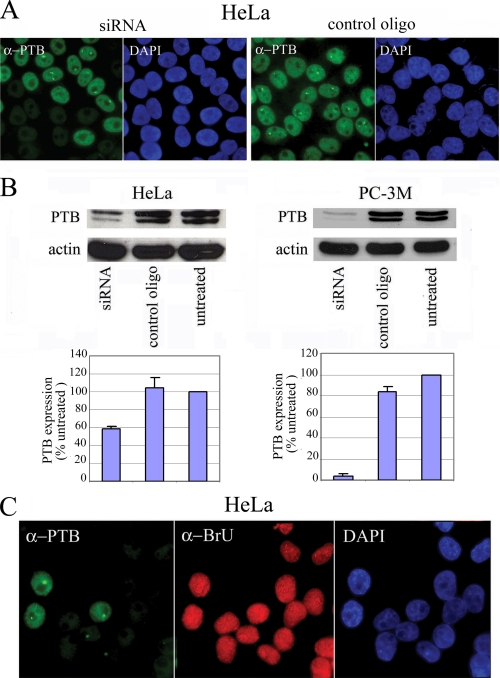
PTB siRNA Down-regulates PTB Expression without Impacting Global Cellular Transcription or Inducing Cell Death—To evaluate the functional significance of increased expression of PTB in the malignant behavior of cells, we knocked down PTB in both malignant cells and nontransformed cells by siRNA. The PTB siRNA oligo used in these experiments targets the 5′ end of the mRNA and effectively eliminates the majority of PTB mRNAs (39). PTB siRNA and control oligos were transfected into PC-3M and HeLa cells. Seventy two hours after transfection, the expression of PTB was evaluated by immunofluorescence labeling and by Western blotting (Fig. 4). Immunolabeling of siRNA-treated HeLa cells demonstrated a significant reduction of PTB expression when compared with cells transfected with the control oligo (Fig. 4A) with a transfection efficiency generally over 50% (data not shown). Western blot analyses show that PTB expression in siRNA transfected cells can be reduced by ∼95% in PC-3M cells (Fig. 4B).
FIGURE 4.
Down-regulation of PTB expression by siRNA. A, immunolabeling demonstrates the reduction of PTB levels in siRNA-transfected HeLa cells, but not in control oligo-transfected cells with a transfection efficiency generally over 50%. B, Western blotting confirms effective siRNA knockdown of PTB in transfected cells. C, BrU incorporation assay with HeLa cells demonstrates that PTB siRNA does not greatly alter the cellular transcription state in the nucleolus (pol I transcription) or in the nucleoplasm (pol II and pol III transcription). DAPI, 4′,6-diamidino-2-phenylindole.
Prior to evaluating how PTB reduction impacts the transformed phenotype, it is important to exclude the possibility that PTB knockdown is detrimental to cells. To do so, we examined the transfected cells for transcriptional activities and apoptotic indices. Our previous studies have shown that PTB knockdown did not significantly change the intra-nuclear distribution of pre-mRNA splicing factors or the nucleolar localization of pre-rRNA processing factors (39). Because localization of these factors is generally sensitive to transcriptional inhibition, those findings suggested that cells with reduced PTB expression remain transcriptionally active and structurally intact in terms of subcellular compartments (39). To directly assess the influence of PTB on cellular transcription, we performed BrU incorporation assays in HeLa cells transfected with PTB siRNA oligos and compared them with the adjacent cells with normal PTB expression. Cells were pulse-labeled with BrU for 5 min, and the newly synthesized RNA incorporated with BrU was detected using a specific antibody recognizing BrU. Cells with severely decreased PTB expression maintain very similar BrU incorporation levels and patterns both in the nucleolus (pol I transcription) and in the nucleoplasm (pol II and pol III transcription) (Fig. 4C), demonstrating that PTB knockdown indeed does not significantly impact global transcriptional activity. To determine whether decreases in PTB levels might induce apoptosis, we compared the apoptotic index in cells treated with PTB siRNA or control oligos. Three days after transfection, the number of cells undergoing apoptosis was evaluated using 4′,6-diamidino-2-phenylindole staining. The apoptotic index (the percentage of apoptotic cells per 100 nonmitotic cells) was not significantly different between cells transfected with PTB siRNA or control oligos (data not shown), which demonstrates that reduction of PTB by siRNA does not induce apoptosis.
