Abstract
Bacterial pathogens are recognized by the innate immune system through pattern recognition receptors, such as Toll-like receptors (TLRs). Engagement of TLRs triggers signaling cascades that launch innate immune responses. Activation of MAPKs and NF-κB, elements of the major signaling pathways induced by TLRs, depends in most cases on the adaptor molecule MyD88. In addition, Gram-negative or intracellular bacteria elicit MyD88-independent signaling that results in production of type I interferon (IFN). Here we show that in mouse macrophages, the activation of MyD88-dependent signaling by the extracellular Gram-positive human pathogen group A streptococcus (GAS; Streptococcus pyogenes) does not require TLR2, a receptor implicated in sensing of Gram-positive bacteria, or TLR4 and TLR9. Redundant engagement of either of these TLR molecules was excluded by using TLR2/4/9 triple-deficient macrophages. We further demonstrate that infection of macrophages by GAS causes IRF3 (interferon-regulatory factor 3)-dependent, MyD88-independent production of IFN. Surprisingly, IFN is induced also by GAS lacking slo and sagA, the genes encoding cytolysins that were shown to be required for IFN production in response to other Gram-positive bacteria. Our data indicate that (i) GAS is recognized by a MyD88-dependent receptor other than any of those typically used by bacteria, and (ii) GAS as well as GAS mutants lacking cytolysin genes induce type I IFN production by similar mechanisms as bacteria requiring cytoplasmic escape and the function of cytolysins.
Group A streptococcus (GAS4; Streptococcus pyogenes) is an important human Gram-positive pathogen responsible for a wide spectrum of infections, ranging from mild diseases (e.g. tonsillitis) to serious illness (e.g. necrotizing fasciitis, sepsis, or severe poststreptococcal sequelae) (1). The persistence of GAS in the human population and the severity of some GAS diseases are the result of activities of a number of virulence factors that enable the pathogen to escape immune surveillance or, on contrary, induce an overreaction of the immune system (2, 3). Although GAS is generally regarded as an extracellular pathogen, recent findings suggest that GAS can survive (although not multiply) within various host cells, such as neutrophils, macrophages, epithelial cells, and fibroblasts (4–7). The surviving bacteria may serve as a reservoir for recurrent GAS diseases.
Immune responses to bacteria are initiated by recognition of bacterial components called pathogen-associated molecular patterns through host cell-encoded pattern recognition receptors (PRRs) (8, 9). Typically, pathogen-associated molecular patterns are components of the bacterial cell wall (e.g. lipopolysaccharide and lipoteichoic acid), but they may also be derived from the inside of bacteria (e.g. DNA). The primary function of PRRs is to trigger signaling cascades that activate antimicrobial defense programs. The best studied class of PRRs is the Toll-like receptor (TLR) family, which consists of 13 transmembrane glycoproteins in mammals (8). Virtually all pathogenic bacteria are recognized by one or more TLRs, with TLR2 being the receptor for lipoteichoic acid of Gram-positive bacteria, TLR4 the receptor for lipopolysaccharide of Gram-negative bacteria, and TLR9 the receptor for bacterial DNA containing unmethylated CpG sequences. Ligand binding to TLR3, TLR4, TLR7, and TLR9 but not to TLR2 launches two distinct signaling pathways that result in the production of proinflammatory cytokines (e.g. TNFα and IL-1) and type I IFNs, respectively. Five TIR domain-containing adaptor molecules are involved in signaling downstream of TLRs: MyD88, TIRAP (or MAL), TRIF (or TICAM1), TRAM (or TICAM2), and SARM, a negative regulator of TRIF (10). MyD88, an essential signaling component of all TLRs except TLR3, is required for activation of MAPKs and NF-κB that cause production of proinflammatory cytokines. Signaling components that are required for the production of type I IFNs are less uniform, and they are partially TLR- and cell type-specific. Recently, much attention has been paid to the elucidation of pathways leading to TLR-independent production of type I IFN in response to intracellular bacteria and/or intracellular DNA (11–14). Although the identity of the critical PRR remains unknown, it is generally believed that it is a cytoplasmic molecule that recognizes bacterial products (possibly bacterial DNA) in the cytoplasm. Signaling downstream of this unknown PRR employs the serine threonine kinase TBK1 or, less frequently, its relative IKK (15–18), which phosphorylates primarily the transcription factor IRF3, causing the activation of the IFN-β gene, the first type I IFN to be expressed (19, 20). Other cytoplasmic PRRs, such as the NOD proteins, recognize bacterial products in the cytoplasm and activate MAPKs and NF-κB but not IFN production (21).
Despite a considerable knowledge of GAS virulence factors, the PRRs that are responsible for recognition of this pathogen by the host are unknown. Remarkably little information is available also about the signaling pathways triggered by GAS in infected innate immune cells that are known to be required for defense against GAS (22). Here we show that GAS activates p38 MAPK, NF-κB, TNFα, and IL-6 production in infected bone marrow-derived mouse macrophages (BMDMs). Similar to other bacteria, these responses depend on MyD88. However, in contrast to most other pathogenic bacteria, the MyD88-mediated signaling was independent of TLR2, the receptor for Gram-positive bacteria, and the other bacterial receptors TLR4 and TLR9. We also rule out the involvement of IL-1 receptor signaling. We further demonstrate that GAS also elicits MyD88-independent signaling that results in type I IFN production. The IFN production did not require the presence of the GAS-encoded cytolysins SLO and SLS. This finding is surprising, since during infections with other Gram-positive bacteria, either the cytolysin itself or cytolysin-mediated cytoplasmic escape of bacteria from phagocytic vesicles was implicated in triggering IFN production (12, 23–25). However, the requirement for IRF3 in the IFN production suggested that other aspects of the GAS-induced IFN production resembled the well established TBK1/IRF3 signaling pathway. This is the first description of activation of MyD88-dependent and -independent pathways upon GAS infection. Our data implicate that GAS is recognized by a yet unknown receptor upstream of MyD88 and establish GAS as a Gram-positive pathogen capable of inducing type I IFN synthesis without molecules usually required for entry of bacterial products into the cytoplasm of infected cells.
