Abstract
During human immunodeficiency virus type 1 (HIV-1) assembly, the nucleocapsid (NC) and the PTAP motif in p6 of Gag play important roles in RNA encapsidation and virus release, respectively. We have previously demonstrated that functional complementation occurs between an NC mutant and a PTAP mutant to rescue viral replication. In this report, we examined the amounts of functional NC and PTAP motif that are required during virus replication. When NC and PTAP mutants were coexpressed at 5:1, 5:5, and 1:5 ratios, virus titers were rescued at 5%, 51%, and 86% of the wild-type level, respectively. These results indicate that HIV-1 requires a small amount of functional PTAP motif but far more functional NC to complete efficient replication. Further analyses reveal that RNA packaging can be significantly rescued in viruses containing a small amount of functional NC. However, most of the NC proteins must be functional to generate the wild-type level of R-U5 DNA product. Once the R-U5 product is generated, viruses containing half of the functional NC can complete reverse transcription and DNA integration at near-wild-type efficiency. These results define the quantitative requirements of NC and p6 during HIV-1 replication and provide insights into the requirement for the development of anti-HIV strategies using NC and p6 as targets.
Keywords: HIV, Nucleocapsid, PTAP late domain, complementation, replication
Introduction
The major structural proteins of retroviruses are encoded in the gag gene (Goff, 2007). The gene product, Gag, is first translated as a polyprotein; in most retroviruses, viral protease (PR) cleaves the polyprotein to yield the mature proteins during or soon after virus assembly (Swanstrom, 1997). Gag polyproteins contain matrix (MA), capsid (CA), and nucleocapsid (NC) domains. In some retroviruses, the Gag polyprotein has additional domains. For example, the Gag polyprotein of human immunodeficiency virus type 1 (HIV-1) has three additional regions, spacer peptide 1 (SP1 or p2), located between CA and NC; SP2 (or p1), located at the C terminus of NC; and p6, located at the C terminus of Gag (Freed, 2007).
The Gag polyprotein plays a central role in virus assembly. Expression of Gag in cells is sufficient to form virus-like particles (Bennett et al., 1991; Gheysen et al., 1989; Wills and Craven, 1991); similarly, purified Gag proteins can self-assemble under suitable conditions (Campbell and Rein, 1999; Campbell and Vogt, 1997). Although in most retroviruses the assembly of virus-like particles can take place without the involvement of other viral and cellular components, the assembly of an infectious virus is far more complex. In this process, Gag interacts with various viral components, such as viral RNA, to specifically encapsidate the viral genetic blueprint (Berkowitz, Fisher, and Goff, 1996), and other viral proteins, such as Gag-Pol and Env, to incorporate the viral proteins that are essential for replication steps (Freed, 1998; Freed and Martin, 1996; Lu et al., 1995; Murakami and Freed, 2000; Srinivasakumar, Hammarskjold, and Rekosh, 1995). Additionally, Gag interacts with the host cell machinery to allow proper protein trafficking and release of the virions (Garrus et al., 2001; Strack et al., 2003; VerPlank et al., 2001).
Different Gag domains have various functions. MA targets the Gag proteins to the cellular membrane. In HIV-1, myristoylation of Gag (Bryant and Ratner, 1990; Gottlinger, Sodroski, and Haseltine, 1989; Hill et al., 1996; Ono and Freed, 1999) and the interaction between MA and cellular factor AP-3 are thought to be important in this targeting (Dong et al., 2005). CA mediates Gag-Gag interactions, and mutations in CA often cause assembly defects (Craven et al., 1995; Mammano et al., 1994; Reicin et al., 1995; Swanstrom, 1997; von Schwedler et al., 2003). The NC domain is important in mediating interactions between Gag-Gag (Bennett, Nelle, and Wills, 1993; Bowzard et al., 1998; Derdowski, Ding, and Spearman, 2004; Ott et al., 2003; Sandefur et al., 2000; Sandefur, Varthakavi, and Spearman, 1998) and Gag-RNA (Berkowitz et al., 1995; Kaye and Lever, 1996; Meric and Spahr, 1986), and as the mature NC protein, also plays a role in events during the early phase of viral replication (Buckman, Bosche, and Gorelick, 2003; Thomas et al., 2006). The p6 domain of HIV-1 contains motifs that interact with viral protein Vpr (Lu et al., 1995) and the members of the human endosomal sorting pathways. It has been well documented that the PTAP motif in p6 interacts with host protein TSG101 (Garrus et al., 2001; Martin-Serrano, Zang, and Bieniasz, 2001; VerPlank et al., 2001). Interference of the Gag-TSG101 interaction, such as by mutating the PTAP motif or expressing dominant-negative forms of TSG101, can cause virion release defects (Demirov et al., 2002; Demirov, Orenstein, and Freed, 2002; Dettenhofer and Yu, 1999; Garrus et al., 2001; Huang et al., 1995; Martin-Serrano, Zang, and Bieniasz, 2001; Martin-Serrano, Zang, and Bieniasz, 2003; VerPlank et al., 2001). Additionally, sequences near the C terminus of p6 can interact with host factor ALIX/AIP1 (Chen et al., 2005; Fisher et al., 2007; Strack et al., 2003). These interactions allow Gag to utilize the host pathway for virion release.
NC proteins of most retroviruses contain two conserved features: a region(s) rich in basic residues, and one or two zinc-binding (zinc finger) motifs. Although the number of zinc finger motifs varies among retroviruses, they all have the conserved sequence CX2CX4HX2C (CCHC) (Berg, 1986; Gorelick et al., 1988). Destroying the conserved zinc finger motifs is detrimental to viral replication (Bowles, Damay, and Spahr, 1993; Gorelick et al., 1999a; Gorelick et al., 1993; Gorelick et al., 1988; Gorelick et al., 1990; Meric and Goff, 1989; Meric, Gouilloud, and Spahr, 1988). Even replacement of the CCHC motifs with another conserved zinc finger motif, such as CCCC or CCHH (Gorelick et al., 1996; Gorelick et al., 1999b), or another CCHC motif (Bowles, Damay, and Spahr, 1993; Gorelick et al., 1993; McGrath et al., 2003) can severely affect viral replication. Although relatively small in size, NC plays multiple roles in viral replication. In addition to its role in Gag-Gag interactions, the NC domain in the Gag polyprotein mediates specific recognition between Gag and viral RNA (Berkowitz et al., 1995; Certo et al., 1999; Kaye and Lever, 1998; Zhang and Barklis, 1995). Mutations in NC can lead to a drastic decrease of viral RNA encapsidation into virions (Bowles, Damay, and Spahr, 1993; Gorelick et al., 1999a; Gorelick et al., 1993; Gorelick et al., 1999b; Gorelick et al., 1988; Gorelick et al., 1990; Meric and Goff, 1989; Meric, Gouilloud, and Spahr, 1988). In vitro experiments indicated that the mature NC protein has a chaperone activity (Levin et al., 2005; Rein, Henderson, and Levin, 1998; Tsuchihashi and Brown, 1994), and can affect nucleic acid annealing, DNA synthesis and integration (Brule et al., 2002; Carteau, Gorelick, and Bushman, 1999; Cen et al., 2000; Heath et al., 2003; Lapadat-Tapolsky et al., 1997; Lener et al., 1998; Levin et al., 2005; Rodriguez-Rodriguez et al., 1995; Rong et al., 2001). In vivo studies indicate that mutations in NC can affect the efficiency of reverse transcription (Buckman, Bosche, and Gorelick, 2003; Gorelick et al., 1999b; Thomas et al., 2006) and frequency of reverse transcriptase (RT) template switching (Zhang et al., 2002). Additionally, NC plays a role during the integration of viral DNA to form a provirus; although the exact role and mechanism of how NC affects integration are unclear, it is thought that NC protects the ends of viral DNA, thereby allowing efficient integration to occur (Buckman, Bosche, and Gorelick, 2003; Thomas et al., 2006).
