Abstract
The Saccharomyces cerevisiae DGK1 gene encodes a diacylglycerol kinase enzyme that catalyzes the formation of phosphatidate from diacylglycerol. Unlike the diacylglycerol kinases from bacteria, plants, and animals, the yeast enzyme utilizes CTP, instead of ATP, as the phosphate donor in the reaction. Dgk1p contains a CTP transferase domain that is present in the SEC59-encoded dolichol kinase and CDS1-encoded CDP-diacylglycerol synthase enzymes. Deletion analysis showed that the CTP transferase domain was sufficient for diacylglycerol kinase activity. Point mutations (R76A, K77A, D177A, and G184A) of conserved residues within the CTP transferase domain caused a loss of diacylglycerol kinase activity. Analysis of DGK1 alleles showed that the in vivo functions of Dgk1p were specifically due to its diacylglycerol kinase activity. The DGK1-encoded enzyme had a pH optimum at 7.0-7.5, required Ca2+ or Mg2+ ions for activity, was potently inhibited by N-ethylmaleimide, and was labile at temperatures above 40 °C. The enzyme exhibited positive cooperative (Hill number = 2.5) kinetics with respect to diacylglycerol (apparent Km = 6.5 mol %) and saturation kinetics with respect to CTP (apparent Km = 0.3 mm). dCTP was both a substrate (apparent Km = 0.4 mm) and competitive inhibitor (apparent Ki = 0.4 mm) of the enzyme. Diacylglycerol kinase activity was stimulated by major membrane phospholipids and was inhibited by CDP-diacylglycerol and sphingoid bases.
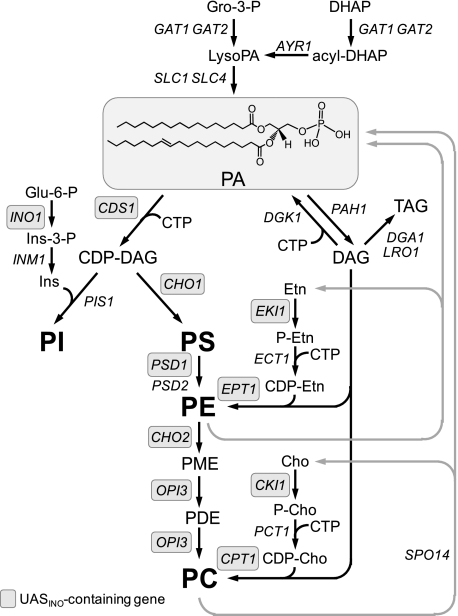
In the yeast Saccharomyces cerevisiae, PA2 is an important phospholipid intermediate in the synthesis of membrane phospholipids and the neutral lipid triacylglycerol (see Fig. 1) (1-5). The major phospholipids phosphatidylinositol, phosphatidylserine, phosphatidylethanolamine, and phosphatidylcholine are derived from PA via the liponucleotide intermediate CDP-DAG (1-3). The mitochondrial phospholipids phosphatidylglycerol and cardiolipin are similarly derived from PA via CDP-DAG (not shown in Fig. 1) (6). Alternatively, phosphatidylethanolamine and phosphatidylcholine may be derived from PA via DAG, which is also utilized for the synthesis of the storage lipid triacylglycerol (1-3, 5). In the de novo biosynthetic pathway, PA is made from lyso-PA that is derived from either glycerol 3-phosphate or dihydroxyacetone phosphate (1-4). Besides the de novo pathway, PA is produced from the phospholipase D-mediated turnover of phosphatidylethanolamine and phosphatidylcholine (1-4). In bacteria, plants, and animals, PA may be produced from DAG via an ATP-dependent DAG kinase reaction (7-10). However, an enzyme catalyzing this reaction has not been identified from S. cerevisiae, and there are no yeast genes that encode a homologous protein in the superfamily of DAG kinase enzymes from bacteria, plants, and animals.
FIGURE 1.
Role of PA in lipid synthesis. The PA structure shown with fatty acyl groups of 16:0 (sn-1) and 18:1 (sn-2) is highlighted by gray shading. The pathways shown for the synthesis of phospholipids and triacylglycerol (TAG) include the relevant steps discussed in this work. The genes that are known to encode enzymes catalyzing individual steps in the lipid synthesis pathways are indicated. The UASINO-containing genes that are subject to regulation by the Ino2p-Ino4p activation complex and the Opi1p repressor are highlighted by gray shading. Gro, glycerol; DHAP, dihydroxyacetone phosphate; Glu, glucose; Ins, inositol; PI, phosphatidylinositol; PS, phosphatidylserine; PE, phosphatidylethanolamine; PME, phosphatidylmonomethylethanolamine; PDE, phosphatidyldimethylethanolamine; PC, phosphatidylcholine; Etn, ethanolamine; Cho, choline.
In addition to its role as an intermediate of lipid metabolism, PA plays a central role in the transcriptional regulation of phospholipid synthesis in S. cerevisiae (1). PA, along with the Scs2p protein at the nuclear/ER membrane, binds and inactivates the transcriptional repressor Opi1p (11, 12). When PA levels are reduced, Opi1p translocates into the nucleus, where it interacts with Ino2p to repress the expression of UASINO-containing genes that encode many of the enzymes responsible for the synthesis of membrane phospholipids (1) (Fig. 1). Maximum expression of the UASINO-containing genes is mediated by the interaction of an Ino2p-Ino4p activation complex with a UASINO element that is present in their promoters (2, 3, 13-18). The most highly regulated UASINO-containing gene is INO1, which encodes the inositol biosynthetic enzyme inositol-3-phosphate synthase (19-21). Abnormally high levels of INO1 expression give rise to an inositol excretion phenotype, whereas abnormally low levels of INO1 expression give rise to an inositol auxotrophic phenotype (1, 3, 16).
The importance of controlling the cellular levels of PA is highlighted by phenotypes associated with mutations that affect the activity of the PAH1-encoded PA phosphatase. This enzyme catalyzes the Mg2+-dependent dephosphorylation of PA to yield DAG and Pi (22-24). Loss-of-function mutations for PAH1-encoded PA phosphatase activity cause elevated levels of PA and the concomitant derepression of UASINO-containing genes (e.g. INO1 and OPI3) (25, 26). These mutations also cause cells to exhibit a nuclear/ER membrane expansion phenotype (25, 26). On the other hand, the overexpression of PAH1-encoded PA phosphatase activity causes inositol auxotrophy that is due to the repression of the INO1 gene (27).
The diacylglycerol kinase gene DGK13 (YOR311C) has been identified as a gene whose function counteracts that of the PAH1-encoded PA phosphatase (28). Specifically, the overexpression of DGK1 complements the inositol auxotrophy caused by the overexpression of PAH1-encoded PA phosphatase activity (28). In addition, a dgk1Δ mutation bypasses the phenotypes caused by the pah1Δ mutation, which include an elevated level of PA, the derepression of the INO1 gene, and the nuclear/ER membrane expansion phenotype (28). Moreover, the overexpression of DGK1 causes the nuclear/ER membrane expansion phenotype (28) that is exhibited by cells that carry mutations in PAH1-encoded PA phosphatase activity (25, 26). This work led to the identification of Dgk1p as a DAG kinase enzyme (28). However, unlike the ATP-dependent DAG kinase enzymes that exist in bacteria, plants, and animals (7-10, 29), the S. cerevisiae enzyme utilizes CTP, instead of ATP, as the phosphate donor in the reaction (see Fig. 2). In this work, we characterized the basic enzymological properties of the yeast CTP-dependent DAG kinase. Through a mutational analysis of the enzyme, we show that phenotypes associated with the overexpression of the DGK1 gene are specifically due to the DAG kinase activity of Dgk1p.
FIGURE 2.
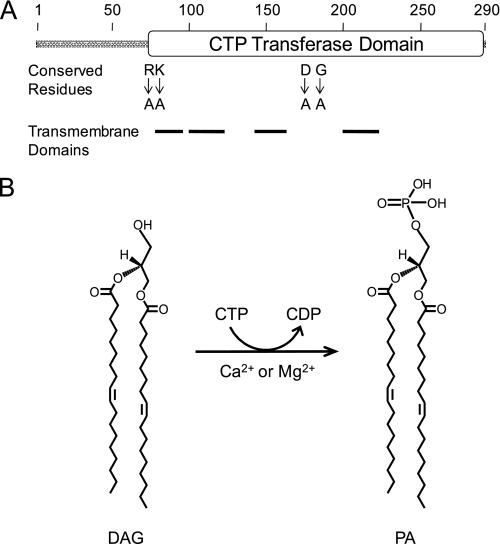
Domain structure and the reaction catalyzed by the DGK1-encoded CTP-dependent DAG kinase. A, the diagram shows the positions of the CTP transferase domain, the transmembrane-spanning domains, and conserved residues of Dgk1p that are found in the cytidylyltransferase domain of Cds1p and Sec59p. B, shown are the structures of DAG and PA and the reaction catalyzed by CTP-dependent DAG kinase.