EXPERIMENTAL PROCEDURES
Bacterial Strains—Escherichia coli DH5-α and TOP10 were used as hosts for cloning. S. pyogenes serotype M1 (ATCC 700294) is a clinical strain originally isolated from an infected wound. The isogenic sagA- and slo-deficient mutants were constructed using a thermosensitive strategy described previously (26). First, using wild-type genomic DNA as template and primers containing flanking restriction sites, a 1111-bp sagA upstream fragment (primers OLEC120/OLEC143), a 1200-bp sagA downstream fragment (primers OLEC123/OLEC144), a 1159-bp slo upstream fragment (primers OLEC248/OLEC249), and a 1033-bp slo downstream fragment (primers OLEC250/OLEC251) were amplified. After digestion with the respective restriction enzymes, the upstream and downstream fragments were cloned into thermo-sensitive plasmids pRDN18 (sagA-deficient mutant) and pEC84 (slo-deficient mutant) (26). After introduction of the recombinant plasmids into S. pyogenes ATCC 700294, a series of temperature shifts with appropriate antibiotic selection was performed, thus leading to the final deficient mutants (strain EC548 (sagA-deficient mutant) and EC997 (slo-deficient mutant)) in which the entire coding sequence of the gene was deleted in a nonpolar fashion. To create the sagA/slo-deficient strain (EC1142), the deletion of slo was performed as mentioned above in the background of the sagA-deficient strain. The correct deletion event in the mutants was checked by PCR, Southern blot, and sequencing analysis. SLS and SLO hemolysis assays further confirmed that the sagA-, slo-, and sagA/slo-deficient mutants were defective in SLS and/or SLO activity as described (27, 28). DNA manipulations are described in the supplemental materials.
Bacterial Culture—S. pyogenes strains were grown at 37 °C with 5% CO2 without agitation in Todd-Hewitt broth (BD Biosciences) supplemented with 0.2% yeast extract and on tryptic soy agar supplemented with 3% defibrinogated sheep blood. Transformation of E. coli and S. pyogenes was performed as previously described (29, 30). Whenever required, antibiotics were added to the medium at the following final concentrations: erythromycin, 300 μg/ml for E. coli and 3 μg/ml for S. pyogenes; spectinomycin, 100 μg/ml for both E. coli and S. pyogenes. Bacterial cell growth was turbidimetrically monitored at 620 nm with a microplate reader.
Macrophage Cell Culture—Primary BMDMs were obtained from the femur bone marrow of 6–10-week-old mice. Cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum in the presence of L cell-derived CSF-1, as described (31). MyD88–/–, TLR2–/–, TLR4–/–, TLR9–/–, TLR2/4/9–/–, IFNAR1–/–, IRF3–/–, IL1-RI–/–, and control WT mice, all on a C57Bl/6 background, were housed under specific pathogen-free conditions (32–36).
GAS Infections—For infection assays, BMDMs were seeded at 5 × 106 cells/dish in 10-cm dishes containing medium without antibiotics. The next day, S. pyogenes cultures grown in THY were harvested at midlogarithmic phase, washed with phosphate-buffered saline, and added to the BMDM monolayers at a multiplicity of infection (MOI) of 100. After 30 min of incubation at 37 °C, nonadherent extracellular bacteria were eliminated by removing the culture medium, and adherent extracellular bacteria were subsequently killed by incubation with fresh medium (without fetal calf serum) containing 60 μg/ml penicillin. At specific time points after infection, supernatants were collected for ELISA, and whole cell extracts were prepared for Western blot analysis. At least three mice of each genotype were used in all infection experiments.
Antibodies—Antibodies to Tyr701-phosphorylated Stat1 (pY701-S1) and phosphorylated p38 (pp38) were purchased from Cell Signaling (Frankfurt/Main, Germany). Antibodies to IκB-α and p38 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibody to Stat1-α C terminus was previously described (37).
ELISA and Western Blot Analysis—For ELISAs, supernatants of infected macrophages were collected and diluted 1:5 in reagent diluent. TNFα and IL-6 were assayed using DuoSET ELISA kits (R&D Systems, Minneapolis, MN). Western blot analysis was performed using fluorophore-linked secondary antibodies (Molecular Probes-Invitrogen (Lofer, Austria) and Rockland (Gilbertsville, PA)) and an Odyssey infrared imaging system (LI-COR Bioscience, Lincoln, NE).
qRT-PCR—Total RNA was isolated using Trizol® LS reagent (Invitrogen). Reverse transcription of total RNA was performed using oligo(dT)18 as primer and Moloney murine leukemia virus reverse transcriptase (Fermentas, St. Leon-Rot, Germany). The cDNA of the IFN-β and Mx2 genes was subsequently analyzed by qRT-PCR as described in the supplemental materials.
RESULTS
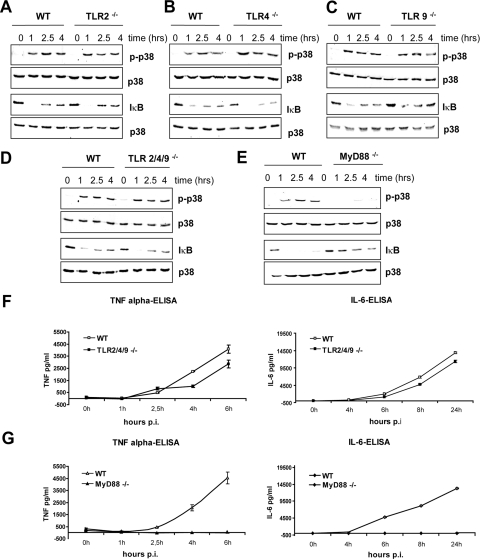
GAS Activates Inflammatory Signaling Independently of TLR2, TLR4, and TLR9—To investigate the role of bacteria-recognizing TLRs in responses to GAS infection, we examined the activation of the transcription factor NF-κB by assaying the degradation of its inhibitor (IκB) and the phosphorylation of p38 MAPK in infected BMDMs. Activation of both NF-κB and p38 MAPK was shown in numerous studies to be essential for production of proinflammatory cytokines (38–42). Furthermore, macrophages express all components of the TLR signaling cascade and are known to be required for defenses against GAS infection (22). We first compared the activation of p38 MAPK and NF-κB in WT and TLR2–/– BMDMs, since many Gram-positive pathogens are recognized by this receptor. GAS induced in both WT and TLR2–/– BMDMs a rapid and sustained activation of p38 MAPK (Fig. 1A). The degradation of IκB was maximal after 1 h of infection. IκB gradually reappeared at later time points but did not reach the original level during the time frame of observation (up to 4 h). The internalization of GAS (monitored by fluorescence microscopy) was equally efficient in cells of both genotypes (data not shown). These data demonstrate that TLR2 was not required for GAS-induced signaling. Other TLRs frequently engaged by bacteria are TLR4 and TLR9. TLR4 is the receptor for lipopolysaccharide of Gram-negative bacteria, but it has been shown to recognize also cell wall components of mycobacteria and several cytolysins of Gram-positive bacteria (25, 43, 44). TLR9, a CpG DNA receptor localized in endosomal and lysosomal membranes of macrophages and, most prominently, dendritic cells, can bind DNA from both Gram-positive and Gram-negative species (35, 45). Similar to TLR2–/– BMDMs, the activation of p38 MAPK and degradation of IκB in TLR4–/– and TLR9–/– BMDMs was indistinguishable from WT BMDMs (Fig. 1, B and C). To examine whether a combination of TLR2, TLR4, and TLR9 was used for GAS recognition, we analyzed GAS-induced signaling in TLR2/4/9 triple-deficient BMDMs. As depicted in Fig. 1D, the signaling events in triple-deficient BMDMs were comparable with WT cells. These data exclude a redundant function of TLR2, TLR4, and TLR9 in GAS recognition and demonstrate that macrophages sense GAS using a PRR other than the most common bacterial receptors.