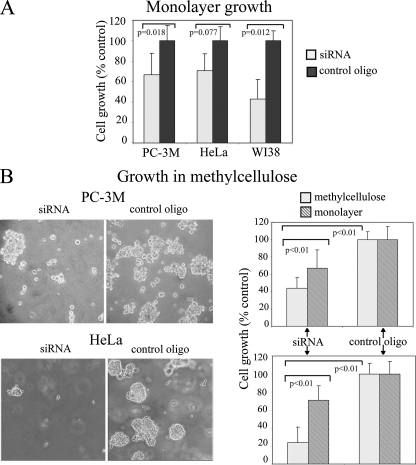
PTB Knockdown Reduces Cellular Proliferation and Anchorage-independent Growth—PTB is involved in the RNA metabolism of a large number of transcripts in several different capacities, including polyadenylation, RNA stability, alternative splicing, and translational regulation of mRNA. Therefore, knockdown of PTB expression may have significant impacts on many fundamental cellular activities. To determine the impact of PTB knockdown on cell proliferation, we compared the growth rate of cells transfected with PTB siRNA and control oligo. Seventy two hours after transfection, cells were trypsinized, and an equal number of cells from both groups were reseeded and allowed to grow for an additional 5 days. PTB knockdown significantly reduced the growth of two tumor cell lines (HeLa and PC-3M) and a normal cell line (WI-38) (Fig. 5A). The reduction appears to be more severe in normal cells than in the two tumor cell lines. These findings demonstrate that PTB is likely ubiquitously important to maintain cell proliferation.
FIGURE 5.
PTB knockdown reduces cellular proliferation and anchorage-independent growth. A, PTB down-regulation causes growth inhibition in tumor cell lines (HeLa and PC-3M) and a normal cell line (WI-38) in monolayer culture. B, cells transfected with PTB siRNA show a significant reduction in anchorage-independent growth in semi-solid media (methylcellulose). Left panel, photographs of plates on a bright field microscope with a ×10 objective. Right panel, quantification of growth in semi-solid media, and comparison with growth in monolayer culture demonstrates that PTB inhibits anchorage-independent growth to a greater extent than monolayer growth. Both parameters in siRNA transfected cells are compared with the cells treated with control oligos (n = 3 and error bars =+S.D.).
To evaluate whether the increased PTB expression observed in tumor cells contributes to their malignant behavior, we examined the impact of PTB knockdown on the growth of cancer cells in semi-solid media and compared it with the results from monolayer culture. The semi-solid media assay is indicative of the ability of cells to grow (form colonies) in an anchorage-independent manner, a trait unique to malignantly transformed cells. Three days following transfection with either PTB siRNA or control oligo, an equal number of cells were seeded in media with 1.5% methylcellulose and cultured for 10 days. The total cell number for the two experimental groups was measured, and the results demonstrate that cells transfected with PTB siRNA show a significant growth reduction in methylcellulose as compared with those transfected with control oligos in two tumor cell lines tested (HeLa and PC-3M) (Fig. 5B). To determine whether the growth reduction is specifically because of suppression of anchorage-independent growth rather than to the general proliferation reduction observed in monolayer culture, we standardized the data to control values for each condition to allow for comparisons. When compared with controls, the growth rate of cells transfected with PTB siRNA was decreased to a greater extent in methylcellulose media than in monolayer culture, demonstrating that PTB reduction not only reduces proliferation in general, but it also reduces anchorage-independent growth, which is an in vitro indicator of transformation.