Retroviruses can interact with each other through several different mechanisms, one of which is complementation following pseudotyping. Pseudotyping refers to the generation of viral particles containing components from more than one virus and the use of these components for viral replication. A common form of pseudotyping is the use of Env proteins from different viruses. Pseudotyping can occur using other viral components as well; for example, Gag proteins from two viruses can coassemble into the same virus particles. Previously, using an NC mutant and a PTAP mutant, we have shown that these two HIV-1 Gag mutants can complement each other via coassembly and complete virus replication (Boyko et al., 2006). In the current study, we used this established system to address the quantitative requirements of functional NC and PTAP motif during HIV-1 assembly and replication. Viral preparations from NC and PTAP mutants coexpressed at different ratios were generated and analyzed. We conclude that to achieve the wild-type virus level of replication efficiency, HIV-1 requires most of its encapsidated NC proteins but only a small amount of PTAP motif. NC affects multiple steps of the infection process, blocking any of these steps can cause a decrease in viral titer. Additionally, different amounts of functional NC may be required at various replication steps. To determine the amounts of NC required, we analyzed viral products during various stages of the infection process. We conclude that HIV-1 requires less functional NC proteins during the RNA encapsidation assembly process than during the events leading to the completion of viral DNA synthesis. To our knowledge, this is the first quantitative study of the NC requirement during HIV-1 replication.
Results
Determination of the ratio of functional Gag proteins required for virus replication
We sought to delineate the amounts of functional NC and PTAP motif required for efficient HIV-1 replication. To approach this experimental question, we used two mutants that each has significant replication defects but at an equal molar ratio can complement each other’s function to complete the steps required for viral replication. To define the amounts of functional NC and PTAP motif required for efficient HIV-1 replication in a single round assay, we coexpressed these mutants at different molar ratios and examined the infectivity of the resulting viruses.
The general structures of the vectors used are shown in Fig. 1A. All of the vectors contained LTRs, cis-acting elements essential for virus replication, and they express gag-pol, tat, rev, and a marker gene. The CCHC/CCHC motifs of the NC mutant (NC*) were changed to CCHH/CCCC; this mutation compromises the specific encapsidation of the viral RNAs and causes a drastic reduction in the viral titers (Gorelick et al., 1999b). The PTAP motif of the p6 mutant (PTAP*) was altered to LIRL; this mutation leads to a defect in virion release and a severe decrease in viral titer (Boyko et al., 2006; Demirov, Orenstein, and Freed, 2002). The NC* and PTAP* vectors also express a mouse thy1.2 (thy) and a green fluorescence protein (gfp) gene, respectively. Vectors WTt and WTg have the same structures as NC* and PTAP*, respectively, except they express the wild-type gag-pol.
Fig. 1.
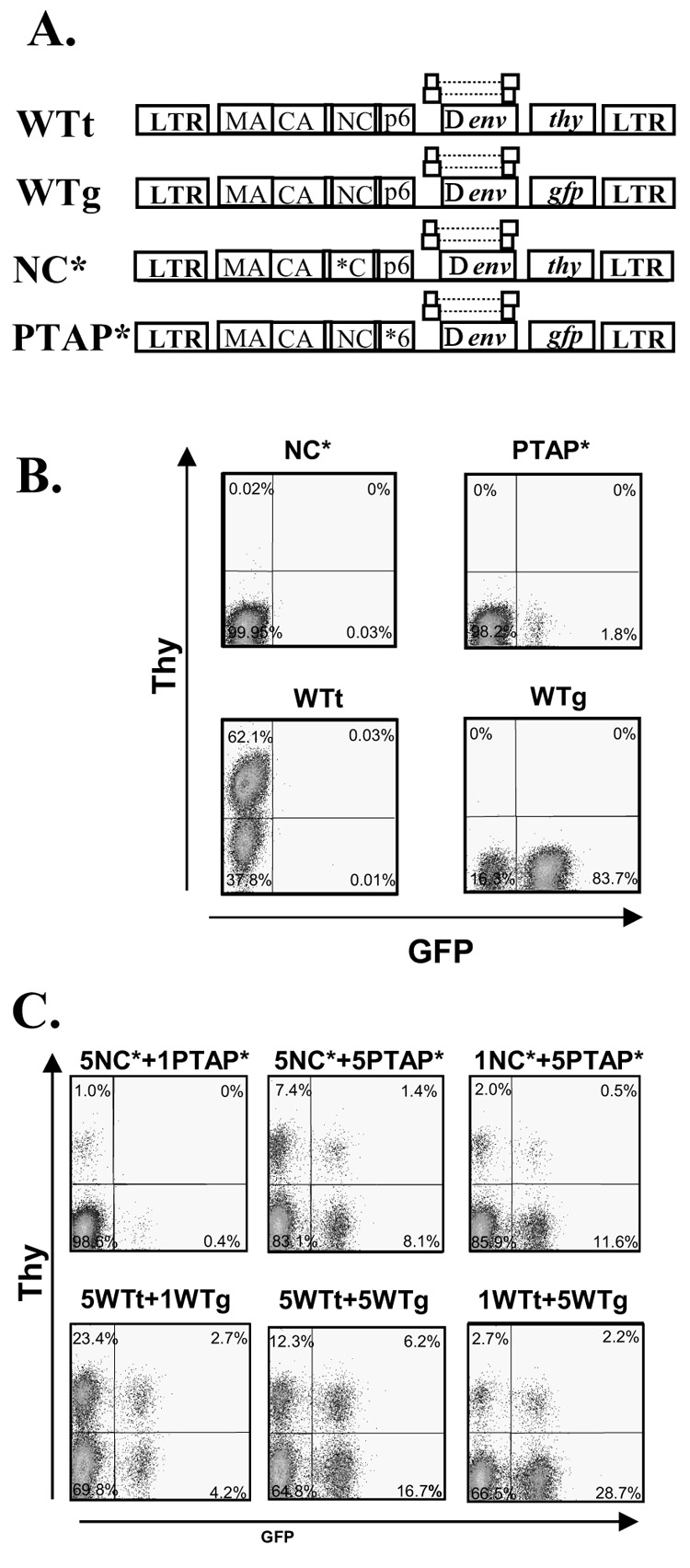
Complementation of two HIV-1 Gag mutants. (A) General structures of the HIV-1 vectors used for the complementation studies. All vectors are similar in their general structures but contain different mutations in gag (indicated by the asterisk) or marker genes. (B) Representative flow cytometry analyses of Hut78/CCR5 cells infected with wild-type or Gag mutant virus. (C) Representative flow cytometry analyses of Hut78/CCR5 cells infected with viruses generated by cotransfection of two mutants or two wild-type vectors at different ratios.
To measure the viral titers generated by these vectors, they were transfected into 293T cells along with a plasmid that expresses CCR5-tropic HIV-1 envelope; the resulting viruses were used to infect a T-cell line, Hut78/CCR5. Infected cells were analyzed by flow cytometry and viral titers were determined by the number of cells expressing either the GFP or Thy marker. As previously observed, compared with vectors carrying the wild-type gag-pol, the NC* and PTAP* mutants generated undetectable and drastically decreased viral titers, respectively (Fig. 1B). We then coexpressed the two mutants, NC* and PTAP*, by transfecting the two plasmids into 293T cells at 5:1, 5:5, and 1:5 molar ratios. As controls, the two wild-type vectors, WTt and WTg, were transfected into 293T cells at 5:1, 5:5, and 1:5 ratios. The resulting viruses were harvested and used to infect Hut78/CCR5 cells; representative flow cytometry analyses are shown in Fig. 1C. As expected, mixing the wild-type vectors at any of the three ratios resulted in the generation of infectious virus that infected large amounts of cells (Fig. 1C, lower panels). Consistent with our previous observation, at an equal molar ratio (5:5), the NC* and PTAP* mutants complemented each other and generated infectious viruses that infected a large number of target cells. Viruses generated from the 1:5 ratio of NC* and PTAP* mutants also infected a large portion of the target cells. However, viruses generated from the 5:1 ratio of NC* and PTAP* mutants, referred to as 5NC*+1PTAP*, infected only a small portion of the target cells (Fig. 1C, top left panel). The multiplicities of infection (MOIs) of these viral preparations were compared with those of the wild-type controls, and the relative viral titers generated from more than seven independent experiments are summarized in Fig. 2A.