EXPERIMENTAL PROCEDURES
Materials—Growth medium components were purchased from Difco. Restriction endonucleases, modifying enzymes, and Vent DNA polymerase were from New England Biolabs. DNA purification kits were from Qiagen. Nucleotides, oligonucleotides, Triton X-100, nucleoside-5′-diphosphate kinase (S. cerevisiae), phytosphingosine, 1-oleoyl-rac-glycerol, DAG kinase inhibitors (R59022 and R59949), and protease inhibitors (phenylmethylsulfonyl fluoride, benzamidine, aprotinin, leupeptin, and pepstatin) were from Sigma. Dioleoyl-DAG, dioctanoyl-DAG, dioleoyl-PA, phosphatidylinositol, ceramide, dolichol, d-erythro-sphingosine, and d-erythro-sphinganine were from Avanti Polar Lipids. Polyethyleneimine-cellulose plates were from EM Science. Protein assay reagents, electro-phoretic reagents, and DNA and protein size standards were from Bio-Rad. Polyvinylidene difluoride membranes and the enhanced chemifluorescence Western blot reagent were from GE Healthcare. Goat anti-rabbit IgG antibodies conjugated with alkaline phosphatase were from Pierce. Radiochemicals were from PerkinElmer Life Sciences. Scintillation counting supplies were from National Diagnostics.
Strains and Growth Conditions—The bacterial and yeast strains used in this work are listed in Table 1. S. cerevisiae strain SS1144 is a dgk1Δ::HIS3 derivative of strain RS453 (28). This strain contained plasmid YEplac181-GAL1/10-DGK1 and was used for the massive overexpression of DAG kinase activity. The dgk1Δ mutant containing YCplac111-GAL1/10-DGK1 alleles was used to examine the effects of DGK1 expression on nuclear/ER morphology. Yeast cells were grown at 30 °C in 1% yeast extract, 2% peptone, and 2% glucose or in synthetic complete medium (30). Plasmid-bearing yeast cells were selected in synthetic complete medium lacking the appropriate amino acid. Cells containing the galactose-inducible DGK1 alleles were grown to exponential phase (A600 ∼ 0.5) in synthetic medium with 2% raffinose as a carbon source. To induce expression of the DGK1 gene, the culture was added with galactose to a final concentration of 2% and incubated for 24 h. Escherichia coli cells were grown at 37 °C in LB medium (1% Tryptone, 0.5% yeast extract, and 1% NaCl (pH 7.4)). Plasmid-bearing E. coli cells were selected in growth medium containing ampicillin (100 μg/ml). Solid growth medium for yeast and E. coli cells contained agar at final concentrations of 2 and 1.5%, respectively. Yeast cell numbers in liquid growth medium were determined spectrophotometrically at an absorbance of 600 nm.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or Ref. |
|---|---|---|
| E. coli | ||
| DH5α | F ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk−mk+) phoA supE44I−thi-1 gyrA96 relA1 | Ref. 31 |
| S. cerevisiae | ||
| RS453 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-52 | Ref. 87 |
| SS1144 | dgk1Δ::HIS3 derivative of strain RS453 | Ref. 28 |
| Plasmid | ||
| YEplac181 | High copy number E. coli/yeast shuttle vector with LEU2 | Ref. 34 |
| YEplac181-GAL1/10-DGK1 | DGK1 under control of GAL1/10 promoter in YEplac181 | This study |
| YCplac111 | Low copy E. coli/yeast shuttle vector with LEU2 | Ref. 34 |
| YCplac111-GAL1/10-DGK1 | DGK1 under control of GAL1/10 promoter in YCplac111 | Ref. 28 |
| YCplac111-GAL1/10-DGK1Δ66 | DGK1Δ66 derivative of YCplac111-GAL1/10-DGK1 | This study |
| YCplac111-GAL1/10-DGK1Δ70 | DGK1Δ70 derivative of YCplac111-GAL1/10-DGK1 | This study |
| YCplac111-GAL1/10-DGK1Δ77 | DGK1Δ77 derivative of YCplac111-GAL1/10-DGK1 | This study |
| YCplac111-GAL1/10-DGK1(R76A) | DGK1(R76A) derivative of YCplac111-GAL1/10-DGK1 | This study |
| YCplac111-GAL1/10-DGK1(K77A) | DGK1(K77A) derivative of YCplac111-GAL1/10-DGK1 | This study |
| YCplac111-GAL1/10-DGK1(D177A) | DGK1(D177A) derivative of YCplac111-GAL1/10-DGK1 | Ref. 28 |
| YCplac111-GAL1/10-DGK1(G184A) | DGK1(G184A) derivative of YCplac111-GAL1/10-DGK1 | This study |
| YCplac33 | Low copy number E. coli/yeast shuttle vector with URA3 | Ref. 34 |
| YCplac33-SEC63-GFP | SEC63-GFP fusion gene in YCplac33 | Ref. 28 |
DNA Manipulations and Plasmid Constructions—Standard methods were used for isolation and manipulation of DNA (31). Transformations of yeast (32, 33) and bacteria (31) with plasmids were performed as described previously. The DGK1 promoter was substituted with the inducible GAL1/10 promoter that was cloned into the low copy YCplac111 and high copy YEplac181 vectors (34). The DGK1 truncation mutants were constructed by ligating a 5′-fragment containing the GAL1/10 promoter followed by the first two codons of DGK1 and a BamHI site to a 3′-fragment containing a BamHI site followed by the codon encoding DGK1Δ66, DGK1Δ70, and DGK1Δ77. The DGK1(R76A), DGK1(K77A), and DGK1(G184A) mutants were constructed by PCR-mediated mutagenesis of YCplac111-GAL1/10-DGK1 using the appropriate primers for each mutation. All constructs were verified by DNA sequencing.
Preparation of Anti-Dgk1p Antibodies and Immunoblot Analysis—The Dgk1p peptides GALMRKKEIHTYN (residues 133-145) and GHLTPKVARNKSLA (residues 188-201) were synthesized and conjugated to carrier protein at Bio-Synthesis Inc. (Lewisville, TX). Antibodies were raised against a 50:50 mixture of these peptides in New Zealand White rabbits by standard procedures (35) at Bio-Synthesis Inc. The IgG fraction was isolated from antisera by protein A-Sepharose chromatography (35). SDS-PAGE (36) and immunoblotting (37) were performed as described previously. Polyvinylidene difluoride membrane was used for the protein blotting. The membrane was probed with anti-Dgk1p antibodies at a dilution of 1 μg/ml, followed by alkaline phosphatase-conjugated anti-rabbit IgG at a dilution of 1:5000. The immune complexes were detected using enhanced chemifluorescence reagents, and the fluorescent signal was processed with a FluoroImager. The immunoblot signals were in the linear range of detection.
Preparation of Cell Extracts and Membranes—All steps were performed at 4 °C. Yeast cultures were harvested by centrifugation at 1500 × g for 5 min, and the resulting cell pellets were washed once with water. Cells were then resuspended in 50 mm Tris-HCl (pH 7.5) containing 0.3 m sucrose, 1 mm EDTA, 10 mm 2-mercaptoethanol, and a mixture of protease inhibitors (0.5 mm phenylmethanesulfonyl fluoride, 1 mm benzamidine, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin). The cell suspension was mixed with glass beads (0.5-mm diameter) and disrupted using a Mini-BeadBeater-16 (BioSpec Products, Inc.) as described previously (38). After removal of unbroken cells and glass beads by centrifugation at 1500 × g for 10 min, cell extracts were fractionated into the cytosolic and total membrane fractions by centrifugation at 100,000 × g for 1 h (23). The membrane fraction was resuspended in the same buffer lacking EDTA at a protein concentration of 1 mg/ml and stored at -80 °C. Protein concentration was measured by the method of Bradford (39) using bovine serum albumin as the standard.
Preparation of Labeled CTP, dCTP, and PA—[γ-32P]CTP was synthesized enzymatically from CDP and [γ-32P]ATP with nucleoside-5′-diphosphate kinase (40). The reaction mixture contained 10 mm MOPS-NaOH (pH 7.6), 10 μm CDP, 75 μCi of [γ-32P]ATP (3000 Ci/mmol), 5 mm MgCl2, and 1 unit of nucleoside-5′-diphosphate kinase in a final volume of 30 μl. The reaction was terminated after 10 min by the addition of 10 mm EDTA. The product [γ-32P]CTP was analyzed by TLC on a polyethyleneimine-cellulose plate using 0.75 m KH2PO4/H3PO4 (pH 3.65) as the solvent (40) and contained >95% of the radioactivity used in the reaction. [γ-32P]dCTP was synthesized by the same method except that dCDP was used instead of CDP. [32P]PA was synthesized from DAG and [γ-32P]ATP using E. coli DAG kinase and purified by TLC as described by Carman and Lin (41).