FIGURE 1.
GAS-induced inflammatory signaling and the production of TNFα and IL-6 depends on MyD88 but not on TLR2, TLR4, and TLR9. A–E, BMDMs from TLR2–/– (A), TLR4–/– (B), TLR9–/– (C), TLR2/4/9–/– (D), MyD88–/– (E), and WT control mice were infected with GAS (MOI 100). At the indicated time points, whole cell extracts were prepared and analyzed by Western blotting using antibodies to phosphorylated p38 MAPK (pp38; top) and to IκB (bottom). Equal loading was controlled by reprobing the membrane with antibody to total p38 MAPK. F and G, BMDMs derived from TLR2/4/9 triple-deficient (TLR2/4/9–/–) (F), MyD88–/– (G), and control mice were infected with GAS (MOI 100). At the indicated time points, supernatants were collected, and TNFα (left) and IL-6 release (right) was measured by ELISA. The data represent one of at least three infection experiments carried out independently with different mice of each genotype.
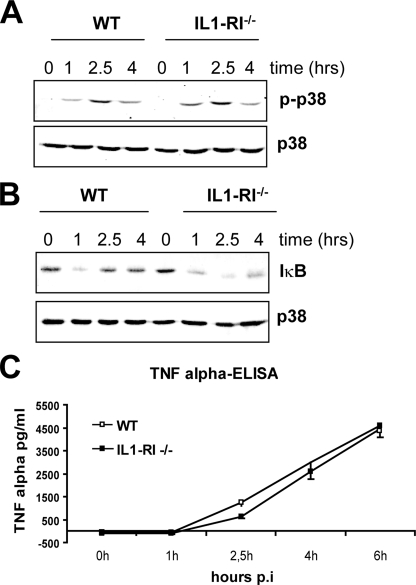
GAS-induced Inflammatory Signaling Depends on MyD88—All TLRs except for the double-stranded RNA receptor TLR3 require MyD88 for activation of MAPKs, NF-κB, and the subsequent proinflammatory cytokine production. To investigate whether MyD88 was involved in GAS-induced inflammatory signaling, we examined the activation of p38 MAPK and degradation of IκB in infected MyD88–/– BMDMs. The experiment revealed that GAS-induced activation of p38 MAPK was almost completely abolished, and the degradation of IκB was strongly diminished in MyD88–/– cells when compared with WT cells (Fig. 1E). A weak p38 MAPK activation and IκB degradation were observed in MyD88–/– cells at later time points of infection. Thus, the GAS-induced inflammatory signaling was markedly reduced and delayed in MyD88–/– cells. The activation of MAPKs and NF-κB plays a fundamental role in induction of proinflammatory cytokines, such as TNFα or IL-6. To estimate the effect of reduced GAS-induced inflammatory signaling in MyD88–/– BMDMs, we determined the amounts of secreted TNFα and IL-6 in supernatants of GAS-infected MyD88–/– and control WT cells. In parallel, we also measured TNFα and IL-6 production by infected TLR2/4/9 triple-deficient cells. Infection of WT BMDMs with GAS resulted in a robust cytokine production that was only slightly reduced (by less than 20%) in TLR2/4/9 triple-deficient cells (Fig. 1F). In marked contrast, TNFα and IL-6 production was completely abolished in GAS-infected MyD88–/– BMDMs (Fig. 1E). These data show that GAS-induced proinflammatory cytokine production is absolutely dependent on MyD88. The slight but reproducible reduction of cytokine production by TLR2/4/9 triple-deficient BMDMs indicates that one of these TLRs or a combination of them plays a minor role in GAS-induced proinflammatory cytokine production. However, the largest part of GAS-induced inflammatory cytokine production is mediated by a receptor (or receptors) different from the most common bacteria-specific TLRs. Importantly, this receptor signals via MyD88. The only other known pathways that use the adaptor MyD88 for signaling are the IL-1/IL-18 pathways (32). Although IL-18 is most relevant for the activation and IFN-γ production by Th1 cells (46), IL-1 has been reported be released by innate immune cells upon infection with various pathogens that are sensed in the cytoplasm by components of the inflammasome (47–49). To investigate whether IL-1 release by GAS-infected macrophages is responsible for the MyD88-dependent inflammatory signaling, we infected cells deficient in IL1-RI, the most important IL-1 receptor chain (50). As shown in Fig. 2, the activation of p38 MAPK, the degradation of IκB, and TNFα production were not affected by the deficiency in IL-1 signaling. These data further strengthen our notion that GAS is sensed by a yet unidentified receptor upstream of MyD88.
FIGURE 2.
IL-1 signaling is not required for responses of BMDM to GAS infection. A and B, BMDMs from IL-1RI–/– and control (WT) mice were infected with GAS (MOI 100) for the indicated periods. p38 MAPK activation (pp38) (A) and degradation of IκB (B) were determined by Western blot analysis. Equal protein loading was confirmed by reprobing the membrane with antibody to total p38 MAPK. C, BMDMs derived from IL-1RI–/– and control mice were infected with GAS (MOI 100). At the indicated time points, supernatants were collected, and TNFα release was measured by ELISA. The data represent one of at least three infection experiments carried out independently with different mice of each genotype.