Fig. 2.
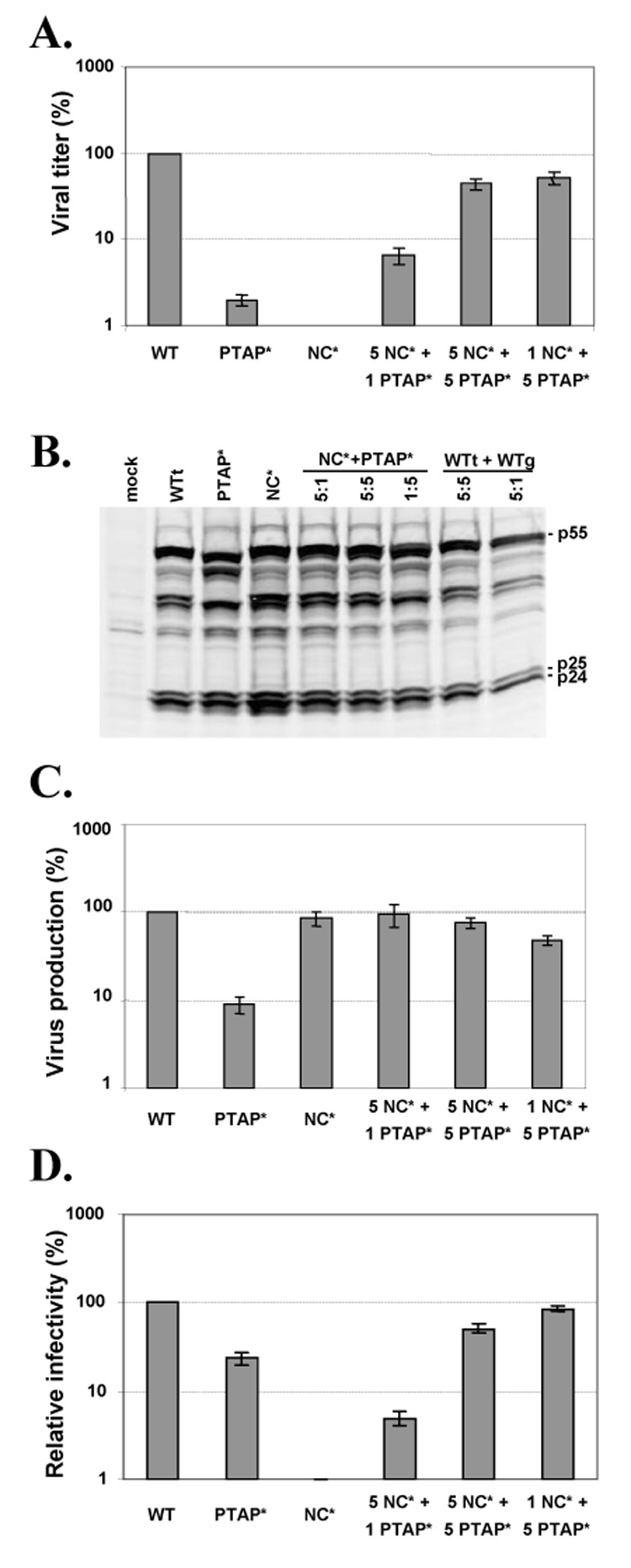
Virus production, infection titers, and relative infectivities of viruses containing wild-type or mutant Gag proteins. (A) Infection titers of wild-type or Gag mutant viruses. Infection was detected by the expression of the marker genes. The MOIs of the wild-type controls were set as 100%. (B) Western analysis of Gag expression in the transfected cells. (C) The level of virus produced from transfected cells determined by RT activity. RT activities of the wild-type controls were set as 100%. (D) Relative infectivity of wild-type or Gag mutant viruses. The MOI of each virus was normalized to the RT activity of the virus preparation, and the values for the wild-type controls were set as 100%. Results from more than seven independent experiments are summarized; error bars indicate standard errors.
To probe the mechanisms that caused the low titer generated by the 5NC*+1PTAP* virus, we examined Gag expression in transfected cells by Western analyses (Fig. 2B) and virion production in all of the samples by quantifying the amounts of and the RT activities (Fig. 2C). Compared with the wild-type controls, the 5NC*+1PTAP* samples have similar Gag expression and virion production. Virion production in these samples was also measured by the amounts of CA antigen (p24), which yielded similar conclusions (data not shown). Therefore, virus production was not the main cause for the low virus titers; instead, the 5NC*+1PTAP* viruses had lower infectivities compared with wild-type viruses. To quantify the relative infectivities of viruses, the MOIs of these viral preparations were normalized to the RT activities. The infectivity of each combination of the mutant viruses was compared with the infectivity of its corresponding wild-type control, which was defined as 100%. The relative infectivities of these viruses generated from at least seven independent experiments are summarized in Fig. 2D. The infectivities of the viruses generated from 5:1, 5:5, and 1:5 ratios of NC*:PTAP* were 5%, 51%, and 86% of the corresponding wild-type viruses. Therefore, a small portion of the functional PTAP motif is sufficient for rescuing virus replication, whereas a small amount of functional NC is not sufficient for rescuing virus replication.
Quantitative requirements of Gag polyprotein with a functional NC domain for encapsidation of viral RNA during the assembly process
To delineate the block(s) in the viral replication that caused the low infectivities of the 5NC*+1PTAP* virus, we examined the properties of these viruses. The NC mutant used in this experiment has a defect in viral RNA encapsidation. It is possible that the 5NC*+1PTAP* viruses have defects in packaging viral RNAs. We isolated virion RNAs and measured their amounts by quantitative RT-PCR, then normalized the amounts of viral RNA to the RT activity (Fig. 3A) or p24 value (Fig. 3B). In general, the results from the two normalization procedures are in good agreement, except for the PTAP* mutant samples. The PTAP* mutants have been shown to have lower RT activity (Dettenhofer and Yu, 1999; Yu et al., 1998), which could have caused the differences.
Fig. 3.