Enzyme Assays—DAG kinase activity was measured for 20 min by following the phosphorylation of 0.1 mm dioleoyl-DAG with 1 mm [γ-32P]CTP (5000 cpm/nmol) in the presence of 50 mm Tris-HCl (pH 7.5), 1 mm Triton X-100, 1 mm CaCl2, 10 mm 2-mercaptoethanol, and enzyme protein in a total volume of 0.1 ml at 30 °C. The reaction was terminated by the addition of 0.5 ml of 0.1 n HCl in methanol. The radioactive chloroform-soluble product PA was separated from the radioactive water-soluble substrate CTP by a chloroform/methanol/water phase partition (42). The radioactive chloroform phase was subjected to liquid scintillation counting. Alternatively, the chloroform-soluble product was subjected to TLC using a solvent system of chloroform/methanol/water (65:25:4, v/v) and visualized by phosphorimaging. The product PA was confirmed by comigration with standard PA (visualized by iodine staining) and with radiolabeled PA that was synthesized by the E. coli ATP-dependent DAG kinase reaction. The DAG kinase assay was conducted in triplicate, and the average S.D. of the assay was ±5%. The lower limit of detection for activity was 2 pmol/min/mg. One unit of DAG kinase activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of product/min. Specific activity was defined as units/mg of protein. The activities of CDP-DAG synthase (43), phosphocholine cytidylyltransferase (44), phosphoethanolamine cytidylyltransferase (44), and dolichol kinase (45) were measured as described previously.
Fluorescence Microscopy—Cells were examined with a Zeiss Axiophot fluorescence microscope equipped with a CCD camera, and images were recorded using IP Labs software. The expression of the SEC63-GFP fusion gene was used to visualize the nuclear/ER membrane (27). Two independent transformants for each strain were examined, and in all cases, the results were same for both.
Data Analyses and Calculation of Lipid Mole Percent—Kinetic data were analyzed according to the Michaelis-Menten and Hill equations using the EZ-FIT enzyme kinetic model-fitting program of Perrella (46). Student's t test (SigmaPlot software) was used to determine statistical significance, and p values <0.05 were taken as a significant difference. The mole percent of a lipid in a Triton X-100/lipid-mixed micelle was calculated using the following formula: mol %lipid = 100 × [lipid (molar)]/([lipid (molar)] + [Triton X-100 (molar)]).
RESULTS
The DGK1 Gene Encodes a CTP-dependent DAG Kinase Activity—DGK1 encodes a 32.8-kDa protein (Dgk1p) with a short N-terminal hydrophilic region followed by four putative transmembrane domains (Fig. 2). The latter portion of the protein contains a predicted CTP transferase domain that is characteristic of enzymes that utilize CTP as a substrate. This CTP transferase domain is found in two other S. cerevisiae proteins, viz. the CDS1-encoded CDP-DAG synthase (47) and the SEC59-encoded dolichol kinase (48). Dgk1p does not catalyze either the forward or reverse reaction of the CDP-DAG synthase enzyme (49, 50) or the reaction catalyzed by the dolichol kinase enzyme (51). Moreover, the enzyme does not catalyze the CTP-dependent reactions that are catalyzed by the PCT-encoded phosphocholine cytidylyltransferase (52) or the ECT-encoded phosphoethanolamine cytidylyltransferase (53) enzyme. Instead, Dgk1p catalyzes the CTP-dependent conversion of DAG to PA.
The specific activity of DAG kinase in cell extracts of wild-type cells was typically 0.018 ± 0.002 units/mg. This level of enzyme activity was ∼20-30-fold lower than those of most phospholipid synthesis enzymes (e.g. phosphatidylserine synthase, phosphatidylinositol synthase, and phosphatidylethanolamine methyltransferase) in S. cerevisiae (54-56). To measure DAG kinase activity in cell extracts, it was necessary to use the relatively high radioactive specific activity of [γ-32P]CTP (50,000 cpm/nmol). As predicted from the presence of transmembrane-spanning regions in Dgk1p (Fig. 2), DAG kinase activity was associated with the membrane fraction of the cell. The galactose-inducible GAL1/10 promoter was used for the overexpression of the DGK1 gene from a high copy number plasmid (YEplac181-GAL1/10-DGK1). The induction of DGK1 gene expression resulted in a massive increase in DAG kinase activity (Table 2). The specific activity of DAG kinase in the membrane fraction of galactose-grown cells bearing the plasmid was typically 130 ± 2.7 units/mg, which was equivalent to a 7222-fold purification of DAG kinase relative to the activity of the enzyme in the cell extract of wild-type glucose-grown cells (Table 2). The DAG kinase activity in this membrane preparation was ∼100-fold greater than the activities of other membrane-associated phospholipid synthesis enzymes (e.g. CDP-DAG synthase (50), phosphatidylserine synthase (57), and phosphatidylinositol synthase (58)) that are found in the membrane fraction of S. cerevisiae. Thus, this membrane fraction provided us with a highly enriched preparation of DAG kinase activity to study the basic enzymological properties of the enzyme.
TABLE 2.
Enrichment of CTP-dependent DAG kinase activity by overexpression of the DGK1 gene
CTP-dependent DAG kinase activity was measured under standard assay conditions with the indicated enzyme preparations.
| Preparation | DAG kinase activity | Enrichment |
|---|---|---|
| nmol/min/mg | -fold | |
| Cell extract (wild-type) | 0.018 ± 0.003 | 1 |
| Membranes (dgk1Δ + YEplac181-GAL1/10-DGK1) | 130 ± 2.7 | 7222 |
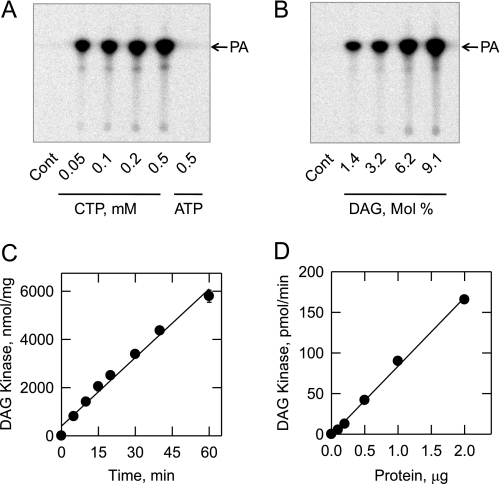
The DAG kinase reaction was carried out with [γ-32P]CTP as the phosphate donor and dioleoyl-DAG as the lipid acceptor. Following the reaction, the chloroform-soluble product of the reaction was analyzed by TLC. Phosphorimaging analysis of thin layer chromatograms showed that the enzyme catalyzed the dose-dependent formation of PA from CTP (Fig. 3A) and DAG (Fig. 3B). Under standard reaction conditions, the DAG kinase reaction was dependent on time (Fig. 3C) and enzyme protein (Fig. 3D). ATP did not serve as a substrate for the DGK1-encoded DAG kinase reaction (Fig. 3A). The addition of 1 mm ATP, GTP, or UTP to the assay system did not affect the activity of the CTP-dependent DAG kinase (data not shown). That GTP and UTP did not inhibit the reaction indicated that these nucleotides were not substrates for the enzyme. Glycerol, PA, phosphatidylinositol, dolichol, sphinganine, sphingosine, phytosphingosine, and ceramide were tested as lipid acceptor substrates for the DGK1-encoded kinase using [γ-32P]CTP as the phosphate donor. None of these lipids served as a substrate for the enzyme (data not shown). The enzyme utilized 0.1 mm dioctanoyl-DAG as a substrate (without Triton X-100 solubilization). However, the activity (50 ± 2.2 units/mg) using this short chain substrate was ∼40% of that using the more physiological long chain substrate dioleoyl-DAG.
FIGURE 3.
Identification of PA as the product of the CTP-dependent DAG kinase reaction and the time course and enzyme dependence of the reaction. DAG kinase activity was measured with the indicated concentrations of [γ-32P]CTP or [γ-32P]ATP (A) and the indicated surface concentrations of DAG (B). CTP was used as the nucleotide substrate for the experiment shown in B. For the experiment shown in A, the molar ratio of Triton X-100 to DAG was maintained at 10:1 (9.1 mol % DAG). For the experiment shown in B, the molar concentration of DAG was held constant at 0.1 mm, and the Triton X-100 concentration was varied to obtain the indicated surface concentrations. The concentration of CTP was maintained at 1 mm. Following the reactions, the chloroform-soluble 32P-labeled products were analyzed by TLC and visualized by phosphorimaging. The position of the PA standard is indicated. In separate experiments, DAG kinase activity was measured under standard assay conditions except for the variation with time (C) and enzyme concentration (D). The data shown in A and B are representative of three independent experiments, and the data shown in C and D are means ± S.D. from triplicate enzyme determinations. Cont, control.