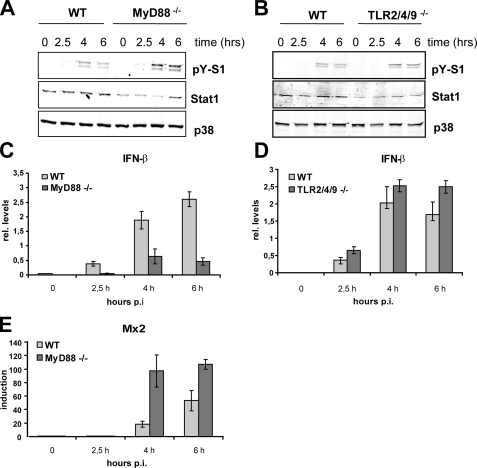
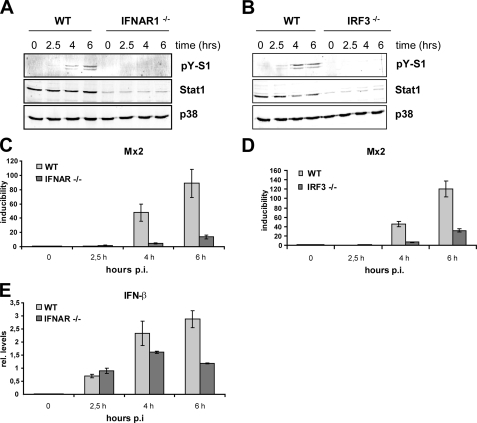
GAS Induces Type I Interferon—Gram-negative and intracellular bacteria are known to elicit MyD88-independent signaling that causes activation of the transcription factor IRF3 and subsequent transcription of the type I IFN-β. The mechanisms of IFN production induced by these two types of bacteria differ upstream of IRF3. Whereas Gram-negative bacteria use TLR4 to stimulate IFN production, intracellular bacteria are recognized by a still unknown cytoplasmic receptor. Engagement of TLR9 can also trigger IRF3/IRF7-mediated type I IFN expression. GAS has been previously reported to induce IFN production in primary human macrophages (51). We asked whether GAS can exert similar effects in mouse macrophages and what the role of MyD88 or the bacteria-specific TLRs therein is. First, tyrosine phosphorylation of the crucial IFN-activated transcription factor Stat1 was analyzed. Tyrosine phosphorylation of Stat1 is a good indicator for autocrine IFN-β production, since this cytokine is the first IFN to be synthetized upon challenge of macrophages with bacteria or their products (52–54). Infection of BMDMs with GAS revealed induction of Stat1 tyrosine phosphorylation after 4 h of infection of WT BMDMs (Fig. 3, A and B). This late tyrosine phosphorylation is similar to challenges with other bacteria (e.g. Listeria monocytogenes) that do not induce IFN synthesis through direct engagement of an IFN-inducing TLR (23). Consistently, Stat1 activation proceeded independently of MyD88 and TLR2, TLR4, and TLR9 signaling, as shown by GAS infection of BMDMs from MyD88–/– (Fig. 3A), TLR2–/–, TLR4–/–, TLR9–/– (all in Fig. S1), or TLR2/4/9 triple-deficient (Fig. 3B) mice. In fact, Stat1 tyrosine phosphorylation was slightly but reproducibly increased in MyD88–/– and TLR2/4/9 triple-deficient BMDMs (Fig. 3, A and B) when normalized to total Stat1 levels. Since the Stat1 levels were generally slightly lower in BMDMs derived from the gene-targeted mice compared with WT controls, the Western blots were reprobed with p38 antibody to allow a more accurate normalization. Reduced Stat1 levels have been already previously observed in cells that are deficient in IFN or TLR/MyD88 signaling (11, 55, 56). To directly assess IFN-β expression, we performed qRT-PCR to determine IFN-β mRNA levels in cells infected with GAS. The data shown represent relative IFN-β mRNA levels normalized to GAPDH mRNA. We did normalize to uninfected cells, since the IFN-β mRNA was very often below the levels that could be reliably detected by qRT-PCR. The experiment confirmed that the IFN-β expression was independent of TLR2, TLR4, and TLR9 (Fig. 3D). The IFN-β mRNA was also induced upon infection of MyD88–/– cells with GAS, albeit the amount was reduced if compared with WT cells (Fig. 3C). The reduced IFN-β mRNA levels in MyD88–/– did not result in diminished IFN signaling, since Stat1 activation was increased in these cells if compared with the controlled BMDMs (Fig. 3A). The functional activation of IFN/Jak/Stat signaling in MyD88–/– and WT cells was further confirmed by the induction of the type I IFN target gene Mx2 (57). The transcription of Mx2 was induced in both the control and MyD88–/– BMDMs with a kinetics that correlated well with the activation profile of Stat1 (Fig. 3E). Moreover, Mx2 transcription was more strongly induced in MyD88–/– cells that also display increased Stat1 activation. Thus, despite the decreased IFN-β mRNA, the IFN/Jak/Stat1 signaling was more efficiently turned on in MyD88–/– cells, suggesting that the increased Stat1 phosphorylation was caused by a more efficient phosphorylation or a lower dephosphorylation rate. To prove the autocrine/paracrine role of the type I IFN in Stat1 activation, we examined Stat1 activation and IFN responses using BMDMs lacking the type I IFN receptor chain 1 (IFNAR1–/–). As shown in Fig. 4A, Stat1 tyrosine phosphorylation was completely abolished in GAS-infected IFNAR1–/– cells despite robust induction of IFN-β gene transcription in these cells (Fig. 4A). Consistently, the induction of the type I IFN-responsive gene Mx2 was entirely dependent on signaling by the IFNAR1 receptor (Fig. 4C). In most cell types, activation of the transcription factor IRF3 launches a feed forward type I IFN amplification loop through induction of IFN-β that drives the expression of IFN-αs (19, 20, 58). Thus, a failure in activation of IRF3 results in a deficient production of both the immediate IFN-β and the late phase IFN-αs. To further elucidate the mechanism of GAS-induced IFN production, we infected IRF3–/– BMDMs with GAS and examined the activation of the transcription factor Stat1 and induction of the Stat1 target gene Mx2. Both the tyrosine phosphorylation of Stat1 and the transcription of the type I IFN-inducible gene Mx2 were diminished in IRF3–/– (Fig. 4, B and D), whereas the MyD88 pathway was not affected by the IRF3 deletion, as revealed by p38 MAPK activation. These data demonstrate that GAS has the ability to induce IFN-β synthesis and IFN signaling in a way that mechanistically resembles the IFN-β activation by intracellular Gram-positive pathogens.
FIGURE 3.
GAS induces IFN-β production and Stat1 activation independently of MyD88, TLR2, TLR4, and TLR9. A and B, BMDMs from control (WT), MyD88–/– (A), and TLR2/4/9 triple-deficient (TLR2/4/9–/–) (B) mice were infected with GAS (MOI 100), and whole cell extracts were prepared after the indicated time periods. Stat1 Tyr701 phosphorylation and expression were determined by Western blotting using antibody to phosphorylated Stat1 (pY-S1) and total Stat1. Differences in Stat1 expression levels were revealed by reprobing the membrane with antibody to total p38 MAPK (p38). Note the double band on the pY-S1 blot represents the phosphorylated forms of both Stat1 splicing isoforms Stat1-α and Stat1-β. Loading control (S1) was performed with antibody directed to the C terminus of Stat1, which is absent in the Stat1-β isoform. C and D, BMDMs from wild type, MyD88–/– (C), and TLR2/4/9 triple-deficient (TLR2/4/9–/–) mice (D) were exposed to GAS (MOI 100). After the indicated time points, total mRNA was extracted, reverse-transcribed, and analyzed by qRT-PCR for IFN-β and GAPDH (for normalization) expression. Note that the data show relative IFN-β mRNA levels normalized to GAPDH but not to uninfected samples. E, GAS-induced IFN-β stimulates transcription of type I IFN target gene Mx2. BMDMs from MyD88–/– and control (wild type) mice were exposed to GAS, and 2.5, 4, and 6 h postinfection (hours p.i.), total mRNA was extracted, reverse-transcribed, and analyzed by qRT-PCR for Mx2 after normalizing to GAPDH and uninfected samples. Bars, S.D. of three experiments.