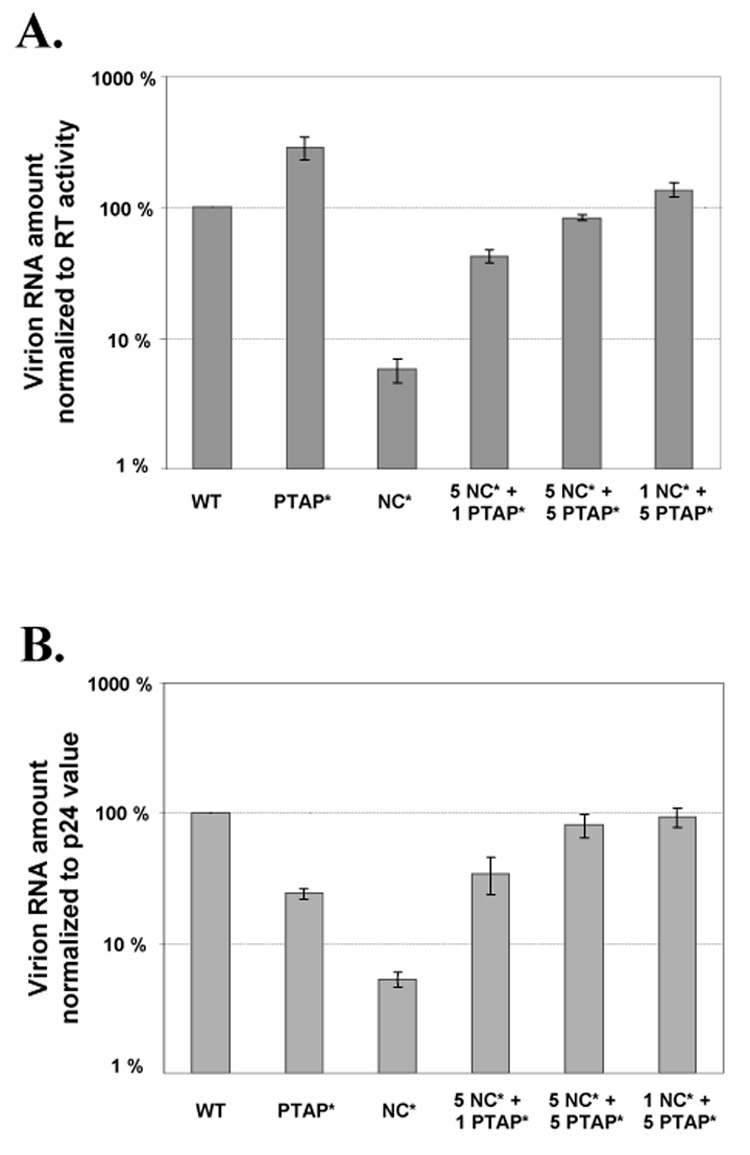
Virion RNA encapsidation by particles containing wild-type or mutant Gag proteins. The amount of viral RNA in each virus preparation was normalized to the values of RT activity (A) or p24 level (B). RNA encapsidation values for wild-type controls were set as 100%. Results from more than three independent experiments are summarized; error bars indicate standard errors.
The NC* mutant, as previously described (Gorelick et al., 1999b), encapsidated only approximately 5% of the RNA compared with wild-type viruses. The 5NC*+1PTAP* virus packaged viral RNAs at approximately 40% of the wild-type level, indicating that a small portion of the Gag with functional NC is sufficient for rescuing the RNA packaging to this level. When the amounts of Gag with functional NC increased to 50% or higher, viral RNAs were packaged at efficiencies equivalent to that of wild-type viruses.
Strategy used to examine the efficiencies of reverse transcription in Gag mutant particles
Although RNA packaging was significantly recovered, 5NC*+1PTAP* virus had very low viral titers, indicating that there are additional blocks to viral replication. To explore the possible blocks in these viruses, we examined their reverse transcription products. Virus preparations were used to infect Hut78/CCR5, cellular DNAs were harvested 24 h later, and the amounts of various viral DNA products were measured by quantitative real-time PCR. The amounts of a cellular gene, CCR5, were also measured in each sample as an internal control; these values were used to determine the levels of DNA recovery in various samples. Additionally, a negative control was included in each set of experiments; in this control, a wild-type virus sample was incubated at 68°C for 60 min (heat-inactivated sample) prior to infection. The inactivated viral sample can not infect cells nor initiate DNA synthesis, and thus served as a control for any possible plasmid DNA contamination in the input virus that may compromise the real-time PCR quantification. We observed very little DNA contamination; all negative controls had < 6% of the wild-type signals.
Like all retroviruses, HIV-1 uses a tRNA primer annealed to the primer-binding site near the 5' end of the viral RNA genome to initiate reverse transcription (Fig. 4A). Therefore, the first DNA fragment synthesized is the R-U5 region. After minus-strand DNA transfer, RT copies U3 and joins the U3-R-U5 regions to reconstitute the LTR, then continues to copy the viral genome. After plus-strand DNA synthesis, reverse transcription continues on both strands to complete DNA synthesis. Using different primer and probe sets, we analyzed and quantified three DNA products: R-U5, U3-U5, and R-5'UTR. The R-U5 region is in all viral DNA that successfully initiated DNA synthesis and copied the first piece of the viral genome. The U3-U5 region is in viral DNA that carried out minus-strand DNA transfer and copied the U3, whereas the R-5' UTR region is in viral DNA that carried out plus-strand DNA transfer and continued DNA synthesis to join the LTR and 5’ UTR. Therefore, by analyzing and comparing these three DNA products, we can examine the progression of reverse transcription.
Fig. 4.
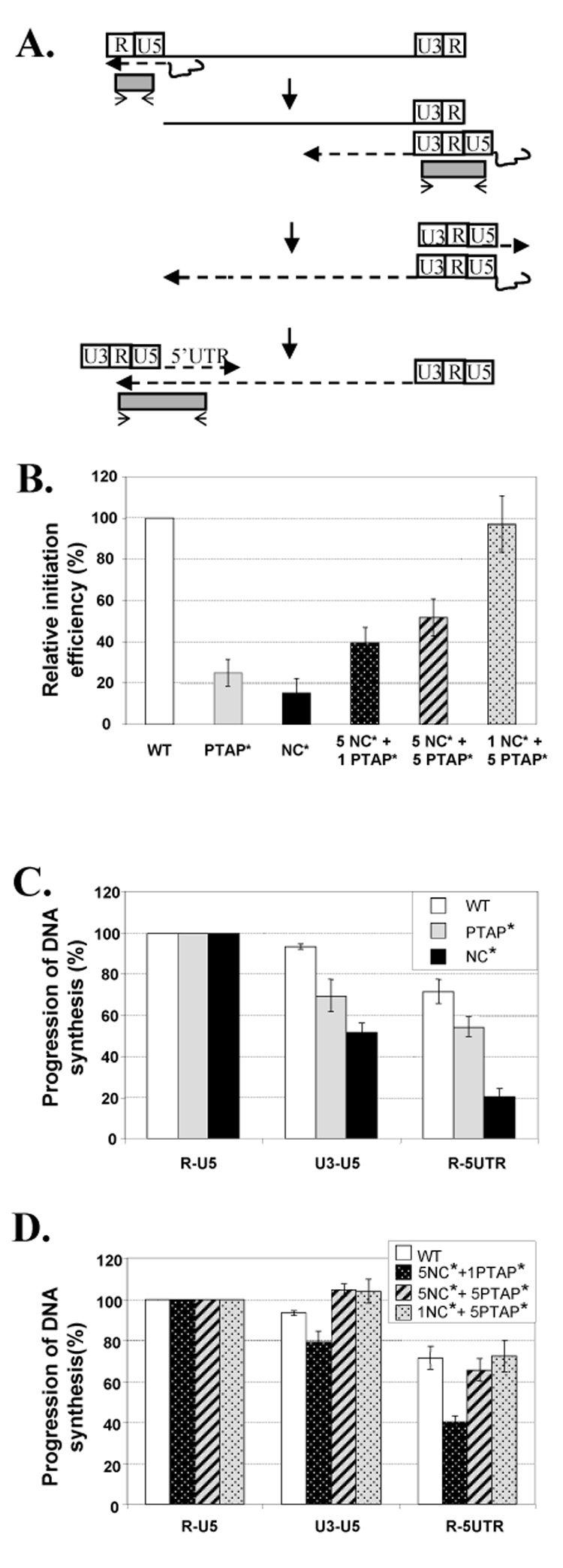
Effects of mutant Gag proteins on the initiation and progression of HIV-1 reverse transcription. (A) Schematic representation of the reverse transcription process. A dashed line with an arrow represents the direction of nascent DNA synthesis; the thin solid line with an arrow and the shaded box represents the real-time PCR primer and product, respectively. (B) Relative efficiencies of reverse transcription initiation in mutant viruses. Initiation efficiency was determined by comparing the amount of viral RNAs in the virion sample with the amount of R-U5 DNA generated after infection. The initiation efficiency of wild-type virus was defined as 100%. The initiation efficiency of mutant virus was compared with that of the wild-type virus to generate the relative efficiency. The progression of viral DNA synthesis of wild-type, PTAP*, and NC* viruses (C) and of phenotypically mixed viruses containing Gag proteins from two viruses (D) was analyzed. In the DNA progression studies, the amount of R-U5 product of each sample was defined as 100%, and the amounts of U3-U5 and the R-5’UTR products were expressed as a percentage of the R-U5 product for each sample. Results from more than three independent experiments are summarized; error bars indicate standard errors.