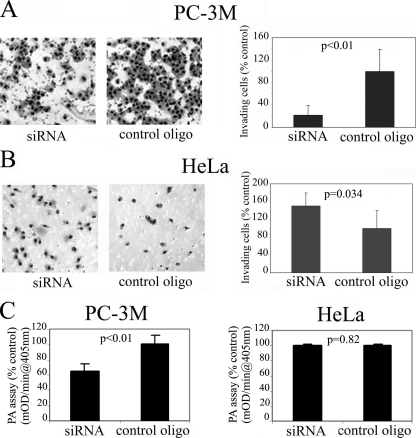
PTB Knockdown Differentially Affects the Invasive Behavior of Cancer Cells—To further evaluate the role of PTB in the malignancy, we examined the ability of PTB siRNA to inhibit the invasive behavior of cancer cells. The Matrigel invasion (Boyden chamber) assay is a well accepted in vitro assay that determines the ability of tumor cells to penetrate a proteinaceous matrix resembling the tumor basement membrane. Equal numbers of cells transfected with siRNA against PTB or control oligo were seeded onto the upper chamber of a transwell and incubated at 37 °C for 24 h. Cells remaining at the upper chamber were removed, whereas cells that penetrated into the lower chamber were fixed, stained, quantified, and compared among the experimental groups. The results from these experiments (n = 6) consistently show that PTB knockdown in both PC-3M (Fig. 6A) and T84 cells (data not shown) significantly reduces the number of cells that are capable of invading through the Matrigel. To further examine the mechanism by which PTB might contribute to the invasive behavior of these cells, we compared the extracellular protease activities in PC-3M cells transfected with either PTB siRNA or control oligos. Plasminogen activators are serine proteases that cleave the inactive zymogen, plasminogen, into active plasmin. Once activated, plasmin can cleave almost all extracellular matrix proteins either directly or indirectly by activating zymogens belonging to other proteases classes like the matrix metalloproteases. Plasminogen activators play a critical role in invasion and metastasis of virtually all cancer types studied. In this study we analyzed the level of plasminogen activator activity in the supernatant of cells treated with PTB siRNA or control oligos using a published protocol (49). PC-3M cells transfected with PTB siRNA produce significantly less active extracellular protease than cells transfected with control oligos (Fig. 6C). This suggests that PTB may play a role in the production of active extracellular proteases in these cells. In contrast to the inhibition of invasion by PTB knockdown in PC-3M and T84 cells, PTB knockdown in HeLa cells increased the invasive behavior of these cells (Fig. 6B). Correspondingly, the production of extracellular plasmin is not reduced in these cells (Fig. 6C). The finding is reproducible and consistent in several independent experiments. Together, these findings demonstrate that PTB differentially influences malignant traits depending on the cell lines.
FIGURE 6.
PTB knockdown differentially affects the invasive capacity of cancer cells. A, PTB knockdown in PC-3M cells significantly reduces Matrigel invasion; B, but it increases cell invasion in HeLa cells. C, PTB knockdown significantly reduces plasminogen activator production in PC-3M cells but does not in HeLa cells (n = 3 and error bars =+S.D.).
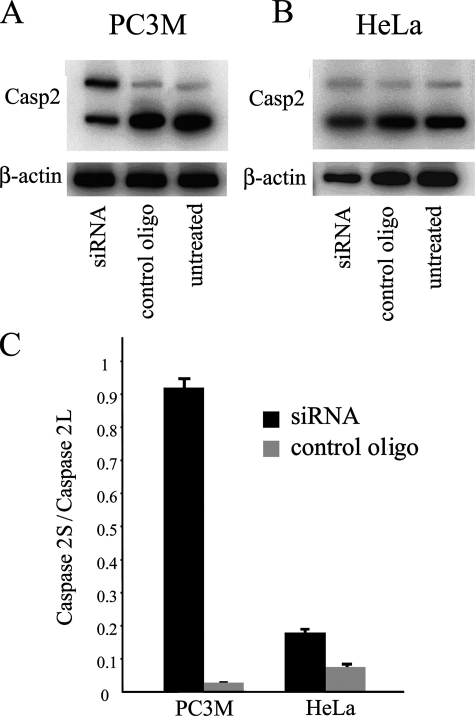
PTB Knockdown Differentially Affects the Alternative Splicing of the Same Substrates in Different Cells—To begin to address the mechanism behind the differential role PTB plays in various cancer cell lines, we examined the influence of PTB knockdown on the same alternatively spliced substrate in HeLa and PC-3M cells. Caspase 2 exon 9 inclusion (47) was assayed as described in Fig. 3. As shown in Fig. 3, overexpression of PTB reduces the inclusion of exon 9 in HEK-293 cells, leading to the increases of the caspase 2L form. Correspondingly, PTB reduction dramatically increases exon 9 inclusion in PC-3M cells, resulting in increases of caspase 2S form (Fig. 7, A and B). However, PTB reduction has little effect on the alternative splicing pattern of this substrate in HeLa cells (Fig. 7, B and C). These observations further demonstrate that PTB acts differentially upon the same cellular function, probably dependent on the genetic and epigenetic background of the specific cells.
FIGURE 7.