FIGURE 4.
GAS-induced IFN signaling depends on IRF3 and IFNAR1. A and B, BMDMs from control (WT), IFNAR1–/– (A), and IRF3-deficient (IRF3–/–) (B) mice were infected with GAS (MOI 100), and whole cell extracts were prepared 2.5, 4, and 6 h postinfection. Stat1 Tyr701 phosphorylation and expression were determined by Western blotting using antibody to phosphorylated Stat1 (pY-S1) and total Stat1. Differences in Stat1 expression levels were revealed by reprobing the membrane with antibody to total p38 MAPK (p38). C and D, BMDMs from IFNAR1–/– (C), IRF3–/– (D), and control (WT) mice were exposed to GAS for the indicated times, and total mRNA was extracted, reverse-transcribed, and analyzed by qRT-PCR for Mx2 after normalization to GAPDH. E, BMDMs from IFNAR1–/– and control (WT) mice were infected with GAS (MOI 100). IFN-β expression was analyzed by qRT-PCR after normalization to GAPDH. Bars, S.D. of three experiments.
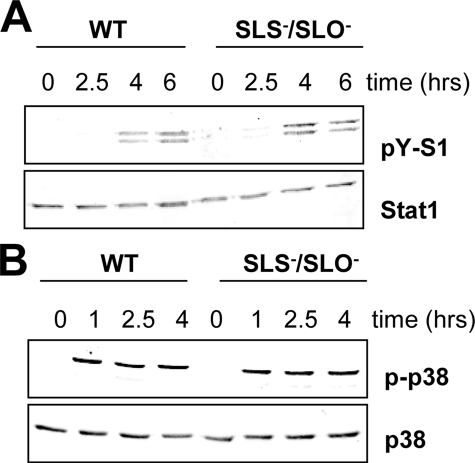
GAS Stimulates IFN Signaling Independently of Cytolysins—The TLR-independent IFN-β induction by GAS was surprising, since GAS is generally regarded as an extracellular pathogen that has only a limited capacity to survive in host cells (4–7). For the so far characterized induction of IFN-β production by Gram-positive bacteria, the expression of cytolysins was required (12, 23, 24). Cytolysins are thought to enable cytoplasmic escape of phagocytosed bacteria. In addition, cytolysins were also shown to directly stimulate IFN production (25). The GAS genome contains genes for two cytolysins. The slo gene encodes SLO (streptolysin O), which resembles other known cytolysins (e.g. listeriolysin, pneumolysin, and anthrolysin) in terms of sequence, sensitivity to oxygen, and cholesterol binding (59, 60). The other cytolysin, SLS (streptolysin S), is encoded by the sagA gene, which is unrelated to the cholesterol-binding cytolysins (60). To examine whether the GAS-derived cytolysins play a role in GAS-induced IFN signaling, sagA (SLS)-deficient, slo (SLO)-deficient, and double sagA/slo-deficient mutants were generated and used in infection assays. Analysis of Stat1 activation revealed that neither the deletion of the single cytolysin genes (data not shown) nor the double deletion (Fig. 5A) caused a reduction of GAS-induced IFN signaling. The inflammatory signaling was also not dependent on the expression of the GAS cytolysins, as shown by p38 MAPK activation (Fig. 5B). These findings suggest that the ability of GAS to induce IFN-β synthesis may not require a contact of GAS components with the still unknown cytoplasmic receptor that is used by other Gram-positive bacteria for IFN-β induction.
FIGURE 5.
GAS-derived cytolysins SLO and SLS are not required for induction of IFN and stress signaling. A and B, BMDMs from wild-type mice were infected with either S. pyogenes WT strain or the isogenic mutant strain sls/sagA lacking both sagA (SLS) and slo (SLO) genes. At the indicated time points, activation of Stat1 (A) and p38 MAPK (B) was detected by Western blot analysis using antibodies to Tyr701-phosphorylated Stat1 (pY-S1) and phosphorylated p38 MAPK (pp38). Equal protein loading was confirmed by reprobing the membranes with antibody to total Stat1 and p38 MAPK (p38), respectively. The data shown represent one of at least three independent infection experiments.
DISCUSSION
GAS is the etiological agent of a variety of human diseases. The heterogeneity of GAS diseases arises in part from the high diversity of GAS-mediated host-pathogen interactions in which virulence factors, GAS genome composition, prophage DNA, and plasticity of the GAS transcriptome play key roles (2, 61–64). A major factor influencing the severity of GAS infections is also the genetic inventory of the host. In humans, the differences in susceptibility to severe GAS diseases were mapped to the major histocompatibility complex locus (65). More severe infections and a generalized toxic shock syndrome appear to be associated with higher expression of inflammatory cytokines in both humans and mice (66). Cytokine production in GAS responses is regulated by T cells through GAS superantigens and the as yet poorly understood activation of innate immune cells. Our findings demonstrate that GAS is able to induce cytokines in macrophages through MyD88-dependent and -independent pathways. The identity of the GAS-recognizing receptor upstream of MyD88 poses an intriguing question, since our study rules out the exclusive involvement of the most prominent bacteria-recognizing receptors, TLR2, TLR4, and TLR9, or their combination. Other MyD88-dependent TLRs are the flagellin receptor TLR5 and the single-stranded RNA receptors TLR7 and TLR8 (8). Involvement of these receptors in GAS-induced MyD88-dependent signaling is not likely, since flagellin or a similar protein has not been found in GAS, and single-stranded RNA is associated with viral recognition. However, the role of these receptors cannot be entirely excluded, because PRRs are, in general, not specific for a single molecule. For example, TLR2 was reported to cooperate with dectin-1 in recognition of fungi (67). We ruled out the role of signaling through the IL-1R, which is known to be MyD88-dependent. IL-1 can be released by infected cells as a result of inflammasome-mediated caspase 1 activation, which is required for IL-1β processing (68). Although we did not address the GAS-mediated IL-1 production, a role for IL-1 as a possible mediator of the MyD88-dependent responses can be excluded. A function of IL-18 as another molecule requiring MyD88 for signaling is rather unlikely, since the IL-18 receptor is predominantly expressed on Th1 cells, whereas a prolonged treatment with e.g. IL-18 plus IL-12 is required for stronger expression in monocytic cells (69, 70). The ability of GAS to induce MyD88-dependent signaling independently of TLR2/4/9 may not be unique, since other Gram-positive pathogens have been reported to initiate inflammatory responses in the absence of multiple TLRs. For example, heat-inactivated group B streptococci and Streptococcus pneumoniae as well as spores of Bacillus anthracis induce MyD88-dependent responses in TLR2-, TLR4-, and TLR9-deficient macrophages or splenocytes (71–73). Despite the fact that TLR2/4/9 triple-deficient cells were not used in these studies, they support our hypothesis that some Gram-positive pathogenic bacteria are recognized by an as yet unidentified receptor upstream of MyD88.
