Summary
Na,K-ATPase is a hetero-oligomer of α- and β-subunits. The Na,K-ATPase β-subunit (Na,K-β ) is involved in both the regulation of ion transport activity, and in cell-cell adhesion. By structure prediction and evolutionary analysis, we identified two distinct faces on the Na,K-β transmembrane domain (TMD) that could mediate protein-protein interactions: a glycine zipper motif and a conserved heptad repeat. Here, we show that the heptad repeat face is involved in the hetero-oligomeric interaction of Na,K-β with Na,K-α , and the glycine zipper face is involved in the homo-oligomerization of Na,K-β . Point mutations in the heptad repeat motif reduced Na,K-β binding to Na,K-α , and Na,K-ATPase activity. Na,K-β TMD homo-oligomerized in biological membranes, and mutation of the glycine zipper motif affected oligomerization and cell-cell adhesion. These results provide a structural basis for understanding how Na,K-β links ion transport and cell-cell adhesion.
Keywords: transmembrane domain, heptad repeat motif, GxxxG, glycine zipper, Na, K-ATPase, cell adhesion
Introduction
Na,K-ATPase is a ubiquitously expressed, plasma membrane bound enzyme that mediates the ATP-dependent transport of three sodium ions out and two potassium ions into the cell. The ion transduction function of Na,K-ATPase is critical for the maintenance of ion homeostasis within the cell, and has been well studied.1 Na,K-ATPase is an oligomeric enzyme consisting of two essential subunits, the α-subunit (Na,K-α ), which is the catalytic subunit, and a regulatory β-subunit (Na,K-β ), which is required for the translation, stability, and membrane insertion of Na,K-α.2,3 Thus, the primary role of Na,K-β is to assist the Na,K-ATPase ion transport function via its interaction with Na,K-α . Recently, we and others have shown that Na,K-β also functions as a cell-cell adhesion molecule.4–7 The structural features of Na,K-β involved in the regulation of Na,K-ATPase enzyme activity and cell-cell adhesion have not been well characterized.
Na,K-β is a type II transmembrane protein with a short cytoplasmic domain and a large glycosylated extracellular domain. Three different isoforms of Na,K-β are known so far, viz. β1, β2, and β3. The transmembrane domain (TMD) of β1 (the β isoform used in this study) is highly conserved (99%) throughout the animal kingdom ranging from human to chicken. Amongst the different isoforms of Na,K-β , there is only a 30–35% identity over the entire protein sequence, however their TMDs are 57–61% identical. This extensive sequence conservation within the TMD of Na,K-β suggests that Na,K-β TMD must play an important role in the structure and function of Na,K-ATPase.
A type II membrane protein typically spans the membrane as a stable α-helix, a conformation that internally satisfies the backbone hydrogen bonding groups in the hydrophobic lipid environment.8 Folding and assembly of membrane proteins is often supported by specific interactions between the preformed helical transmembrane segments.9–13 Sequence motifs have been identified in transmembrane helices that are strongly associated with helix-helix interactions.14–18 In this study, we show that the TMD of Na,K-β has both, a conserved heptad repeat, and a glycine zipper motif on two different faces, suggesting the possibility that Na,K-β could interact with two different partners. To test this hypothesis, we introduced mutations into the two faces of the Na,K-β TMD to disrupt lateral association with its binding partners. We find that one face of the Na,K-β TMD interacts with Na,K-α , and regulates enzyme activity, while the other face is involved in Na,K-β self-association, which is important for the cell-cell adhesion function of this protein. Thus, Na,K-β is not only a key structural component of the Na,K ATPase monomer, but it also serves to oligomerize the protein.