A and C, PTB reduction dramatically increases exon 9 inclusion in PC-3M cells; B and C, but it minimally affects the alternative splicing pattern of caspase (Casp) 2 in HeLa cells (n = 3 and error bars =+S.D.).
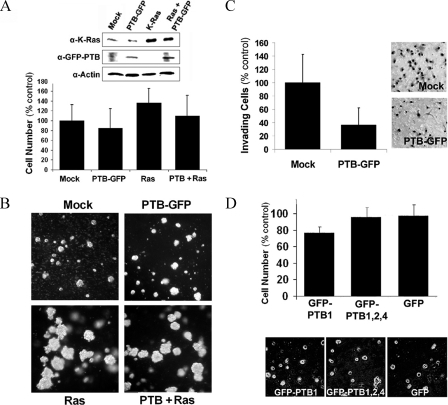
Overexpression of PTB Is Not Sufficient to Induce Transformation—To further characterize the role of PTB in malignancy and to determine whether PTB is oncogenic, we examined if increasing PTB levels can induce transformation in NIH-3T3 cells, an immortalized cell line classically used for in vitro transformation assays. Cells were transfected with GFP-PTB1 (the small isoform that is overexpressed in cancer cells), oncogenic K-Ras, or both. Fusion to GFP did not affect PTB1 function as demonstrated in Fig. 2, in which overexpression of GFP-PTB effectively shifts the alternative splicing pattern of caspase 2. After transfections, nonclonal stable expressing populations were created using the appropriate selection. The results show that overexpression of GFP-PTB1 did not significantly affect cellular proliferation rate (Fig. 8A), but the Rastransformed cells did proliferate slightly more rapidly than the mock-transfected cells (Fig. 8A). In addition, co-expression of PTB1-GFP with Ras slightly decreases the proliferation rate compared with the Ras-transformed cells (Fig. 8A), indicating that a high level of PTB1 protein alone is not sufficient to stimulate cellular growth and may even antagonize Ras-stimulated growth. Furthermore, overexpression of GFP-PTB1 does not induce anchorage-independent growth in a semi-solid media assay. In contrast, cells expressing oncogenic K-Ras formed large colonies. Expression of PTB in the co-transfected 3T3 cells did not prevent colony formation (Fig. 8B); however, it did decrease the overall number of colonies when cell number was determined (data not shown), which further suggests that PTB may be antagonistic to the transformation by the oncogenic Ras. Additionally, the effect of PTB overexpression on the invasive behavior of 3T3 cells was determined with the Matrigel invasion assay. The results show a significant (p < 0.002) decrease in invasive capacity in GFP-PTB1-expressing cells as compared with mock-transfected cells (Fig. 8C). These findings demonstrate that PTB does not cause malignant transformation in immortalized cells and even antagonizes the transformed phenotype by these in vitro criteria. To further test if PTB overexpression could influence the growth of normal human primary cells, WI-38 cells were transfected with GFP-PTB, and their growth in monolayer and methylcellulose media was examined. There is no significant change in cell proliferation in monolayer between the control and PTB-transfected cells (Fig. 8D). In addition, PTB overexpression did not induce growth in semi-solid media (Fig. 8D). Therefore, PTB overexpression alone does not stimulate cellular growth or induce a transformed phenotype in vitro.
FIGURE 8.
Overexpression of PTB is not sufficient to cause transformation in normal cells. A, Western blot analysis of Ras and GFP-PTB expression in stable cell lines. Monolayer growth of stable cell lines shows that PTB overexpression does not affect proliferation rate (n = 3, error bars =+S.D.). B, anchorage-dependent growth in methylcellulose media 14 days after plating. Co-expression of PTB1-GFP with Ras did not prevent colony formation but did decrease the total number of colonies formed. C, overexpression of PTB1-GFP significantly (p < 0.002) decreased the invasive behavior NIH-3T3 cells in the Matrigel invasion assay (n = 6, error bars = +S.D.). D, overexpression of PTB in WI-38 cells (primary normal human fibroblasts) does not significantly (p > 0.2) change their growth in monolayer culture (top panel; n = 3, error bars =+S.D.) nor does it stimulate anchorage-independent growth.