Efficiencies of reverse transcription initiation and DNA strand transfer events for the Gag mutant particles
R-U5 is the first piece of DNA synthesized by RT. The measurement of R-U5 estimates the combined efficiencies of events that lead to the initiation of reverse transcription. To determine the efficiencies of reverse transcription initiation, we compared the amounts of R-U5 DNA generated after infection with the amounts of the virion RNA in each virus preparation. The abilities of the mutant viruses to initiate DNA synthesis were compared with those of the corresponding wild-type viruses. NC* and PTAP* mutants initiated their DNA synthesis at 15% and 25% of the wild-type virus level, respectively (Fig. 4B). DNA synthesis was initiated at 40% of the wild-type efficiency for the 5NC*+1PTAP* viruses, and at 52% and 97% of the wild-type efficiency for the 5NC*+5PTAP* and the 1NC*+5PTAP* viruses, respectively (Fig. 4B). Therefore, both PTAP* and NC* mutants have significant defects in generating the R-U5 product; these defects were gradually recovered when more functional NC proteins were present in the virus.
To examine whether Gag mutants encounter further blocks during DNA synthesis after the initiation of reverse transcription, we also quantified the U3-U5 and R-5'UTR products. Gag mutants initiated reverse transcription at different efficiencies; to determine the efficiencies of DNA synthesis progression, the amount of R-U5 DNA of each virus was defined as 100% and the amounts of U3-U5 and R-5'UTR of each virus were expressed as a percentage of R-U5 products (Fig. 4C and D). Additionally, R-5’UTR copy numbers were multiplied by two prior to comparison with R-U5 copy numbers. This modification was necessary because LTR sequences are duplicated in viral DNA; thus there are two R-U5 and U3-U5 template sequences for PCR detection. In contrast, each viral DNA only has one template sequence for R-5'UTR detection.
The results generated from more than three independent experiments demonstrated that, once initiated, the progression of reverse transcription was very efficient in wild-type viruses. Of the R-U5 DNA detected, 93% contain U3-U5 sequences and 70% contain R-5'UTR sequences, indicating that these products have progressed beyond successful minus-strand and plus-strand DNA transfer events and further elongation, respectively (Fig. 4C). The progression of reverse transcription was less efficient in the two Gag mutants. Of the R-U5 DNA detected in the PTAP* mutant infection, 70% and 54% contained U3-U5 and R-5'UTR sequences, respectively. Of the R-U5 DNA detected in the NC* mutant infection, only 52% and 20% contained U3-U5 and R-5'UTR products, respectively (Fig. 4C).
Although both NC* and PTAP* mutants have defects in completing reverse transcription, these defects are recovered in some of the mixed particles. The completion of reverse transcription was still blocked in 5NC*+1PTAP* viruses; 79% and 40% of R-U5 DNA progressed to contain U3-U5 and R-5’UTR regions, respectively. In contrast, most of the reverse transcription defects were recovered in 5NC*+5PTAP* and 1NC*+5PTAP* viruses and DNA synthesis had efficiencies similar to those of wild-type viruses (Fig. 4D).
The effects of mutant Gag proteins on the integration of viral DNA into the host genome
After reverse transcription, HIV-1 DNA is integrated into the host genome and forms a provirus. Although integrase (IN) is the only viral protein required to perform integration reaction in vitro, it is recognized that other viral proteins, such as NC, also play important role in this process (Buckman, Bosche, and Gorelick, 2003). To examine the effects of the mutant Gag proteins on viral integration, we determined the integration efficiencies during HIV-1 infection by using an Alu-PCR procedure similar to that described previously (Butler, Hansen, and Bushman, 2001). In this assay, one primer anneals to the human repetitive element Alu, and the other primer anneals to viral sequences. Thus, this procedure only amplifies integrated DNA – specifically, those proviruses that integrate close to an Alu element. To evaluate the integration efficiency between different viruses, we compared the copy numbers of the Alu-PCR product with those of the R-5’UTR product to generate an approximate integration efficiency of each virus preparation. To compare the integration efficiencies between different viruses, the integration efficiency of the wild-type viruses was defined as 100%, and the relative integration efficiencies of the mutant viruses were expressed as a percentage of the wild-type efficiency. Results from more than three independent experiments are summarized in Fig. 5.
Fig. 5.
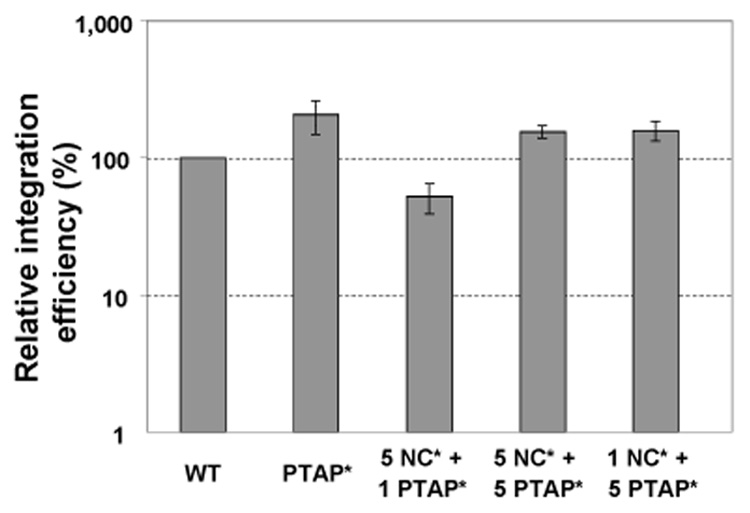
Relative integration efficiency of mutant viruses. Integration efficiency was calculated by comparing the amount of R-5’UTR product with that of the Alu-PCR product. The integration efficiency of the wild-type virus was defined as 100% and that of mutant viruses was expressed as a portion of the wild-type efficiency. Due to the replication defects of NC* mutant and the sensitivity limitation of the Alu-PCR assay, the integration efficiency of the NC* mutant could not be reliably determined. Results from more than three independent experiments are summarized; error bars indicate standard errors.
Of the two mutant viruses we used in this study, we found that the PTAP* mutants did not exhibit integration defects; DNA containing R-5’UTR sequences was integrated at frequencies similar to those of the wild-type viruses. As shown in Fig. 3 and Fig. 4, the NC* mutant has multiple defects and thus generated very little R-5’UTR products during infection. Compounded with the detection limitation of the Alu-PCR assay, the amounts of integrated proviruses could not be reliably determined. However, many of the NC* mutant defects were at least partially recovered in the NC*+PTAP* viruses; therefore, we were able to measure the integration efficiencies of these viruses. The 5NC*+1 PTAP* viruses integrated at 52% of the wild-type level, whereas the 5NC*+5PTAP* and 1NC*+5PTAP* viruses integrated at the wild-type level. These results suggest that, similar to many HIV-1 NC mutants, the NC* mutant exhibits defects in integration. These defects can be rescued by the presence of the wild-type NC proteins.