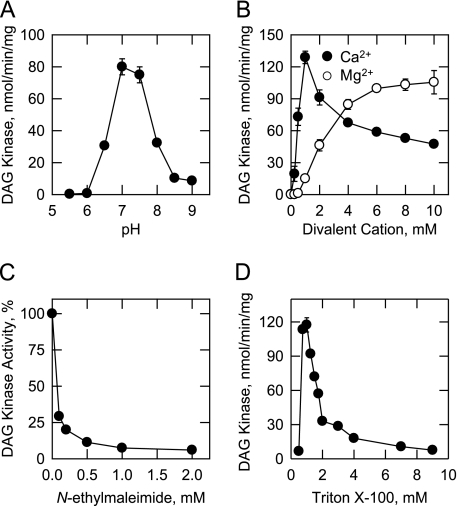
Effects of pH, Cations, Triton X-100, N-Ethylmaleimide, and DAG Kinase Inhibitors on CTP-dependent DAG Kinase Activity—The effect of pH on DAG kinase activity is shown in Fig. 4A. Maximum DAG kinase activity was obtained at pH 7.0-7.5. Because DAG kinase activity was essentially the same at both pH values, we arbitrarily measured activity with Tris-HCl at pH 7.5. DAG kinase activity was dependent on the addition of a divalent cation to the assay system (Fig. 4B). The enzyme exhibited a dose-dependent requirement for Ca2+ or Mg2+ ions. The maximum DAG kinase activity obtained with 1 mm Ca2+ ions was slightly greater than the maximum activity obtained with 10 mm Mg2+ ions (Fig. 4B). Accordingly, we measured DAG kinase activity using Ca2+ ions as cofactor. Ca2+ concentrations of > 1 mm resulted in an inhibition of DAG kinase activity (Fig. 4B). The effects of Ca2+ and Mg2+ ions on activity were not additive. The addition of the chelating agent EDTA or EGTA at 5 mm to the assay system resulted in the loss of detectable DAG kinase activity, but activity was not affected by Na+, K+, or Li+ ions at 100 mm. At 5 mm, Mn2+ and Zn2+ ions abolished detectable DAG kinase activity. R59022 (59) and R59949 (60) are commonly used inhibitors of ATP-dependent DAG kinase activities in mammalian cells (10). These reagents were not particularly potent inhibitors of the CTP-dependent DAG kinase from S. cerevisiae. At 50 μm, R59022 and R59949 inhibited activity by 25 and 15%, respectively. The alkylating reagent N-ethylmaleimide, which reacts with sulf-hydryl groups (61), was an inhibitor (IC50 = 0.1 mm) of DAG kinase activity (Fig. 4C).
FIGURE 4.
Effects of pH, divalent cations, N-ethylmaleimide, and Triton X-100 on CTP-dependent DAG kinase activity. DAG kinase activity was measured at the indicated pH values with 50 mm Tris maleate/glycine buffer (A), the indicated concentrations of CaCl2 or MgCl2 (B), the indicated concentrations ofN-ethylmaleimide (C), and the indicated concentrations of Triton X-100 (D). The data shown are means ± S.D. from triplicate enzyme determinations.
The effect of the nonionic detergent Triton X-100 on DAG kinase activity is shown in Fig. 4D. The addition of Triton X-100 to the assay system stimulated DAG kinase activity to a maximum at a concentration of 1 mm (10:1 molar ratio of Triton X-100 to DAG), followed by an apparent inhibition of activity at concentrations above 1 mm. This property is characteristic of surface dilution kinetics (62). The function of Triton X-100 in the assay is to form a mixed micelle with the lipid substrate to provide a surface for catalysis (62). The apparent inhibition of activity is simply due to the dilution of substrate in the Triton X-100/lipid-mixed micelle (62).
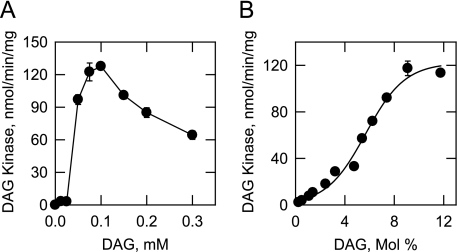
Dependence of CTP-dependent DAG Kinase Activity on the Molar and Surface Concentrations of DAG—Because the enzyme exhibited surface dilution kinetics, the analysis of enzyme activity was performed using Triton X-100/DAG-mixed micelles. According to the surface dilution kinetic model (62), the activity of a lipid-dependent enzyme should be dependent on the molar (e.g. number of micelles containing lipid) and surface (e.g. number of lipid molecules on a micelle surface) concentrations of lipid. In the experiment shown in Fig. 5A, DAG kinase activity was measured as a function of the molar concentration of DAG in a Triton X-100/DAG-mixed micelle maintained at a molar ratio of 10:1. Maximum activity was obtained at 0.1 mm. Concentrations above 0.1 mm resulted in the inhibition of DAG kinase activity. In the experiment shown in Fig. 5B, DAG kinase activity was measured as a function of the surface concentration of DAG using a Triton X-100/DAG-mixed micelle in which the molar concentration of DAG was maintained at 0.1 mm. The enzyme exhibited positive cooperative kinetics with respect to the surface concentration of DAG and reached maximum activity at a surface concentration of 9.1 mol % (e.g. 10:1 molar ratio of Triton X-100 to DAG) (Fig. 5B). Analysis of the kinetic data according to the Hill equation yielded a Hill number of 2.5 and an apparent Km for DAG of 6.5 mol %. Because DAG is not soluble and the enzyme exhibited surface dilution kinetics, the concentration of DAG was expressed as a surface concentration (i.e. mole percent).
FIGURE 5.
Dependence of CTP-dependent DAG kinase activity on the molar and surface concentrations of DAG. DAG kinase activity was measured as a function of the indicated molar concentrations of DAG (A) and as a function of the indicated surface concentrations of DAG (B). The amounts of 32P-labeled PA produced were quantified by scintillation counting. For the experiment shown in A, the molar ratio of Triton X-100 to DAG was maintained at 10:1. For the experiment shown in B, the molar concentration of DAG was held constant at 0.1 mm, and the Triton X-100 concentration was varied to obtain the indicated surface concentrations. The concentration of CTP in both experiments was maintained at 1 mm. The data shown in A and B are means ± S.D. from triplicate enzyme determinations.
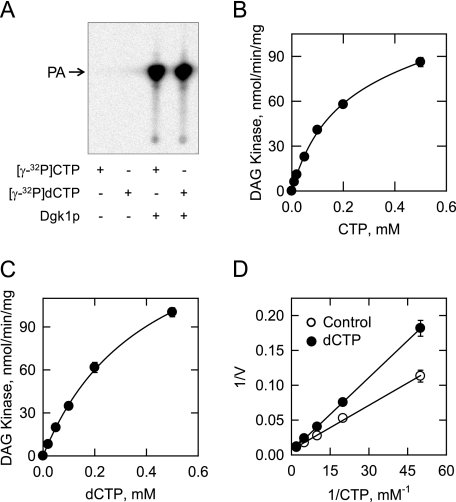
Dependence of CTP-dependent DAG Kinase Activity on the Concentrations of CTP and dCTP and Inhibition of Activity by dCTP—The dependence of DAG kinase activity on CTP was examined using a saturating surface concentration of DAG. In contrast to the cooperative kinetic pattern exhibited by DAG kinase with respect to DAG, the enzyme followed saturation kinetics with respect to CTP (Fig. 6B). Analysis of the data according to the Michaelis-Menten equation yielded an apparent Km for CTP of 0.3 mm. We questioned whether DAG kinase would utilize dCTP as a substrate. Indeed, the enzyme catalyzed the formation of PA from DAG and [γ-32P]dCTP (Fig. 6A). DAG kinase activity followed saturation kinetics with respect to dCTP (apparent Km = 0.4 mm) (Fig. 6C). In addition, dCTP inhibited the CTP-dependent DAG kinase activity of the enzyme. The addition of 0.5 mm dCTP to the assay system for CTP-dependent DAG kinase activity did not affect the apparent Vmax of the reaction but instead caused an increase in the Km for CTP (Fig. 6D). This result was consistent with the conclusion that dCTP was a competitive inhibitor with respect to CTP. The analysis of the data using the EZ-FIT computer program yielded an apparent Ki for dCTP of 0.4 mm.
FIGURE 6.
Dependence of CTP-dependent DAG kinase activity on the concentrations of CTP and dCTP and the inhibition of activity by dCTP. DAG kinase activity was measured with either 1 mm CTP or dCTP (A). Following the reactions, the chloroform-soluble 32P-labeled products were analyzed by TLC and visualized by phosphorimaging. The position of the PA standard is indicated. DAG kinase activity was measured as a function of the indicated concentrations of CTP (B), the indicated concentrations of dCTP (C), and the indicated concentrations of CTP in the absence and presence of 0.5 mm dCTP (D). The surface concentration of DAG was maintained at 9.1 mol % (molar concentrations of Triton X-100 and DAG of 1 and 0.1 mm, respectively). For the experiments shown in B-D, the amounts of 32P-labeled PA produced were quantified by scintillation counting, and the data shown are means ± S.D. from triplicate enzyme determinations.