DISCUSSION
RNA processing is one of the key regulatory mechanisms that control gene expression in normal and cancer cells. PTB plays critical roles in several aspects of RNA processing, particularly alternative splicing, which can significantly alter gene expression patterns in given cells. As the human genome contains a surprisingly small number of genes, isoforms derived from the same genes through alternative splicing offers diversity in gene function and provides a mechanism to specify cellular activities based on their roles in multicellular organisms. Including or excluding specific exons generates functionally different proteins that cater to specific requirements of cells depending on their function (8, 10, 51, 52). There is growing evidence demonstrating the correlation between specific alternatively spliced variants and the malignant phenotype as well as drug resistance to chemotherapy in cancer patients (31). Comprehensive mapping of cancer-specific alternative spliced genes is underway to resolve the complex patterns of spliced variants in cancer cells (53). PTB is one of over 100 known splicing factors and regulators that influence alternative splicing decisions. Although some evidence indicates that the fine balance of different factors is important for splice sites choices, how these factors generate the final protein variant pattern in each cell population remains unclear. In this study, we provide evidence that echoes the complexity of RNA processing and alternative splicing decisions in cancer cells by demonstrating the different phenotypic changes resulting from the alternation of PTB expression level in different cell lines.
PTB has been shown to be a repressive splicing regulator that generally leads to the exclusion of targeted exons (8, 10, 12). As a splicing repressor, the level of PTB expression can potentially influence the splicing of a large number of substrates and impact malignancy in transformed cells through a variety of cellular pathways. In fact, PTB levels have been shown to increase in transformed cell lines and ovarian cancer (27, 28, 34). Our findings that PTB is significantly increased in endometrial cancer tissues and in transformed and cancer cell lines of various origins are consistent with these previous observations (26–29), further demonstrating that PTB expression increases are generally associated with malignant transformation and are not specific to certain tumor types.
Although the elevated levels of PTB in tumor cells have been documented, its role in malignancy was not examined until recently. The increased expression of PTB is thought to potentiate the malignant behaviors in cancer cells. Two examples include the PTB-based alternative splicing of the fibroblast growth factor receptor 1 and the multidrug resistance protein 1/ATP-binding cassette transporter (MDR1). These alternatively spliced proteins promote malignant growth of tumor cells (27) and confer drug resistance (28), respectively. A recent report showed that knockdown of PTB suppresses ovarian tumor cell growth and invasiveness in vitro (34), which supports the promoting role of PTB in the transformed phenotype and led to the consideration of PTB as a drug target for cancer chemotherapy. However, our observations paint a more complex picture when looking into more cell lines, in which PTB is not always a promoter for the transformed phenotype in vitro.
Our studies show that although down-regulation of PTB significantly reduces the invasive capacity of a prostate cancer cell line, PC-3M, and a colon cancer cell line, T84, it consistently increases the invasion capacity of HeLa cells. Interestingly, overexpression of PTB1 in NIH-3T3 cells reduces their invasive capacity. These differential changes in invasive capacity upon the alternation in PTB expression in different cell lines demonstrate that PTB can promote or antagonize the malignant behavior of cells dependent upon the specific intracellular environment. Molecularly, the differential alternative splicing pattern of caspase 2 in various cell lines induced by PTB siRNA knockdown mechanistically supports that the intracellular environment directly affects the role of PTB in functional cellular processes. Although PTB knockdown leads to inhibition of cell growth, it is not selective for the transformed cells. Rather, the inhibition of growth is more profound in normal human fibroblasts than in tumor cells. These observations together support the hypothesis that PTB acts in concert with complex cellular mechanisms to regulate gene expression. Alternative splicing of specific pre-mRNAs has been shown previously to be dependent upon the concentration of splicing promoters and splicing repressors. For example, excess ASF/SF2 over-heterogeneous nuclear ribonucleoprotein A1 prevents improper exon skipping of β-tropomyosin pre-mRNA (50). Thus, we speculate that our data can be explained by the idea that there is differential expression of endogenous PTB (including isoform ratios), differential expression of PTB substrate mRNAs, and differential expression of factors that promote or antagonize PTB function among different cell lines, which leads to different phenotypic changes upon alterations in PTB expression.