Taken together, the 5NC*+1PTAP* viruses can only partially overcome multiple defects of the two mutant parents. However, the 5NC*+5PTAP* and 1NC*+5PTAP* viruses were able to efficiently undergo most steps of HIV-1 replication. These comparisons reveal the quantitative requirements of the Gag proteins during different stages of viral replication.
Discussion
In this report, we studied the requirements for functional NC and PTAP domains at various stages of viral replication. Like many viral proteins, NC is multifunctional, thereby complicating the analyses of its functions during replication. By expressing an NC and a PTAP HIV-1 mutant at different ratios, we studied the level of functional complementation at various stages of viral replication. Our results indicate that to achieve the wild-type level of viral replication, HIV-1 requires the function of most of NC but only a small amount of PTAP. Furthermore, HIV-1 requires less functional NC during viral RNA packaging than during events leading to efficient DNA synthesis.
The PTAP* mutant described here has been well studied (Demirov, Orenstein, and Freed, 2002; Huang et al., 1995; Yu et al., 1998). The alteration of PTAP to LIRL prevented the Gag polyprotein from interacting with TSG101 and resulted in reduced virus production (Demirov, Orenstein, and Freed, 2002; Garrus et al., 2001; Huang et al., 1995; Martin-Serrano, Zang, and Bieniasz, 2001). Many of the viruses produced have immature virion morphology and lower infectivity (Boyko et al., 2006; Demirov, Orenstein, and Freed, 2002; Fu et al., 2006; Huang et al., 1995). We have previously shown that the viral RNA dimers have lower thermostability than those from wild-type viruses (Fu et al., 2006). In this study, the replication defect of the PTAP* mutant is further characterized. Our analyses indicate that this mutant has defects in events leading to and during the initiation of viral DNA replication; this result is consistent with the immature virion morphology observed by others and us. Interestingly, the PTAP* mutant does not display many defects after the initiation of viral DNA synthesis; those viruses that can initiate DNA synthesis can also complete reverse transcription and integration at near-wild-type efficiency. These results suggest the possibility that two populations of viruses are produced by the PTAP* mutants; most of the viruses have defects in the early steps of viral replication, perhaps during entry and uncoating. A small population of the mutant viruses bypassed the early replication defect and those viruses can continue to complete replication at near-wild-type efficiency.
Late-domain mutations in various retroviruses exhibit different effects of dominance. Although late domain mutants are dominant in some viruses such as human T cell leukemia virus type I (Heidecker et al., 2007), it was previously shown that a late domain mutant is not dominant in Rous sarcoma virus (Bennett and Wills, 1999). Similarly, we and others have shown that a HIV-1 late-domain mutant can be rescued by complementation (Boyko et al., 2006; Martin-Serrano, Zang, and Bieniasz, 2001). In this current report, we further demonstrated that only a small amount of functional PTAP motif is needed to rescue virus release and titer (Fig. 2A and 2C); this is in agreement with a previous study indicating a portion of the functional PTAP is needed for the rescue (Martin-Serrano, Zang, and Bieniasz, 2001). Additionally, we have shown that the released viruses have infectivities similar to those of wild-type viruses (Fig. 2D), indicating that the defects of virus release and those that led to DNA synthesis initiation have been relieved by a low amount of functional PTAP motif.
Much of our knowledge of HIV-1 NC function comes from mutational analyses. Various NC mutations can affect virus assembly, RNA packaging, reverse transcription, and integration. However, how much functional NC is required in each step of the infection process cannot be addressed in these analyses. In the current report, we generated viruses with different ratios of functional NC proteins and analyzed the properties of these viruses and their infection processes. We found that during the assembly process, having a small portion of functional NC (one sixth) in the virus can significantly recover specific viral RNA encapsidation. The specific encapsidation of the viral RNA is mediated by the interactions between Gag and the packaging signal of the viral RNA. In addition to the specific interaction, Gag also interacts with RNA in a nonspecific manner. It has been suggested that RNA serves as a scaffold for Gag multimerization and virion formation (Campbell and Rein, 1999; Campbell and Vogt, 1997; Muriaux et al., 2001). In the situation where both wild-type Gag and NC mutant Gag are present in the cells, the wild-type Gag can interact with viral RNAs in a sequence-specific manner, thereby facilitating the subsequent Gag-Gag and nonspecific Gag-RNA interactions during the virion assembly process. Therefore, it is feasible that RNA packaging can be rescued by the presence of a small number of Gag proteins with functional NC.
Our results indicate that both 5NC*+1PTAP* and 5NC*+5PTAP* viruses appear to have a replication block during very early events that lead to the generation of R-U5. Therefore, much of the functional NC is needed to complete steps required to efficiently initiate reverse transcription and generate R-U5. NC mediates part of the Gag-Gag interactions; therefore, mutations in NC can cause defects in virion assembly, maturation, and stability (Yovandich et al., 2001). Some of the viruses containing significant amounts of mutant NC proteins may have these aforementioned defects, thereby reducing efficiencies in virus entry and uncoating. Similarly, the initiation of the reverse transcription may also be affected. The decrease of the R-U5 product is most likely a combination of the defects in both steps. Due to its chaperone activity, NC has been implicated in minus-strand and plus-strand DNA transfer reactions (Levin et al., 2005; Rein, Henderson, and Levin, 1998). Our results indicated that after the generation of R-U5 products, the 5NC*+5PTAP* virus preparations have little defects in reverse transcription, whereas the 5NC*+ 1PTAP* virus preparations have an approximately two-fold defect. These results suggest that to achieve a wild-type level of elongation of reverse transcription, including the two obligatory strand transfer reactions, HIV-1 requires more than one-sixth, but not more than half, of the functional NC in the virion. Many NC mutants also have defects in the DNA integration step (Buckman, Bosche, and Gorelick, 2003; Thomas et al., 2006). The integration efficiencies of the 5NC*+1PTAP* virus preparations, but not the 5NC*+5PTAP* or 1NC*+5PTAP* virus preparations, appear to be lower than that of wild-type virus, suggesting that the function of more than one-sixth of the NC proteins is needed for the efficient integration reaction.
Despite more than two decades of dedicated efforts, currently we do not have a cure or an effective vaccine for HIV-1 infection. The development of antiviral drugs and treatment regimens has significantly increased life expectancy for those HIV-1 patients who have access to treatments. The emergence of drug-resistant HIV-1 variants makes it imperative to develop new antiviral therapies and drug targets. NC is an essential protein for HIV-1 replication and its functions are important in multiple stages of viral replication, thereby making NC an attractive target for antiviral drug development. Our study indicates that in order for a candidate drug to inhibit HIV-1 replication, it needs to render most of the NC in the virion nonfunctional, as virions having approximately half of the functional NC can still replicate at 50% efficiency. More than 80% of the NC proteins in the virions must lose their functions to achieve 95% inhibition of viral replication. Therefore, our study not only reveals the requirements of NC at various stages of HIV-1 replication, but also provides insights into aspects essential to the development of effective antiviral drugs targeting the HIV-1 NC protein.
Materials and methods
Cell culture, DNA transfection, virus infection, DNA isolation, and flow cytometry analyses
Human embryonic kidney cell line 293T (DuBridge et al., 1987) and human T-cell line Hut78/CCR5 (Wu et al., 2002) were maintained in Dulbecco’s modified Eagle’s medium and Roswell Park Memorial Institute-1640 medium, respectively. Both media were supplemented with 5% fetal calf serum, 5% calf serum, penicillin (50 U/ml), and streptomycin (50 U/ml). Additionally, L-glutamine (2 mM) was included in the 293T cell medium, whereas puromycin (1 µg/ml) and G418 (500 µg/ml) were included in the Hut78/CCR5 medium. All cultured cells were maintained in humidified 37°C incubators with 5% CO2.