Effects of Lipids on CTP-dependent DAG Kinase Activity—DAG kinase activity was assayed in the presence of various lipids (Table 3). A surface concentration of DAG (4.5 mol %) below its Km was used, so we could simultaneously observe stimulatory or inhibitory effects of lipids on activity. DAG kinase activity was stimulated by phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, phosphatidylserine, phosphatidylglycerol, and PA by 48-134%. DAG pyrophosphate, cardiolipin, CDP-DAG, and lyso-PA inhibited activity by 23-66%. In particular, DAG kinase activity was sensitive to inhibition (77-90%) by the sphingoid bases sphinganine, sphingosine, and phytosphingosine.
TABLE 3.
Effect of lipids on CTP-dependent DAG kinase activity
CTP-dependent DAG kinase activity was measured with a subsaturating concentration (4.5 mol %) of DAG in the presence of the indicated lipids (4.5 mol %). The specific activity in the control reaction was 60 ± 2.5 units/mg.
| Lipid | Relative DAG kinase activity |
|---|---|
| % | |
| Control | 100 |
| +Phosphatidylcholine | 217 |
| +Phosphatidylethanolamine | 165 |
| +Phosphatidylinositol | 234 |
| +Phosphatidylserine | 207 |
| +Phosphatidylglycerol | 193 |
| +PA | 148 |
| +DAG pyrophosphate | 77 |
| +Cardiolipin | 35 |
| +CDP-DAG | 34 |
| +Lyso-PA | 74 |
| +Monoacylglycerol | 97 |
| +Triacylglycerol | 87 |
| +Dolichol | 109 |
| +Sphinganine | 23 |
| +Sphingosine | 10 |
| +Phytosphingosine | 11 |
| +Ceramide | 87 |
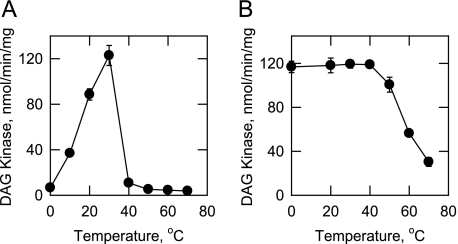
Effect of Temperature on the Activity and Stability of CTP-dependent DAG Kinase—DAG kinase activity was measured at temperatures from 0 to 70 °C in a temperature-controlled water bath (Fig. 7A). Maximum activity was observed at 30 °C. The enzyme was not active at temperatures above 40 °C. The activation energy for the reaction, as calculated from an Arrhenius plot of the data, was 16 kcal/mol. The DAG kinase was examined for its temperature stability by incubating the enzyme for 20 min at various temperatures (Fig. 7B). Activity was 100% stable from 0 to 40 °C, but was unstable at higher temperatures. About 70% of the activity was lost at 70 °C. The enzyme preparation was completely stable for at least 3 months of storage at -80 °C, and it was stable to several cycles of freezing and thawing.
FIGURE 7.
Effect of temperature on the activity and stability of CTP-dependent DAG kinase. A, DAG kinase activity was measured at the indicated temperatures in a temperature-controlled water bath. B, enzyme samples were preincubated at the indicated temperatures for 20 min. After incubation, the samples were cooled in an ice bath for 10 min to allow for renaturation, and DAG kinase activity was measured at 30 °C. The data shown are means ± S.D. from triplicate enzyme determinations.
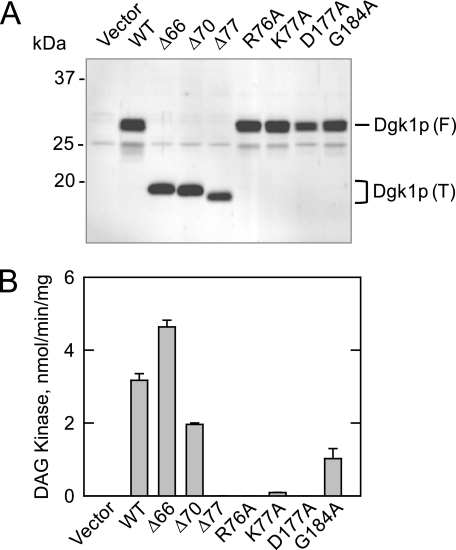
Effects of Truncation and Point Mutations in Dgk1p on CTP-dependent DAG Kinase Activity—Several truncation and point mutations were constructed for the DGK1-encoded enzyme. The Δ66 and Δ70 truncations removed most of the N-terminal hydrophilic region of Dgk1p, whereas the Δ77 truncation removed the entire N-terminal hydrophilic region plus the first two residues contained within the CTP transferase domain (Fig. 2A). The R76A, D177A, and G184A mutations were made at conserved amino acid residues that are found in the CTP transferase domains of Cds1p and Sec59p. K77A is a mutation of a conserved residue found in other fungal homologs of Dgk1p. The mutant alleles were overexpressed in dgk1Δ mutant cells using galactose-inducible YCplac111-GAL1/10-DGK1 (Table 1). The expression of the wild-type and mutant forms of Dgk1p was confirmed by immunoblot analysis using anti-Dgk1p antibodies (Fig. 8A). Although the predicted size of wild-type Dgk1p is 32.8 kDa, the protein migrated on SDS-polyacrylamide gels with a molecular mass that was ∼4 kDa lower than the predicted size of the protein (Fig. 8A). Similarly, the truncation mutant proteins migrated faster than their predicted molecular masses (Fig. 8A). The reason for this observation was unclear. There was no evidence of proteolytic degradation of the full-length and truncated proteins.
FIGURE 8.
Immunoblot analysis and CTP-dependent DAG kinase activities of wild-type and mutant forms of Dgk1p. dgk1Δ mutant cells bearing the indicated wild-type (WT) and mutant galactose-inducible DGK1 alleles on the YCplac111 plasmid (see Table 1) were grown to exponential phase in synthetic medium with 2% raffinose as the carbon source. The expression of the DGK1 alleles was induced by the addition of 2% galactose to the growth medium. The cells were then incubated for 24 h, followed by the preparation of cell extracts and the membrane fraction. A, 10 μg of each membrane fraction was subjected to immunoblot analysis using anti-Dgk1p antibodies (1 μg/ml). The positions of the full-length (F) and truncated (T) forms of Dgk1p are indicated. The blot shown is representative of two independent experiments. B, each cell extract was used for the measurement of DAG kinase activity. The data shown are means ± S.D. from triplicate enzyme determinations from two independent experiments.
The effects of the truncation and point mutations on CTP-dependent DAG kinase activity are presented in Fig. 8B. Deletion of the first 66 amino acid residues of Dgk1p resulted in a 45% increase in CTP-dependent DAG kinase activity, whereas the deletion of the first 70 residues resulted in a 40% decrease in activity. When the truncation (Δ77) included the first conserved amino acid residue (i.e. Arg76) within the CTP transferase domain, the DAG kinase activity of Dgk1p was at the lower limit of detection. DAG kinase activity was also lost with the R76A and K77A mutations and, as described previously (28), with the D177A mutation. The conserved G184A mutation caused a 70% reduction in DAG kinase activity.
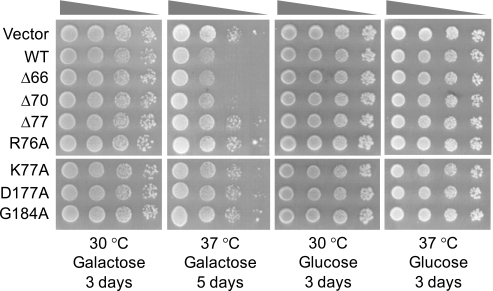
Temperature Sensitivity of dgk1Δ Cells Overexpressing DGK1 and Its Mutant Alleles—Previous studies have shown that loss-of-function mutations in PAH1-encoded PA phosphatase result in a temperature-sensitive phenotype (22, 26). Because the PA phosphatase reaction consumes PA and the DAG kinase reaction produces PA, we questioned whether the overexpression of DGK1 would cause cells to be temperature-sensitive for growth similar to the lack of PAH1 expression. Serial dilutions of cells bearing the wild-type and mutant DGK1 alleles were incubated on galactose-containing agar plates at 30 and 37 °C. Cells that overexpressed the wild-type and truncation mutants that possessed DAG kinase activity (Fig. 8B) were sensitive for growth at 37 °C (Fig. 9). However, the truncation and point mutants that lacked or had low DAG kinase activity (Fig. 8B) were not temperature-sensitive for growth (Fig. 9). Cells that were not induced (glucose-grown cells) for the overexpression of DAG kinase activity did not exhibit the temperature-sensitive phenotype (Fig. 9). These data supported the conclusion that the DAG kinase and PA phosphatase enzymes play reciprocal roles in physiology that impact on the temperature-sensitive phenotype.
FIGURE 9.