The surprising finding that GAS is able to induce IFN-β independently of cytolysins represents another so far unique feature of GAS-induced responses in innate immune cells. In this regard, GAS-derived streptolysins are different from several other cytolysins that have been reported to induce IFN-β synthesis through activation of TLR4 (25). Another proposed mechanism of IFN-β induction by Gram-positive bacteria (e.g. L. monocytogenes) involves listeriolysin O-dependent liberation of bacteria and/or bacterial components from phagosomes (12, 23, 24). Although GAS is a prototype extracellular pathogen, it can be efficiently internalized by many phagocytic and nonphagocytic cells. However, GAS survives only for a short period of time in host cells due to degradation in lysosomal and autophagosomal compartments (5, 6). Interestingly, Bacillus subtilis engineered to express streptococcal SLO was not able to survive or multiply in infected cells, as opposed to listeriolysin O-expressing B. subtilis (74). This observation suggests that SLO, which displays a high degree of similarity with listeriolysin O, cannot mediate cytoplasmic escape. Thus, the reported data together with our findings support the hypothesis that GAS stimulates IFN-β synthesis by a novel mechanism that does not require cytoplasmic escape. Interestingly, the signaling events that culminate in Stat1 activation and transcription of IFN-responsive genes follow the well established pathway involving the transcription factor IRF3 and the type I IFN receptor but independent of MyD88. The increased IFN signaling in MyD88–/– cells may be explained by reduced expression of the inhibitor of IFN signaling SOCS1 (75). SOCS1 expression is known to be dependent on p38 MAPK (76), which we show in our study to be strongly reduced in GAS-infected MyD88–/– cells.
Our work demonstrates the ability of GAS to induce multiple inflammatory responses via in part novel recognition mechanisms. MyD88-dependent TLR2/TLR4/TLR9-independent signaling and cytolysin-independent IFN-β induction are so far exceptional characteristics of host cell responses to infection. Since GAS induces proinflammatory cytokine and IFN production in human macrophages (51), we assume that our findings are not restricted to the murine system, which, in the context of the whole organism, is considerably more resistant to GAS infection (66). The efficient stimulation of multiple signaling pathways may contribute to the known high capability of GAS to cause severe inflammatory diseases.
Supplementary Material
Acknowledgments
We thank Silvia Stockinger for critically reading the manuscript and Birgit Rapp for help with bacterial cultures.
Author's Choice—Final version full access.
This work was supported in part by Austrian Research Foundation Grants P16726-B14, I27-B03, and SFB F28 (to P. K.) and Grant P17238-B09 (to E. C.); the University Jubilee Foundation of the City of Vienna (Hochschuljubiläumsstiftung der Stadt Wien, HJST) Grant H-1020-2004 (to E. C.); and the European Science Foundation under the EUROCORES Programme Euro-DYNA through Contract ERAS-CT-2003-980409 of the European Commission, DG Research, FP6 (to the laboratory of P. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: GAS, group A streptococcus; BMDM, bone marrow-derived macrophages; IFN, interferon; IRF, interferon regulatory factor; MAPK, mitogen-activated protein kinase; PRRs, pattern-recognition receptors; TLR, Toll-like receptor; TNFα, tumor necrosis factor α; IL, interleukin; MOI, multiplicity of infection; ELISA, enzyme-linked immunosorbent assay; qRT, quantitative reverse transcription.
References
- 1.Carapetis, J. R., Steer, A. C., Mulholland, E. K., and Weber, M. (2005) Lancet 5 685–694 [DOI] [PubMed] [Google Scholar]
- 2.Bisno, A. L., Brito, M. O., and Collins, C. M. (2003) Lancet 3 191–200 [DOI] [PubMed] [Google Scholar]
- 3.Chhatwal, G. S., and McMillan, D. J. (2005) Trends Mol. Med. 11 152–155 [DOI] [PubMed] [Google Scholar]
- 4.Staali, L., Morgelin, M., Bjorck, L., and Tapper, H. (2003) Cell. Microbiol. 5 253–265 [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa, I., Amano, A., Mizushima, N., Yamamoto, A., Yamaguchi, H., Kamimoto, T., Nara, A., Funao, J., Nakata, M., Tsuda, K., Hamada, S., and Yoshimori, T. (2004) Science 306 1037–1040 [DOI] [PubMed] [Google Scholar]
- 6.Hakansson, A., Bentley, C. C., Shakhnovic, E. A., and Wessels, M. R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5192–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thulin, P., Johansson, L., Low, D. E., Gan, B. S., Kotb, M., McGeer, A., and Norrby-Teglund, A. (2006) PLoS Med. 3 e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783–801 [DOI] [PubMed] [Google Scholar]
- 9.Lee, M. S., and Kim, Y. J. (2007) Annu. Rev. Biochem. 76 447–480 [DOI] [PubMed] [Google Scholar]
- 10.O'Neill, L. A., and Bowie, A. G. (2007) Nat. Rev. 7 353–364 [DOI] [PubMed] [Google Scholar]
- 11.Stockinger, S., Reutterer, B., Schaljo, B., Schellack, C., Brunner, S., Materna, T., Yamamoto, M., Akira, S., Taniguchi, T., Murray, P. J., Muller, M., and Decker, T. (2004) J. Immunol. 173 7416–7425 [DOI] [PubMed] [Google Scholar]
- 12.Stetson, D. B., and Medzhitov, R. (2006) Immunity 24 93–103 [DOI] [PubMed] [Google Scholar]
- 13.Takaoka, A., Wang, Z., Choi, M. K., Yanai, H., Negishi, H., Ban, T., Lu, Y., Miyagishi, M., Kodama, T., Honda, K., Ohba, Y., and Taniguchi, T. (2007) Nature 448 501–505 [DOI] [PubMed] [Google Scholar]