Although there is a general increase of PTB expression in malignant cells, the levels are highly variable among different types of cancer cells and are not directly correlated with the degree of malignancy in the cells tested. For example, PTB expression in a grade 3 endometrial carcinoma is equivalent to that of a grade 1 tumor. Moreover, the PC-3M cell line and its derivatives, which are from the same origin but selected for different metastatic capacity in vivo (45), all show a very similar expression level of PTB. In addition, overexpression of PTB1, which is elevated in many cancer cell lines, does not enhance cellular proliferation, anchorage-independent growth, or invasion capacity in NIH-3T3 cells, indicating that PTB alone is not sufficient to induce transformation in vitro. Together with the findings summarized in the previous paragraph, we conclude that PTB itself is not sufficient to induce a transformed phenotype, and it may support or antagonize malignant phenotype dependent upon a specific cellular environment. Thus, we believe it is premature to consider PTB as a ubiquitous or general potentiator of malignancy, particularly as an anti-cancer drug target.
Although PTB is generally increased in malignant cells, there is a great heterogeneity of PTB isoform expression among tumor cell lines and tissues of different origins. PTB1 increases greatly in most transformed cells and tissues examined, although it is hardly detectable in normal tissues, causing a switch in the ratio of PTB1:PTB2 in some of the cancer cells; however, other cancer cells maintain a greater expression of the larger isoforms. A previous study showed that the three PTB isoforms have differential activities over β-tropomyosin exon 3 skipping but are equally active in α-actinin exon skipping, suggesting a substrate specific splice site selection by the three isoforms (32). We evaluated the activity of the three isoforms on another substrate, caspase 2 minigene (47). Overexpression of each of the three PTB isoforms individually represses exon 9 inclusion. All three PTB isoforms show similar effects on caspase 2 alternative splicing. Thus far, only a few of the splicing substrates of PTB have been examined for the regulatory effects by different PTB isoforms, and a comprehensive splicing substrate list that details the effect of all three PTB isoforms on alternative splicing of all PTB target genes remains to be established. The alternative splicing patterns that are influenced by the three PTB isoforms could potentially provide profiles that differentiate one cell population from another.
In summary, we have shown that altering PTB expression in various cancer and nontransformed cell lines has differential effects, either promoting or suppressing a malignant trait in vitro. In addition, reduction of PTB has a differential impact on the alternative splicing selection of the same substrate in various cell lines, which provides a mechanistic explanation of the differences observed in functional assays. PTB alone is not sufficient to induce transformation in NIH-3T3 or normal human cells, and there is no clear correlation between PTB levels or isoform ratios with the degree of malignancy. These findings demonstrate the complex role of PTB in cancer cells and lead to our working model; the differential effects of PTB upon cellular behavior observed here are due to the tremendous number of possible gene expression outcomes affected by overall PTB expression level, relative isoform ratios, expression level of PTB substrates, and concentrations of factors that influence PTB function in different cell populations. Therefore, PTB likely contributes to the malignant phenotype as an integrated component of a complex mechanism.
Acknowledgments
We thank Dr. Christopher Smith, University of Cambridge, and Dr. Douglas Black, UCLA, for generously supplying reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants GM070967 (to J. Y. W.), CA 097761, and GM078555-01 (to S. H.). This work was also supported by a Young Investigators Award and the Northwestern Memorial Foundation and Friends of Prentice (to J. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PTB, polypyrimidine tract-binding protein; RRM, RNA recognition motif; siRNA, small interfering RNA; pol, polymerase; Br, bromo; RT, reverse transcription; PBS, phosphate-buffered saline; GFP, green fluorescent protein; oligo, oligonucleotide.