DNA transfection was performed using the standard calcium phosphate method (Sambrook, 1989). Salmon sperm DNA was added to some samples so that each transfection cocktail contained the same amount of DNA. After incubation at 37°C with 3% CO2 for 6–8 h, the transfection mixture was removed by gently washing the cells four times, each time with 10 ml of phosphate-buffered saline (PBS) without calcium or magnesium (BioWhittaker). Supernatants from the transfected cells were harvested 48 h posttransfection, clarified through a 0.22-µm-pore-size polyvinylidene fluoride filter to remove cellular debris, and used for infection or stored at −80°C for future analysis. Prior to infection, supernatants were treated with 10 U/ml of RNase-free DNase I (Roche) for 2 h at 37°C in the presence of 10 mM MgCl2 to decrease contamination with plasmid DNA.
Hut78/CCR5 cells were used as target cells for infection; at 24 h postinfection, a portion of the sample was removed for DNA isolation and the rest of the cells remained in culture for an additional 48 h prior to flow cytometry analyses. For the portion that underwent DNA isolation, cells were passed through a Ficoll-Pague™ Plus (Amersham Biosciences) cushion by centrifugation to remove debris, and washed twice with PBS, and DNA was isolated using the QIAamp DNA Blood mini kit (Qiagen) according to the manufacturer’s instructions. To determine the level of virus infection, cells were stained with allophycocyanin (APC)-conjugated α-Thy1.2 antibody (eBioscience), flow cytometry analyses were performed on a FACSCalibur system (Becton Dickinson Biosciences), and data were analyzed using the FlowJo software (Tree Star). Infected cells were identified by the expression of GFP or Thy 1.2 marker. The number of total live cells (Y) and infected cells (Z) were used to calculate multiplicities of infection (MOI) as log (1 − Z/Y) /log [(Y − 1)/Y]/Y, based on Poisson distribution as previously described (Rhodes et al., 2005).
Viral RNA quantification and Real-time PCR analyses of viral DNA
Virion RNA was isolated as previously described with minor modifications (Thomas et al., 2006). Briefly, virions were incubated with lysis buffer (50 mM Tris, pH 7.4, 10 mM EDTA, 1% w/v SDS, 100 mM NaCl, 50 µg/ml tRNA, 100 µg/ml proteinase K), extracted twice with phenol/chloroform/isoamyl alcohol (25:24:1), and precipitated with ethanol. RNA was pelleted at 21,000 × g for 30 min and treated with RQ1 RNase-free DNase (Promega) to remove plasmid DNA contamination. After the DNase was inactivated by adding 4 M guanidinium isothiocyanate, RNA was precipitated again with ethanol, and resuspended in diethylpyrocarbonate-treated water with 1 mM dithiothreitol and 1 U/µl RNaseOUT (Invitrogen).
Quantitative reverse transcription PCR (RT-PCR) was performed as previously described (Cline et al., 2005). Viral RNA was serially diluted to ensure the linearity of RT-PCR quantification. RT-PCR was carried out using a two-step protocol in 96-well optical plates. The RT reaction was incubated at 25°C for 15 min, 42°C for 40 min, 85°C for 10 min, and 25°C for 30 min. After the reverse transcription step, PCR was carried out by first incubating the reaction at 95°C for 10 min, then 45 cycles of 95°C for 15 sec and 60°C for 1 min. RNA transcripts generated from a KpnI-digested pRB1334 plasmid were used as a standard for RNA quantification. In this assay, primers and probe anneal to sequences in the gag gene of the viral RNA (Buckman, Bosche, and Gorelick, 2003).
Viral DNA synthesis was measured by real-time PCR as previously described (Buckman, Bosche, and Gorelick, 2003). To monitor the possible contamination of plasmid DNA, in each experiment a mock infection was carried out using heat-inactivated wild-type virus and the resulting sample was measured in parallel with the other samples. Cellular DNA was diluted 3-to 10-fold in buffer containing 10 mM Tris and 0.5 mM EDTA (pH 8.3); some samples were digested with 0.5 U of DpnI for 3 h prior to analyses. Relative recovery of cellular DNA was determined by measuring the copy numbers of the CCR5 gene in a given sample. Previously described primers, probes, standards, and protocol were used to quantify R-U5, U3-U5, and R-5’UTR (Buckman, Bosche, and Gorelick, 2003) and CCR5 (Thomas et al., 2006).
The amount of provirus integration was measured by Alu-PCR similar to the method previously described by Bushman and colleagues (Butler, Hansen, and Bushman, 2001). DNA isolated from a HOS cell line, IH624-1, referred to as IS DNA, was used as a standard for Alu-PCR and provirus integration (Thomas et al., 2006). IH624-1 consists of a pool of cells infected with an HIV-1 vector. IS DNA was serially diluted with Hut78/CCR5 cellular DNA such that each dilution had the same copy number of a cellular gene, CCR5, and different numbers of proviruses. Proviral copies in IS DNA dilutions were determined by measuring the amount of R-5’UTR.
RT activity assay and western analyses
RT activity and viral capsid (p24) amounts were determined to measure virion production. For the RT assay, viruses were pelleted at 50,000 × g for 1 h at 4°C and resuspended in PBS. RT activity was determined using the Quan-T-RT scintillation proximity assay (Amersham Biosciences) according to the manufacturer’s instructions. A series of increasing virus amounts was analyzed to ensure the linearity of the assay. The amount of viral p24 was detected using the Alliance HIV p24 ELISA kit (PerkinElmer) according to the manufacturer’s instructions.
To examine cellular Gag expression, transfected 293T cells were washed twice in phosphate-buffered saline, lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 % Triton-100), and clarified by centrifugation. A portion of the lysate from each sample was loaded on a 12% Tris-glycine gel, separated by gel electrophoresis, and transferred to an Immobilon-FL polyvinylidene difluoride membrane (Millipore). The membrane was probed with a human anti-HIV-1 antisera obtained from NIH AIDS Research and Reference Reagents Program, followed by donkey anti-human antibodies labeled with the IRDye800 fluorophore at dilution of 1:10,000 (Rockland). Images of the analyses were captured using the Odyssey infrared imaging system (Li-Cor).