- 14.Ishii, K. J., Kawagoe, T., Koyama, S., Matsui, K., Kumar, H., Kawai, T., Uematsu, S., Takeuchi, O., Takeshita, F., Coban, C., and Akira, S. (2008) Nature 451 725–729 [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S. M., and Maniatis, T. (2003) Nat. Immunol. 4 491–496 [DOI] [PubMed] [Google Scholar]
- 16.Sharma, S., tenOever, B. R., Grandvaux, N., Zhou, G. P., Lin, R., and Hiscott, J. (2003) Science 300 1148–1151 [DOI] [PubMed] [Google Scholar]
- 17.Hemmi, H., Takeuchi, O., Sato, S., Yamamoto, M., Kaisho, T., Sanjo, H., Kawai, T., Hoshino, K., Takeda, K., and Akira, S. (2004) J. Exp. Med. 199 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry, A. K., Chow, E. K., Goodnough, J. B., Yeh, W. C., and Cheng, G. (2004) J. Exp. Med. 199 1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy, D. E. (2002) J. Exp. Med. 195 F15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi, T., and Takaoka, A. (2002) Curr. Opin. Immunol. 14 111–116 [DOI] [PubMed] [Google Scholar]
- 21.Strober, W., Murray, P. J., Kitani, A., and Watanabe, T. (2006) Nat. Rev. 6 9–20 [DOI] [PubMed] [Google Scholar]
- 22.Goldmann, O., Rohde, M., Chhatwal, G. S., and Medina, E. (2004) Infect Immun. 72 2956–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockinger, S., Materna, T., Stoiber, D., Bayr, L., Steinborn, R., Kolbe, T., Unger, H., Chakraborty, T., Levy, D. E., Muller, M., and Decker, T. (2002) J. Immunol. 169 6522–6529 [DOI] [PubMed] [Google Scholar]
- 24.O'Riordan, M., Yi, C. H., Gonzales, R., Lee, K. D., and Portnoy, D. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13861–13866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, J. M., Ng, V. H., Maeda, S., Rest, R. F., and Karin, M. (2004) J. Exp. Med. 200 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangold, M., Siller, M., Roppenser, B., Vlaminckx, B. J., Penfound, T. A., Klein, R., Novak, R., Novick, R. P., and Charpentier, E. (2004) Mol. Microbiol. 53 1515–1527 [DOI] [PubMed] [Google Scholar]
- 27.Ruiz, N., Wang, B., Pentland, A., and Caparon, M. (1998) Mol. Microbiol. 27 337–346 [DOI] [PubMed] [Google Scholar]
- 28.Sierig, G., Cywes, C., Wessels, M. R., and Ashbaugh, C. D. (2003) Infect. Immun. 71 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caparon, M. G., Stephens, D. S., Olsen, A., and Scott, J. R. (1991) Infect. Immun. 59 1811–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, pp. 1.74–1.84, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 31.Baccarini, M., Bistoni, F., and Lohmann-Matthes, M. L. (1985) J. Immunol. 134 2658–2665 [PubMed] [Google Scholar]
- 32.Adachi, O., Kawai, T., Takeda, K., Matsumoto, M., Tsutsui, H., Sakagami, M., Nakanishi, K., and Akira, S. (1998) Immunity 9 143–150 [DOI] [PubMed] [Google Scholar]
- 33.Hoshino, K., Takeuchi, O., Kawai, T., Sanjo, H., Ogawa, T., Takeda, Y., Takeda, K., and Akira, S. (1999) J. Immunol. 162 3749–3752 [PubMed] [Google Scholar]
- 34.Takeuchi, O., Hoshino, K., Kawai, T., Sanjo, H., Takada, H., Ogawa, T., Takeda, K., and Akira, S. (1999) Immunity 11 443–451 [DOI] [PubMed] [Google Scholar]
- 35.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K., and Akira, S. (2000) Nature 408 740–745 [DOI] [PubMed] [Google Scholar]
- 36.Yasuda, K., Yu, P., Kirschning, C. J., Schlatter, B., Schmitz, F., Heit, A., Bauer, S., Hochrein, H., and Wagner, H. (2005) J. Immunol. 174 6129–6136 [DOI] [PubMed] [Google Scholar]
- 37.Kovarik, P., Stoiber, D., Novy, M., and Decker, T. (1998) EMBO J. 17 3660–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alcamo, E., Mizgerd, J. P., Horwitz, B. H., Bronson, R., Beg, A. A., Scott, M., Doerschuk, C. M., Hynes, R. O., and Baltimore, D. (2001) J. Immunol. 167 1592–1600 [DOI] [PubMed] [Google Scholar]
- 39.Senftleben, U., Li, Z. W., Baud, V., and Karin, M. (2001) Immunity 14 217–230 [DOI] [PubMed] [Google Scholar]
- 40.Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M. W., and Ausubel, F. M. (2002) Science 297 623–626 [DOI] [PubMed] [Google Scholar]
- 41.Suzuki, N., Suzuki, S., Duncan, G. S., Millar, D. G., Wada, T., Mirtsos, C., Takada, H., Wakeham, A., Itie, A., Li, S., Penninger, J. M., Wesche, H., Ohashi, P. S., Mak, T. W., and Yeh, W. C. (2002) Nature 416 750–756 [DOI] [PubMed] [Google Scholar]
- 42.Matsuzawa, A., Saegusa, K., Noguchi, T., Sadamitsu, C., Nishitoh, H., Nagai, S., Koyasu, S., Matsumoto, K., Takeda, K., and Ichijo, H. (2005) Nat. Immunol. 6 587–592 [DOI] [PubMed] [Google Scholar]
- 43.Abel, B., Thieblemont, N., Quesniaux, V. J., Brown, N., Mpagi, J., Miyake, K., Bihl, F., and Ryffel, B. (2002) J. Immunol. 169 3155–3162 [DOI] [PubMed] [Google Scholar]
- 44.Ferwerda, G., Girardin, S. E., Kullberg, B. J., Le Bourhis, L., de Jong, D. J., Langenberg, D. M., van Crevel, R., Adema, G. J., Ottenhoff, T. H., Van der Meer, J. W., and Netea, M. G. (2005) PLoS Pathog. 1 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latz, E., Schoenemeyer, A., Visintin, A., Fitzgerald, K. A., Monks, B. G., Knetter, C. F., Lien, E., Nilsen, N. J., Espevik, T., and Golenbock, D. T. (2004) Nat. Immunol. 5 190–198 [DOI] [PubMed] [Google Scholar]
- 46.Wei, X. Q., Leung, B. P., Niedbala, W., Piedrafita, D., Feng, G. J., Sweet, M., Dobbie, L., Smith, A. J., and Liew, F. Y. (1999) J. Immunol. 163 2821–2828 [PubMed] [Google Scholar]