Temperature sensitivity of dgk1Δ cells overexpressing DGK1 and its mutant alleles. dgk1Δ mutant cells bearing the indicated wild-type (WT) and mutant galactose-inducible DGK1 alleles on the YCplac111 plasmid (see Table 1) were grown to exponential phase in synthetic medium with 2% raffinose as the carbon source. The expression of the DGK1 alleles was induced for 24 h by the addition of 2% galactose to the growth medium. After adjustment of the culture to the density of 2 × 107 cells/ml followed by 10-fold serial dilutions to 2 × 104 cells/ml, 10 μl of each diluted culture was spotted onto agar plates containing 2% galactose or glucose. Cells on the galactose medium were incubated for 3 days at 30 °C and 5 days at 37 °C, whereas cells on the glucose medium were incubated for 3 days at 30 °C and at 37 °C.
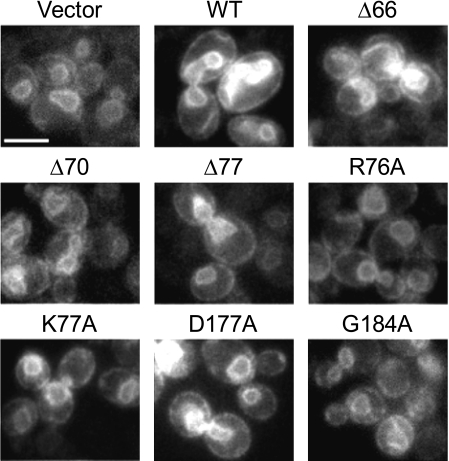
Effects of the Overexpression of DGK1 and Its Mutant Alleles on Nuclear/ER Membrane Growth—We examined the nuclear/ER membrane structure in cells that overexpressed the Dgk1p truncation and point mutants. The nuclear/ER membrane was visualized by fluorescence microscopy of the SEC63-GFP fusion protein that was coexpressed with wild-type and mutant alleles of DGK1. As described previously (28), the overexpression of wild-type Dgk1p resulted in the membrane expansion phenotype, whereas cells that expressed the D177A mutant protein lacking DAG kinase activity did not exhibit this phenotype (Fig. 10). Moreover, cells that overexpressed other Dgk1p mutants (e.g. Δ77, R76A, K77A, and G184A) with low or no DAG kinase activity had regularly shaped nuclear/ER membrane structures, whereas cells that overexpressed Dgk1p mutants (e.g. Δ66 and Δ70) with activity that was comparable with the wild-type control showed the membrane expansion phenotype (Fig. 10).
FIGURE 10.
Effects of the overexpression of DGK1 and its mutant alleles on nuclear/ER membrane growth. Wild-type cells (strain RS453) expressing the indicated wild-type (WT) and mutant galactose-inducible DGK1 alleles on the YCplac111 plasmid (see Table 1) and YCplac33-SEC63-GFP (to label the nuclear/ER membrane) were grown to exponential phase in galactose-containing medium. Cells were viewed with a Zeiss Axiophot fluorescence microscope equipped with a CCD camera, and images were recorded using IP Labs software. Scale bar = 5 μm.
DISCUSSION
The S. cerevisiae DGK1-encoded DAG kinase enzyme differs from other DAG kinase enzymes that exist in bacteria, plants, and animals (7-10, 29) in that the yeast enzyme utilizes CTP, not ATP, as the phosphate donor in the reaction. Yeast Dgk1p does not possess the signature catalytic domain with its ATP-binding site that is essential for the activity of ATP-dependent DAG kinases found in other organisms (9, 10, 29). This fact provides the explanation why an ATP-dependent DAG kinase activity or a putative gene encoding a DAG kinase enzyme has never been identified in S. cerevisiae. An earlier report by Szkopinska et al. (45) showed that yeast membranes contain an activity that catalyzes the transfer of the γ-phosphate of CTP into both dolichol phosphate and PA. It has been unclear whether these reactions are catalyzed by the same or separate enzymes. The identification of SEC59 as the structural gene encoding the dolichol kinase enzyme confirmed that separate enzymes are responsible for the CTP-dependent formation of dolichol phosphate and PA in yeast membranes (48, 51). The identity of the second enzyme that is responsible for the CTP-dependent formation of PA has been elusive until now (28). The CTP-dependent DAG kinase had a pH optimum at 7.0-7.5, required Ca2+ or Mg2+ ions for activity, and was inhibited by N-ethylmaleimide. Like most lipid-dependent enzymes that utilize membrane-associated substrates (62), the CTP-dependent DAG kinase enzyme followed surface dilution kinetics using a Triton X-100/DAG-mixed micellar substrate. These basic enzymological properties provide the foundation for further studies on this novel DAG kinase enzyme from yeast.
Most of Dgk1p is composed of the CTP transferase domain (Fig. 2A) that is also found in the S. cerevisiae SEC59-encoded dolichol kinase (48) and the CDS1-encoded CDP-DAG synthase (47) enzymes. The hydrophilic N-terminal region of Dgk1p is not essential for DAG kinase activity because the N-terminally truncated protein exhibited a sufficient level of DAG kinase activity to be functional in vivo. The primary structures of the mammalian, fly, worm, and plant ATP-dependent DAG kinase enzymes (9, 10, 29) are larger and more complex compared with the yeast enzyme. In addition to their catalytic domain, the ATP-dependent DAG kinases possess various regulatory and localization domains that are responsible for their functions as signaling enzymes (9, 10, 29). It is interesting that a Dgk1p mutant lacking the first 66 amino acid residues exhibited a higher level of DAG kinase activity compared with the wild-type protein. This suggests that the N-terminal region may contain regulatory sequences. The conserved amino acid residues within Dgk1p were essential for CTP-dependent DAG kinase activity. A previous study on the human dolichol kinase indicates that Asp177 and Gly184 are conserved residues that are important for CTP binding (88). Phenotypes (e.g. temperature sensitivity and nuclear/ER membrane expansion) associated with the overexpression of the DGK1 gene were specifically due to CTP-dependent DAG kinase activity and not simply the result of elevated levels of Dgk1p. These studies, along with previous studies on the PAH1-encoded Mg2+-dependent PA phosphatase enzyme (22, 25-27), support the importance of PA in the regulation of phospholipid synthesis and cell physiology in S. cerevisiae.
CTP plays a prominent role in the synthesis of phospholipids (Fig. 1) and in the regulation of their synthesis in S. cerevisiae (2, 63, 64). CTP is the direct precursor of the activated energy-rich phospholipid pathway intermediates CDP-DAG (65), CDP-choline (66), and CDP-ethanolamine (66) (Fig. 1). CDP-DAG is the source of the phosphatidyl moiety of the major phospholipids phosphatidylinositol, phosphatidylserine, phosphatidylethanolamine, and phosphatidylcholine as well as phosphatidylglycerol and cardiolipin (not shown in Fig. 1). The phosphatidylinositol derived from CDP-DAG also serves as the precursor for sphingolipids (67, 68), the D3-, D4-, and D5-phosphoinositides (64, 69-72), and glycosylphosphatidylinositol anchors (73, 74). CDP-choline and CDP-ethanolamine are the sources of the hydrophilic headgroups of phosphatidylcholine and phosphatidylethanolamine that are synthesized by way of DAG (Fig. 1). That the DGK1-encoded enzyme uses CTP as its substrate further emphasizes the importance of CTP in the synthesis of phospholipids in S. cerevisiae.
The cellular levels of CTP are primarily controlled through the biochemical regulation of CTP synthetase by CTP product inhibition (75, 76). CTP is synthesized from UTP via the URA7- and URA8-encoded CTP synthetase enzymes (77, 78). Cells that carry E161K mutations in the URA7- and URA8-encoded CTP synthetase enzymes, which no longer respond to CTP product inhibition, accumulate elevated (6-15-fold) levels of CTP compared with cells that express the wild-type enzyme (79). The elevation in cellular CTP that is brought about by the mutation in the URA7-encoded enzyme leads to an increased rate of PA synthesis and the aberrant regulation of phospholipid synthesis (79). This regulation is typified by the derepression of the INO1 gene and the inositol excretion phenotype (79). Although the basis for this CTP-mediated regulation has been unclear (79), we speculate that the stimulation of DGK1-encoded DAG kinase activity by the elevated levels of CTP is involved with this regulation.
DGK1-encoded CTP-dependent DAG kinase also utilized dCTP as a phosphate donor. Based on its Km and Ki values as both a substrate and competitive inhibitor, respectively, dCTP was as good a substrate as CTP. The utilization of the deoxy derivative of CTP as a substrate is known for several phospholipid biosynthetic enzymes, including CDP-DAG synthase (50), phosphocholine cytidylyltransferase (44, 66, 80, 81), and phosphoethanolamine cytidylyltransferase (66, 80, 81). In addition, the phosphatidylserine synthase from S. cerevisiae utilizes dCDP-DAG as a substrate to synthesize phosphatidylserine (57). Although the deoxyribonucleotide phospholipid pathway intermediates have been identified in cells, no clear function for their existence has been uncovered (82). For the DGK1-encoded DAG kinase, utilization of dCTP may simply indicate that the enzyme lacks specificity between CTP and dCTP. However, this lack of specificity provides an advantage for cells to utilize dCTP when CTP is limiting.