Acknowledgements
We thank Anne Arthur for expert editorial help, James Thomas and Tamyo Mbisa for their advice on the real-time PCR detections, Gisela Heidecker for discussions, Eric Freed for his kind gift of plasmid.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and in part by NCI contract no. N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bennett RP, Nelle TD, Wills JW. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J Virol. 1993;67(11):6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RP, Rhee S, Craven RC, Hunter E, Wills JW. Amino acids encoded downstream of gag are not required by Rous sarcoma virus protease during gag-mediated assembly. J Virol. 1991;65(1):272–280. doi: 10.1128/jvi.65.1.272-280.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RP, Wills JW. Conditions for copackaging rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding. J Virol. 1999;73(3):2045–2051. doi: 10.1128/jvi.73.3.2045-2051.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- Berkowitz RD, Ohagen A, Hoglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69(10):6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles NE, Damay P, Spahr PF. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J Virol. 1993;67(2):623–631. doi: 10.1128/jvi.67.2.623-631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowzard JB, Bennett RP, Krishna NK, Ernst SM, Rein A, Wills JW. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72(11):9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko V, Leavitt M, Gorelick R, Fu W, Nikolaitchik O, Pathak VK, Nagashima K, Hu WS. Coassembly and complementation of Gag proteins from HIV-1 and HIV-2, two distinct human pathogens. Mol Cell. 2006;23(2):281–287. doi: 10.1016/j.molcel.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Brule F, Marquet R, Rong L, Wainberg MA, Roques BP, Le Grice SF, Ehresmann B, Ehresmann C. Structural and functional properties of the HIV-1 RNA-tRNA( Lys)3 primer complex annealed by the nucleocapsid protein: comparison with the heat-annealed complex. Rna. 2002;8(1):8–15. doi: 10.1017/s1355838202010981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A. 1990;87(2):523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol. 2003;77(2):1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7(5):631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73(3):2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Vogt VM. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71(6):4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73(8):6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Khorchid A, Gabor J, Rong L, Wainberg MA, Kleiman L. Roles of Pr55(gag) and NCp7 in tRNA(3)(Lys) genomic placement and the initiation step of reverse transcription in human immunodeficiency virus type 1. J Virol. 2000;74(22):10796–10800. doi: 10.1128/jvi.74.22.10796-10800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo JL, Kabdulov TO, Paulson ML, Anderson JA, Hu WS. The nucleocapsid domain is responsible for the ability of spleen necrosis virus (SNV) Gag polyprotein to package both SNV and murine leukemia virus RNA. J Virol. 1999;73(11):9170–9177. doi: 10.1128/jvi.73.11.9170-9177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Vincent O, Jin J, Weisz OA, Montelaro RC. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J Biol Chem. 2005;280(49):40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34(5–6):303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Craven RC, Leure-duPree AE, Weldon RA, Jr, Wills JW. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69(7):4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002;99(2):955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76(1):105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdowski A, Ding L, Spearman P. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J Virol. 2004;78(3):1230–1242. doi: 10.1128/JVI.78.3.1230-1242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettenhofer M, Yu XF. Proline residues in human immunodeficiency virus type 1 p6(Gag) exert a cell type-dependent effect on viral replication and virion incorporation of Pol proteins. J Virol. 1999;73(6):4696–4704. doi: 10.1128/jvi.73.6.4696-4704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Li H, Derdowski A, Ding L, Burnett A, Chen X, Peters TR, Dermody TS, Woodruff E, Wang JJ, Spearman P. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120(5):663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7(1):379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128(5):841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251(1):1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- Freed EO, Martin MA. Retroviridae: The Retroviruses and their Replication. In: David PMH, Knipes M, editors. Fields Virology. fifth ed. II. II vols. Piladelphia, PA, USA: Lippincott William and Wilkins; 2007. [Google Scholar]
- Freed EO, Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70(1):341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Dang Q, Nagashima K, Freed EO, Pathak VK, Hu WS. Effects of Gag mutation and processing on retroviral dimeric RNA maturation. J Virol. 2006;80(3):1242–1249. doi: 10.1128/JVI.80.3.1242-1249.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59(1):103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- Goff SP. Retroviridae: The Retroviruses and their Replication. In: David PMH, Knipes M, editors. Fields Virology. fifth ed. II. II vols. Piladelphia, PA, USA: Lippincott William and Wilkins; 2007. [Google Scholar]
- Gorelick RJ, Benveniste RE, Gagliardi TD, Wiltrout TA, Busch LK, Bosche WJ, Coren LV, Lifson JD, Bradley PJ, Henderson LE, Arthur LO. Nucleocapsid protein zinc-finger mutants of simian immunodeficiency virus strain mne produce virions that are replication defective in vitro and in vivo. Virology. 1999a;253(2):259–270. doi: 10.1006/viro.1998.9513. [DOI] [PubMed] [Google Scholar]
- Gorelick RJ, Chabot DJ, Ott DE, Gagliardi TD, Rein A, Henderson LE, Arthur LO. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70(4):2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Chabot DJ, Rein A, Henderson LE, Arthur LO. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67(7):4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur LO. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999b;256(1):92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- Gorelick RJ, Henderson LE, Hanser JP, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a "zinc finger-like" protein sequence. Proc Natl Acad Sci U S A. 1988;85(22):8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Nigida SM, Jr, Bess JW, Jr, Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64(7):3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989;86(15):5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MJ, Derebail SS, Gorelick RJ, DeStefano JJ. Differing roles of the N- and C-terminal zinc fingers in human immunodeficiency virus nucleocapsid protein-enhanced nucleic acid annealing. J Biol Chem. 2003;278(33):30755–30763. doi: 10.1074/jbc.M303819200. [DOI] [PubMed] [Google Scholar]
- Heidecker G, Lloyd PA, Soheilian F, Nagashima K, Derse D. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J Virol. 2007;81(18):9769–9777. doi: 10.1128/JVI.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci U S A. 1996;93(7):3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69(11):6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JF, Lever AM. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70(2):880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JF, Lever AM. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J Virol. 1998;72(7):5877–5885. doi: 10.1128/jvi.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M, Gabus C, Rau M, Darlix JL. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268(2):250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix JL. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J Biol Chem. 1998;273(50):33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Lu YL, Bennett RP, Wills JW, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69(11):6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammano F, Ohagen A, Hoglund S, Gottlinger HG. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68(8):4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7(12):1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003;77(8):4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath CF, Buckman JS, Gagliardi TD, Bosche WJ, Coren LV, Gorelick RJ. Human cellular nucleic acid-binding protein Zn2+ fingers support replication of human immunodeficiency virus type 1 when they are substituted in the nucleocapsid protein. J Virol. 2003;77(15):8524–8531. doi: 10.1128/JVI.77.15.8524-8531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric C, Goff SP. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63(4):1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric C, Gouilloud E, Spahr PF. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J Virol. 1988;62(9):3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric C, Spahr PF. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60(2):450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000;74(8):3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci U S A. 2001;98(9):5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol. 1999;73(5):4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Chertova EN, Gagliardi TD, Nagashima K, Sowder RC, 2nd, Poon DT, Gorelick RJ. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J Virol. 2003;77(10):5547–5556. doi: 10.1128/JVI.77.10.5547-5556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reicin AS, Paik S, Berkowitz RD, Luban J, Lowy I, Goff SP. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69(2):642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23(8):297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- Rhodes TD, Nikolaitchik O, Chen J, Powell D, Hu WS. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J Virol. 2005;79(3):1666–1677. doi: 10.1128/JVI.79.3.1666-1677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes GM, Bambara RA, Fay PJ. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270(25):15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- Rong L, Liang C, Hsu M, Guo X, Roques BP, Wainberg MA. HIV-1 nucleocapsid protein and the secondary structure of the binary complex formed between tRNA(Lys.3) and viral RNA template play different roles during initiation of (−) strand DNA reverse transcription. J Biol Chem. 2001;276(50):47725–47732. doi: 10.1074/jbc.M105124200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sandefur S, Smith RM, Varthakavi V, Spearman P. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag) J Virol. 2000;74(16):7238–7249. doi: 10.1128/jvi.74.16.7238-7249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72(4):2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasakumar N, Hammarskjold ML, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69(10):6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114(6):689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology. 2006;353(1):41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68(9):5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98(14):7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77(9):5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills JW, Craven RC. Form, function, and use of retroviral gag proteins. Aids. 1991;5(6):639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- Wu L, Martin TD, Vazeux R, Unutmaz D, KewalRamani VN. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J Virol. 2002;76(12):5905–5914. doi: 10.1128/JVI.76.12.5905-5914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yovandich JL, Chertova EN, Kane BP, Gagliardi TD, Bess JW, Jr, Sowder RC, 2nd, Henderson LE, Gorelick RJ. Alteration of zinc-binding residues of simian immunodeficiency virus p8(NC) results in subtle differences in gag processing and virion maturation associated with degradative loss of mutant NC. J Virol. 2001;75(1):115–124. doi: 10.1128/JVI.75.1.115-124.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XF, Dawson L, Tian CJ, Flexner C, Dettenhofer M. Mutations of the human immunodeficiency virus type 1 p6Gag domain result in reduced retention of Pol proteins during virus assembly. J Virol. 1998;72(4):3412–3417. doi: 10.1128/jvi.72.4.3412-3417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WH, Hwang CK, Hu WS, Gorelick RJ, Pathak VK. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J Virol. 2002;76(15):7473–7484. doi: 10.1128/JVI.76.15.7473-7484.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69(9):5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