References
- 1.Ghetti, A., Pinol-Roma, S., Michael, W. M., Morandi, C., and Dreyfuss, G. (1992) Nucleic Acids Res. 20 3671-3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gil, A., Sharp, P. A., Jamison, S. F., and Garcia-Blanco, M. A. (1991) Genes Dev. 5 1224-1236 [DOI] [PubMed] [Google Scholar]
- 3.Patton, J. G., Mayer, S. A., Tempst, P., and Nadal-Ginard, B. (1991) Genes Dev. 5 1237-1251 [DOI] [PubMed] [Google Scholar]
- 4.Oh, Y. L., Hahm, B., Kim, Y. K., Lee, H. K., Lee, J. W., Song, O., Tsukiyama-Kohara, K., Kohara, M., Nomoto, A., and Jang, S. K. (1998) Biochem. J. 331 169-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez, I., Lin, C. H., McAfee, J. G., and Patton, J. G. (1997) RNA (N. Y.) 3 764-778 [PMC free article] [PubMed] [Google Scholar]
- 6.Valcarcel, J., and Gebauer, F. (1997) Curr. Biol. 7 R705-R708 [DOI] [PubMed] [Google Scholar]
- 7.Perez, I., McAfee, J. G., and Patton, J. G. (1997) Biochemistry 36 11881-11890 [DOI] [PubMed] [Google Scholar]
- 8.Wagner, E. J., and Garcia-Blanco, M. A. (2001) Mol. Cell. Biol. 21 3281-3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner, E. J., and Garcia-Blanco, M. A. (2002) Mol. Cell 10 943-949 [DOI] [PubMed] [Google Scholar]
- 10.Black, D. L. (2003) Annu. Rev. Biochem. 72 291-336 [DOI] [PubMed] [Google Scholar]
- 11.Robinson, F., and Smith, C. W. J. (2006) J. Biol. Chem. 281 800-806 [DOI] [PubMed] [Google Scholar]
- 12.Spellman, R., and Smith, C. W. J. (2006) Trends Biochem. Sci. 31 73-76 [DOI] [PubMed] [Google Scholar]
- 13.Lou, H., Gagel, R. F., and Berget, S. M. (1996) Genes Dev. 10 208-219 [DOI] [PubMed] [Google Scholar]
- 14.Lou, H., Helfman, D. M., Gagel, R. F., and Berget, S. M. (1999) Mol. Cell. Biol. 19 78-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castelo-Branco, P., Furger, A., Wollerton, M., Smith, C., Moreira, A., and Proudfoot, N. (2004) Mol. Cell. Biol. 24 4174-4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellen, C. U., Pestova, T. V., Litterst, M., and Wimmer, E. (1994) J. Virol. 68 941-950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski, A., Hunt, S. L., Patton, J. G., and Jackson, R. J. (1995) RNA (N. Y.) 1 924-938 [PMC free article] [PubMed] [Google Scholar]
- 18.Witherell, G. W., Schultz-Witherell, C. S., and Wimmer, E. (1995) Virology 214 660-663 [DOI] [PubMed] [Google Scholar]
- 19.Schneider, R., Agol, V. I., Andino, R., Bayard, F., Cavener, D. R., Chappell, S. A., Chen, J. J., Darlix, J. L., Dasgupta, A., Donze, O., Duncan, R., Elroy-Stein, O., Farabaugh, P. J., Filipowicz, W., Gale, M., Jr., et al. (2001) Mol. Cell. Biol. 21 8238-8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickering, B. M., Mitchell, S. A., Evans, J. R., and Willis, A. E. (2003) Nucleic Acids Res. 31 639-646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamath, R. V., Leary, D. J., and Huang, S. (2001) Mol. Biol. Cell 12 3808-3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie, J., Lee, J.-A., Kress, T. L., Mowry, K. L., and Black, D. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8776-8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gromak, N., Matlin, A. J., Cooper, T. A., and Smith, C. W. (2003) RNA (N. Y.) 9 443-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rideau, A. P., Gooding, C., Simpson, P. J., Monie, T. P., Lorenz, M., Huttelmaier, S., Singer, R. H., Matthews, S., Curry, S., and Smith, C. W. J. (2006) Nat. Struct. Mol. Biol. 13 839-848 [DOI] [PubMed] [Google Scholar]
- 25.Sauliere, J., Sureau, A., Expert-Bezancon, A., and Marie, J. (2006) Mol. Cell. Biol. 26 8755-8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, W., McCutcheon, I. E., Fuller, G. N., Huang, E. S., and Cote, G. J. (2000) Cancer Res. 60 1221-1224 [PubMed] [Google Scholar]