The yeast DAG kinase was stable for prolonged storage when the enzyme was associated with its native membrane environment. However, the solubilization and purification of Dgk1p led to a great loss (∼30-fold) of its enzyme activity (data not shown). This may be explained by the loss of a specific membrane component that is required for enzyme activity. The membrane-associated E. coli ATP-dependent DAG kinase enzyme is also labile upon solubilization and purification (83). In fact, this membrane-associated enzyme requires phospholipid activators to elicit a maximum turnover number in vitro (84, 85). The requirement of a phospholipid activator for the yeast membrane-associated enzyme was indicated by the stimulation of its activity by major membrane phospholipids (e.g. phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylserine). Additional studies are planned to determine whether the reconstitution of the purified enzyme with phospholipids activates the purified enzyme. Maintaining activity during the purification of mammalian forms of ATP-dependent DAG kinases has not been a critical issue presumably because these enzymes are not integral membrane proteins (9, 10, 29). Instead, the mammalian DAG kinases are cytosolic enzymes that translocate to the membrane to catalyze their reactions (9, 10, 29).
CDP-DAG was among the phospholipids that inhibited CTP-dependent DAG kinase activity. CDP-DAG is the product of the CTP-dependent CDP-DAG synthase enzyme that utilizes PA as its phospholipid substrate (Fig. 1). The inhibition of DAG kinase activity by CDP-DAG might favor the accumulation of DAG that may be used for the synthesis of phosphatidylethanolamine and phosphatidylcholine via DAG or favor the synthesis of TAG (Fig. 1). Alternatively, the accumulation of DAG may regulate some other DAG-mediated function (e.g. signaling through protein kinase C or membrane structure) in S. cerevisiae. It is also interesting that DAG kinase activity was inhibited by sphingoid bases (e.g. sphinganine and phytosphingosine). Sphingoid bases are both precursors and turnover products of sphingolipid metabolism in S. cerevisiae (68). A relationship between the DAG-derived synthesis of phosphatidylethanolamine and sphingolipid synthesis exists through sphingoid base metabolism (2). In addition, DAG is a product of the inositol phosphoceramide synthase enzyme that is responsible for the synthesis of a major yeast sphingolipid inositol phosphoceramide (68). The inhibition of DAG kinase activity by sphingoid bases might favor the utilization of DAG for other cellular functions as discussed above.
In mammalian cells, the function of the ATP-dependent DAG kinases has been attributed to the attenuation of the signaling roles of DAG (e.g. activation of protein kinase C). Moreover, mammalian DAG kinase enzymes appear to be involved in numerous physiological processes (10). As far as we know, there is only one form of CTP-dependent DAG kinase activity in S. cerevisiae. The dgk1Δ mutant lacks detectable DAG kinase activity (28). Current data indicate that the yeast DGK1-encoded DAG kinase functions in concert with the PAH1-encoded PA phosphatase to control the nuclear/ER levels of PA for regulation of phospholipid synthesis and membrane growth (28).
Supplementary Material
Acknowledgments
We thank Avula Sreenivas and Yu-Fang Chang for help during the initial phase of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-28140 (to G. M. C.) from the United States Public Health Service. This work was also supported by a Wellcome Trust Career Development Fellowship in Basic Biomedical Science (to S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: PA, phosphatidate; DAG, diacylglycerol; ER, endoplasmic reticulum; UASINO, upstream activating sequence inositol-responsive element; MOPS, 4-morpholinepropanesulfonic acid; GFP, green fluorescent protein.
DGK1 was formally known as HSD1. It was originally identified as a high copy number suppressor of the sly1 temperature-sensitive mutation in ER-Golgi vesicular transport (86).
References
- 1.Carman, G. M., and Henry, S. A. (2007) J. Biol. Chem. 282 37293-37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carman, G. M., and Henry, S. A. (1999) Prog. Lipid Res. 38 361-399 [DOI] [PubMed] [Google Scholar]
- 3.Henry, S. A., and Patton-Vogt, J. L. (1998) Prog. Nucleic Acid Res. 61 133-179 [DOI] [PubMed] [Google Scholar]
- 4.Sorger, D., and Daum, G. (2003) Appl. Microbiol. Biotechnol. 61 289-299 [DOI] [PubMed] [Google Scholar]
- 5.Czabany, T., Athenstaedt, K., and Daum, G. (2007) Biochim. Biophys. Acta 1771 299-309 [DOI] [PubMed] [Google Scholar]
- 6.Li, G., Chen, S., Thompson, M. N., and Greenberg, M. L. (2007) Biochim. Biophys. Acta 1771 432-441 [DOI] [PubMed] [Google Scholar]
- 7.Raetz, C. R., and Dowhan, W. (1990) J. Biol. Chem. 265 1235-1238 [PubMed] [Google Scholar]
- 8.Testerink, C., and Munnik, T. (2005) Trends Plant Sci. 10 368-375 [DOI] [PubMed] [Google Scholar]
- 9.Sakane, F., Imai, S., Kai, M., Yasuda, S., and Kanoh, H. (2007) Biochim. Biophys. Acta 1771 793-806 [DOI] [PubMed] [Google Scholar]
- 10.Merida, I., Vila-Flores, A., and Merino, E. (2008) Biochem. J. 409 1-18 [DOI] [PubMed] [Google Scholar]
- 11.Loewen, C. J. R., Gaspar, M. L., Jesch, S. A., Delon, C., Ktistakis, N. T., Henry, S. A., and Levine, T. P. (2004) Science 304 1644-1647 [DOI] [PubMed] [Google Scholar]
- 12.Loewen, C. J. R., and Levine, T. P. (2005) J. Biol. Chem. 280 14097-14104 [DOI] [PubMed] [Google Scholar]
- 13.Hirsch, J. P., and Henry, S. A. (1986) Mol. Cell. Biol. 6 3320-3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachhawat, N., Ouyang, Q., and Henry, S. A. (1995) J. Biol. Chem. 270 25087-25095 [DOI] [PubMed] [Google Scholar]
- 15.Loewy, B. S., and Henry, S. A. (1984) Mol. Cell. Biol. 4 2479-2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg, M. L., and Lopes, J. M. (1996) Microbiol. Rev. 60 1-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwank, S., Ebbert, R., Rautenstrauss, K., Schweizer, E., and Schuller, H. J. (1995) Nucleic Acids Res. 23 230-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, M., Hancock, L. C., and Lopes, J. M. (2007) Biochim. Biophys. Acta 1771 310-321 [DOI] [PubMed] [Google Scholar]
- 19.Donahue, T. F., and Henry, S. A. (1981) J. Biol. Chem. 256 7077-7085 [PubMed] [Google Scholar]
- 20.Donahue, T. F., and Henry, S. A. (1981) Genetics 98 491-503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenberg, M., Reiner, B., and Henry, S. A. (1982) Genetics 100 19-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han, G.-S., Wu, W.-I., and Carman, G. M. (2006) J. Biol. Chem. 281 9210-9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, Y.-P., and Carman, G. M. (1989) J. Biol. Chem. 264 8641-8645 [PubMed] [Google Scholar]
- 24.Smith, S. W., Weiss, S. B., and Kennedy, E. P. (1957) J. Biol. Chem. 228 915-922 [PubMed] [Google Scholar]
- 25.Santos-Rosa, H., Leung, J., Grimsey, N., Peak-Chew, S., and Siniossoglou, S. (2005) EMBO J. 24 1931-1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, G.-S., Siniossoglou, S., and Carman, G. M. (2007) J. Biol. Chem. 282 37026-37035 [DOI] [PubMed] [Google Scholar]