- 47.Gavrilin, M. A., Bouakl, I. J., Knatz, N. L., Duncan, M. D., Hall, M. W., Gunn, J. S., and Wewers, M. D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mariathasan, S., Weiss, D. S., Newton, K., McBride, J., O'Rourke, K., Roose-Girma, M., Lee, W. P., Weinrauch, Y., Monack, D. M., and Dixit, V. M. (2006) Nature 440 228–232 [DOI] [PubMed] [Google Scholar]
- 49.Ozoren, N., Masumoto, J., Franchi, L., Kanneganti, T. D., Body-Malapel, M., Erturk, I., Jagirdar, R., Zhu, L., Inohara, N., Bertin, J., Coyle, A., Grant, E. P., and Nunez, G. (2006) J. Immunol. 176 4337–4342 [DOI] [PubMed] [Google Scholar]
- 50.Glaccum, M. B., Stocking, K. L., Charrier, K., Smith, J. L., Willis, C. R., Maliszewski, C., Livingston, D. J., Peschon, J. J., and Morrissey, P. J. (1997) J. Immunol. 159 3364–3371 [PubMed] [Google Scholar]
- 51.Miettinen, M., Lehtonen, A., Julkunen, I., and Matikainen, S. (2000) J. Immunol. 164 3733–3740 [DOI] [PubMed] [Google Scholar]
- 52.Gao, J. J., Filla, M. B., Fultz, M. J., Vogel, S. N., Russell, S. W., and Murphy, W. J. (1998) J. Immunol. 161 4803–4810 [PubMed] [Google Scholar]
- 53.Toshchakov, V., Jones, B. W., Perera, P. Y., Thomas, K., Cody, M. J., Zhang, S., Williams, B. R., Major, J., Hamilton, T. A., Fenton, M. J., and Vogel, S. N. (2002) Nat. Immunol. 3 392–398 [DOI] [PubMed] [Google Scholar]
- 54.Decker, T., Muller, M., and Stockinger, S. (2005) Nat. Rev. 5 675–687 [DOI] [PubMed] [Google Scholar]
- 55.Karaghiosoff, M., Neubauer, H., Lassnig, C., Kovarik, P., Schindler, H., Pircher, H., McCoy, B., Bogdan, C., Decker, T., Brem, G., Pfeffer, K., and Muller, M. (2000) Immunity 13 549–560 [DOI] [PubMed] [Google Scholar]
- 56.Park, C., Li, S., Cha, E., and Schindler, C. (2000) Immunity 13 795–804 [DOI] [PubMed] [Google Scholar]
- 57.Hug, H., Costas, M., Staeheli, P., Aebi, M., and Weissmann, C. (1988) Mol. Cell. Biol. 8 3065–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panne, D., Maniatis, T., and Harrison, S. C. (2007) Cell 129 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alouf, J. E. (1980) Pharmacol. Ther. 11 661–717 [DOI] [PubMed] [Google Scholar]
- 60.Nizet, V. (2002) Trends Microbiol. 10 575–580 [DOI] [PubMed] [Google Scholar]
- 61.Beres, S. B., Sylva, G. L., Sturdevant, D. E., Granville, C. N., Liu, M., Ricklefs, S. M., Whitney, A. R., Parkins, L. D., Hoe, N. P., Adams, G. J., Low, D. E., DeLeo, F. R., McGeer, A., and Musser, J. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11833–11838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beres, S. B., Richter, E. W., Nagiec, M. J., Sumby, P., Porcella, S. F., Deleo, F. R., and Musser, J. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7059–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cho, K. H., and Caparon, M. G. (2005) Mol. Microbiol. 57 1545–1556 [DOI] [PubMed] [Google Scholar]
- 64.Virtaneva, K., Porcella, S. F., Graham, M. R., Ireland, R. M., Johnson, C. A., Ricklefs, S. M., Babar, I., Parkins, L. D., Romero, R. A., Corn, G. J., Gardner, D. J., Bailey, J. R., Parnell, M. J., and Musser, J. M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9014–9019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kotb, M., Norrby-Teglund, A., McGeer, A., El-Sherbini, H., Dorak, M. T., Khurshid, A., Green, K., Peeples, J., Wade, J., Thomson, G., Schwartz, B., and Low, D. E. (2002) Nat. Med. 8 1398–1404 [DOI] [PubMed] [Google Scholar]
- 66.Medina, E., and Lengeling, A. (2005) Brief. Funct. Genomics Proteomics 4 248–257 [DOI] [PubMed] [Google Scholar]
- 67.Rogers, N. C., Slack, E. C., Edwards, A. D., Nolte, M. A., Schulz, O., Schweighoffer, E., Williams, D. L., Gordon, S., Tybulewicz, V. L., Brown, G. D., and Reis e Sousa, C. (2005) Immunity 22 507–517 [DOI] [PubMed] [Google Scholar]
- 68.Delbridge, L. M., and O'Riordan, M. X. (2007) Curr. Opin. Immunol. 19 10–16 [DOI] [PubMed] [Google Scholar]
- 69.Xu, D., Chan, W. L., Leung, B. P., Hunter, D., Schulz, K., Carter, R. W., McInnes, I. B., Robinson, J. H., and Liew, F. Y. (1998) J. Exp. Med. 188 1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerdes, N., Sukhova, G. K., Libby, P., Reynolds, R. S., Young, J. L., and Schonbeck, U. (2002) J. Exp. Med. 195 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Henneke, P., Takeuchi, O., Malley, R., Lien, E., Ingalls, R. R., Freeman, M. W., Mayadas, T., Nizet, V., Akira, S., Kasper, D. L., and Golenbock, D. T. (2002) J. Immunol. 169 3970–3977 [DOI] [PubMed] [Google Scholar]
- 72.Lee, K. S., Scanga, C. A., Bachelder, E. M., Chen, Q., and Snapper, C. M. (2007) Cell. Immunol. 245 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Glomski, I. J., Fritz, J. H., Keppler, S. J., Balloy, V., Chignard, M., Mock, M., and Goossens, P. L. (2007) Cell. Microbiol. 9 502–513 [DOI] [PubMed] [Google Scholar]
- 74.Portnoy, D. A., Tweten, R. K., Kehoe, M., and Bielecki, J. (1992) Infect. Immun. 60 2710–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alexander, W. S., Starr, R., Fenner, J. E., Scott, C. L., Handman, E., Sprigg, N. S., Corbin, J. E., Cornish, A. L., Darwiche, R., Owczarek, C. M., Kay, T. W., Nicola, N. A., Hertzog, P. J., Metcalf, D., and Hilton, D. J. (1999) Cell 98 597–608 [DOI] [PubMed] [Google Scholar]
- 76.Vazquez, N., Greenwell-Wild, T., Rekka, S., Orenstein, J. M., and Wahl, S. M. (2006) J. Leukocyte Biol. 80 1136–1144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.