- 27.Jin, W., Bruno, I. G., Xie, T.-X., Sanger, L. J., and Cote, G. J. (2003) Cancer Res. 63 6154-6157 [PubMed] [Google Scholar]
- 28.He, X., Ee, P. L., Coon, J. S., and Beck, W. T. (2004) Clin. Cancer Res. 10 4652-4660 [DOI] [PubMed] [Google Scholar]
- 29.McCutcheon, I. E., Hentschel, S. J., Fuller, G. N., Jin, W., and Cote, G. J. (2004) Neuro-Oncol. 6 9-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunthert, U., Hofmann, M., Rudy, W., Reber, S., Zoller, M., Haussmann, I., Matzku, S., Wenzel, A., Ponta, H., and Herrlich, P. (1991) Cell 65 13-24 [DOI] [PubMed] [Google Scholar]
- 31.Venables, J. P. (2006) BioEssays 28 378-386 [DOI] [PubMed] [Google Scholar]
- 32.Wollerton, M. C., Gooding, C., Robinson, F., Brown, E. C., Jackson, R. J., and Smith, C. W. (2001) RNA (N. Y.) 7 819-832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faustino, N. A., and Cooper, T. A. (2003) Genes Dev. 17 419-437 [DOI] [PubMed] [Google Scholar]
- 34.He, X., Pool, M., Darcy, K. M., Lim, S. B., Auersperg, N., Coon, J. S., and Beck, W. T. (2007) Oncogene 26 4961-4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, S., Deerinck, T. J., Ellisman, M. H., and Spector, D. L. (1997) J. Cell Biol. 137 965-974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, pp. 7.39-7.52, Cold Spring Harbor, NY
- 37.Cote, J., Dupuis, S., Jiang, Z., and Wu, J. Y. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 938-943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, W.-J., and Wu, J. Y. (1996) Mol. Cell. Biol. 16 5400-5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, C., Politz, J. C., Pederson, T., and Huang, S. (2003) Mol. Biol. Cell 14 2425-2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh, S., Munshi, H. G., Sen, R., Linz-McGillem, L. A., Goldman, R. D., Lorch, J., Green, K. J., Jones, J. C., and Stack, M. S. (2002) Cancer 95 2524-2533 [DOI] [PubMed] [Google Scholar]
- 41.Stack, S., Gonzalez-Gronow, M., and Pizzo, S. V. (1990) Biochemistry 29 4966-4970 [DOI] [PubMed] [Google Scholar]
- 42.Ghosh, S., Brown, R., Jones, J. C., Ellerbroek, S. M., and Stack, M. S. (2000) J. Biol. Chem. 275 23869-23876 [DOI] [PubMed] [Google Scholar]
- 43.Kopp, K., and Huang, S. (2005) J. Cell Biochem. 95 217-225 [DOI] [PubMed] [Google Scholar]
- 44.Kozlowski, J. M., Fidler, I. J., Campbell, D., Xu, Z. L., Kaighn, M. E., and Hart, I. R. (1984) Cancer Res. 44 3522-3529 [PubMed] [Google Scholar]
- 45.Pettaway, C. A., Pathak, S., Greene, G., Ramirez, E., Wilson, M. R., Killion, J. J., and Fidler, I. J. (1996) Clin. Cancer Res. 2 1627-1636 [PubMed] [Google Scholar]
- 46.Wagner, E. J., Carstens, R. P., and Garcia-Blanco, M. A. (1999) Electrophoresis 20 1082-1086 [DOI] [PubMed] [Google Scholar]
- 47.Cote, J., Dupuis, S., and Wu, J. Y. (2001) J. Biol. Chem. 276 8535-8543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, L., Miura, M., Bergeron, L., Zhu, H., and Yuan, J. (1994) Cell 78 739-750 [DOI] [PubMed] [Google Scholar]
- 49.Dano, K., Behrendt, N., Hoyer-Hansen, G., Johnsen, M., Lund, L. R., Ploug, M., and Romer, J. (2005) Thromb. Haemostasis 93 676-681 [DOI] [PubMed] [Google Scholar]
- 50.Mayeda, A., Helfman, D. M., and Krainer, A. R. (1993) Mol. Cell. Biol. 13 2993-3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matlin, A. J., Clark, F., and Smith, C. W. (2005) Nat. Rev. Mol. Cell Biol. 6 386-398 [DOI] [PubMed] [Google Scholar]
- 52.Stamm, S., Ben-Ari, S., Rafalska, I., Tang, Y., Zhang, Z., Toiber, D., Thanaraj, T. A., and Soreq, H. (2005) Gene (Amst.) 344 1-20 [DOI] [PubMed] [Google Scholar]
- 53.Watahiki, A., Waki, K., Hayatsu, N., Shiraki, T., Kondo, S., Nakamura, M., Sasaki, D., Arakawa, T., Kawai, J., Harbers, M., Hayashizaki, Y., and Carninci, P. (2004) Nat. Methods 1 233-239 [DOI] [PubMed] [Google Scholar]