- 27.O'Hara, L., Han, G.-S., Peak-Chew, S., Grimsey, N., Carman, G. M., and Siniossoglou, S. (2006) J. Biol. Chem. 281 34537-34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han, G.-S., O'Hara, L., Carman, G. M., and Siniossoglou, S. (2008) J. Biol. Chem. 283 20433-20442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topham, M. K., and Prescott, S. M. (1999) J. Biol. Chem. 274 11447-11450 [DOI] [PubMed] [Google Scholar]
- 30.Rose, M. D., Winston, F., and Heiter, P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 31.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 32.Ito, H., Yasuki, F., Murata, K., and Kimura, A. (1983) J. Bacteriol. 153 163-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiestl, R. H., and Gietz, R. D. (1989) Curr. Genet. 16 339-346 [DOI] [PubMed] [Google Scholar]
- 34.Gietz, R. D., and Sugino, A. (1988) Gene (Amst.) 74 527-534 [DOI] [PubMed] [Google Scholar]
- 35.Harlow, E., and Lane, D. (1999) Using Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 36.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 37.Haid, A., and Suissa, M. (1983) Methods Enzymol. 96 192-205 [DOI] [PubMed] [Google Scholar]
- 38.Klig, L. S., Homann, M. J., Carman, G. M., and Henry, S. A. (1985) J. Bacteriol. 162 1135-1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 40.Muller, M. O., Meylan-Bettex, M., Eckstein, F., Martinoia, E., Siegenthaler, P. A., and Bovet, L. (2000) J. Biol. Chem. 275 19475-19481 [DOI] [PubMed] [Google Scholar]
- 41.Carman, G. M., and Lin, Y.-P. (1991) Methods Enzymol. 197 548-553 [DOI] [PubMed] [Google Scholar]
- 42.Carman, G. M., and Fischl, A. S. (1992) Methods Enzymol. 209 305-312 [DOI] [PubMed] [Google Scholar]
- 43.Carman, G. M., and Kelley, M. J. (1992) Methods Enzymol. 209 242-247 [DOI] [PubMed] [Google Scholar]
- 44.Nikawa, J., Yonemura, K., and Yamashita, S. (1983) Eur. J. Biochem. 131 223-229 [DOI] [PubMed] [Google Scholar]
- 45.Szkopinska, A., Nowak, L., Swiezewska, E., and Palamarczyk, G. (1988) Arch. Biochem. Biophys. 266 124-131 [DOI] [PubMed] [Google Scholar]
- 46.Perrella, F. (1988) Anal. Biochem. 174 437-447 [DOI] [PubMed] [Google Scholar]
- 47.Shen, H., Heacock, P. N., Clancey, C. J., and Dowhan, W. (1996) J. Biol. Chem. 271 789-795 [DOI] [PubMed] [Google Scholar]
- 48.Heller, L., Orlean, P., and Adair, W. L., Jr. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 7013-7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belendiuk, G., Mangnall, D., Tung, B., Westley, J., and Getz, G. S. (1978) J. Biol. Chem. 253 4555-4565 [PubMed] [Google Scholar]
- 50.Kelley, M. J., and Carman, G. M. (1987) J. Biol. Chem. 262 14563-14570 [PubMed] [Google Scholar]
- 51.Fernandez, F., Shridas, P., Jiang, S., Aebi, M., and Waechter, C. J. (2002) Glycobiology 12 555-562 [DOI] [PubMed] [Google Scholar]
- 52.Tsukagoshi, Y., Nikawa, J., and Yamashita, S. (1987) Eur. J. Biochem. 169 477-486 [DOI] [PubMed] [Google Scholar]
- 53.Min-Seok, R., Kawamata, Y., Nakamura, H., Ohta, A., and Takagi, M. (1996) J. Biochem. (Tokyo) 120 1040-1047 [DOI] [PubMed] [Google Scholar]
- 54.Iwanyshyn, W. M., Han, G.-S., and Carman, G. M. (2004) J. Biol. Chem. 279 21976-21983 [DOI] [PubMed] [Google Scholar]
- 55.Homann, M. J., Poole, M. A., Gaynor, P. M., Ho, C.-T., and Carman, G. M. (1987) J. Bacteriol. 169 533-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carman, G. M., and Belunis, C. J. (1983) Can. J. Microbiol. 29 1452-1457 [DOI] [PubMed] [Google Scholar]
- 57.Bae-Lee, M., and Carman, G. M. (1984) J. Biol. Chem. 259 10857-10862 [PubMed] [Google Scholar]
- 58.Fischl, A. S., and Carman, G. M. (1983) J. Bacteriol. 154 304-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Chaffoy de Courcelles, D. C., Roevens, P., and Van Belle, H. (1985) J. Biol. Chem. 260 15762-15770 [PubMed] [Google Scholar]
- 60.Rodriguez-Linares, B., Walker, T., and Watson, S. (1991) Biochem. Pharmacol. 41 835-838 [DOI] [PubMed] [Google Scholar]
- 61.Dawson, R. M. C., Elliott, D. C., Elliott, W. H., and Jones, K. M. (1986) Data for Biochemical Research, pp. 384-397, Oxford University Press, Oxford
- 62.Carman, G. M., Deems, R. A., and Dennis, E. A. (1995) J. Biol. Chem. 270 18711-18714 [DOI] [PubMed] [Google Scholar]
- 63.Carman, G. M., and Henry, S. A. (1989) Annu. Rev. Biochem. 58 635-669 [DOI] [PubMed] [Google Scholar]
- 64.Paltauf, F., Kohlwein, S. D., and Henry, S. A. (1992) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression (Jones, E. W., Pringle, J. R., and Broach, J. R., eds) pp. 415-500, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 65.Carter, J. R., and Kennedy, E. P. (1966) J. Lipid Res. 7 678-683 [PubMed] [Google Scholar]
- 66.Kennedy, E. P., and Weiss, S. B. (1956) J. Biol. Chem. 222 193-214 [PubMed] [Google Scholar]
- 67.Dickson, R. C. (1998) Annu. Rev. Biochem. 67 27-48 [DOI] [PubMed] [Google Scholar]
- 68.Dickson, R. C., and Lester, R. L. (2002) Biochim. Biophys. Acta 1583 13-25 [DOI] [PubMed] [Google Scholar]
- 69.Fruman, D. A., Meyers, R. E., and Cantley, L. C. (1998) Annu. Rev. Biochem. 67 481-507 [DOI] [PubMed] [Google Scholar]
- 70.Balla, T. (1998) Biochim. Biophys. Acta 1436 69-85 [DOI] [PubMed] [Google Scholar]
- 71.Gehrmann, T., and Heilmayer, L. G., Jr. (1998) Eur. J. Biochem. 253 357-370 [DOI] [PubMed] [Google Scholar]
- 72.Odorizzi, G., Babst, M., and Emr, S. D. (2000) Trends Biochem. Sci. 25 229-235 [DOI] [PubMed] [Google Scholar]
- 73.Leidich, S. D., Drapp, D. A., and Orlean, P. (1994) J. Biol. Chem. 269 10193-10196 [PubMed] [Google Scholar]
- 74.Leidich, S. D., and Orlean, P. (1996) J. Biol. Chem. 271 27829-27837 [DOI] [PubMed] [Google Scholar]
- 75.Yang, W.-L., McDonough, V. M., Ozier-Kalogeropoulos, O., Adeline, M.-T., Flocco, M. T., and Carman, G. M. (1994) Biochemistry 33 10785-10793 [DOI] [PubMed] [Google Scholar]
- 76.Nadkarni, A. K., McDonough, V. M., Yang, W.-L., Stukey, J. E., Ozier-Kalogeropoulos, O., and Carman, G. M. (1995) J. Biol. Chem. 270 24982-24988 [DOI] [PubMed] [Google Scholar]
- 77.Ozier-Kalogeropoulos, O., Fasiolo, F., Adeline, M.-T., Collin, J., and Lacroute, F. (1991) Mol. Gen. Genet. 231 7-16 [DOI] [PubMed] [Google Scholar]
- 78.Ozier-Kalogeropoulos, O., Adeline, M.-T., Yang, W.-L., Carman, G. M., and Lacroute, F. (1994) Mol. Gen. Genet. 242 431-439 [DOI] [PubMed] [Google Scholar]
- 79.Ostrander, D. B., O'Brien, D. J., Gorman, J. A., and Carman, G. M. (1998) J. Biol. Chem. 273 18992-19001 [DOI] [PubMed] [Google Scholar]
- 80.Kennedy, E. P., Borkenhagen, L. F., and Smith, S. W. (1959) J. Biol. Chem. 234 1998-2000 [PubMed] [Google Scholar]
- 81.Spyrou, G., and Reichard, P. (1987) J. Biol. Chem. 262 16425-16432 [PubMed] [Google Scholar]
- 82.Kennedy, E. P. (1986) in Lipids and Membranes: Past, Present and Future (Op den Kamp, J. A. F., Roelofsen, B., and Wirtz, K. W. A., eds) pp. 171-206, Elsevier Science Publishers B.V., Amsterdam
- 83.Loomis, C. R., Walsh, J. P., and Bell, R. M. (1985) J. Biol. Chem. 260 4091-4097 [PubMed] [Google Scholar]
- 84.Walsh, J. P., and Bell, R. M. (1986) J. Biol. Chem. 261 6239-6247 [PubMed] [Google Scholar]
- 85.Walsh, J. P., and Bell, R. M. (1986) J. Biol. Chem. 261 15062-15069 [PubMed] [Google Scholar]
- 86.Kosodo, Y., Imai, K., Hirata, A., Noda, Y., Takatsuki, A., Adachi, H., and Yoda, K. (2001) Yeast 18 1003-1014 [DOI] [PubMed] [Google Scholar]
- 87.Wimmer, C., Doye, V., Grandi, P., Nehrbass, U., and Hurt, E. C. (1992) EMBO J. 11 5051-5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shridas, P., and Waechter, C. J. (2006) J. Biol. Chem. 281 31696-31704 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.