Results
Packing interface of Na,K-β transmembrane domain
Certain sequence motifs are prevalent in TM helix-helix interactions. One of the most well characterized sequence motifs known to mediate helix oligomerization is the GxxxG motif.9,19 As shown in Fig. 1B, the putative TMD of Na,K-β contains two tandem GxxxG sequences (GxxxGxxxG), also known as a glycine zipper.20 Glycine zippers have been shown to strongly drive TM helix oligomerization, hinting at the possibility that the glycine zipper face of Na,K-β could form an interaction surface with other TMDs. However, the perfect glycine zipper is not conserved in all Na,K-β subunits (see Supp Fig. 1). Nevertheless, further analysis revealed that small residues are strongly preferred at the first position (44 has G, A, T or V), and also at the second position (48 has only G or A). Large residues are common at the third position (52 has G, I, L or F), but in all such cases, a small residue is found one position before it, placing it on the same face of the putative helix. Small residues are known to be preferred in transmembrane helix packing.21,22 Thus, in some cases Na,K-β has a perfect glycine zipper, strongly implicated in helix-helix interactions, and in all cases, Na,K-β possesses a face of small residues.
Figure 1.
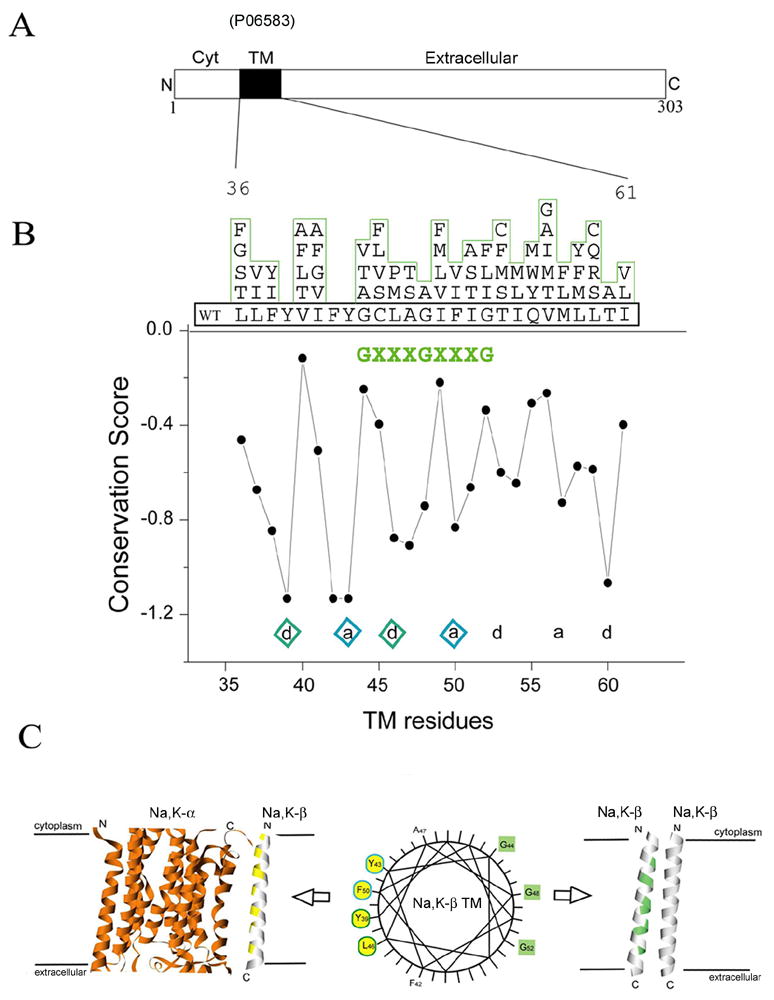
Two faces of the TMD of Na,K-β . (A) Linear arrangements of Na,K-β include cytoplasmic (Cyt), transmembrane (TM) and extra cellular domains. (B) Sequence conservation scoring calculated from Conseq using the multiple sequence alignments of Na,K-β . The residue variety at each position in the aligned sequence is shown above the Na,K-β TMD sequence. Structurally or functionally important residues are usually conserved through evolution. The conservation scores are a relative measure of evolutionary conservation at each sequence site of Na,K-β . The lower score represents the more conserved position in a protein. Well-conserved heptad repeats are indicated with diamonds and the potential homo-oligomeric motif (GxxxGxxxG) is green colored. (C) Wheel representation shows two distinct faces of Na,K-β TMD sequences. Heptad repeat residues (left) are yellow colored and the GxxxGxxxG motif (right) is green colored. The model represents the TMD interactions of Na,K-β with Na,K-α (hetero-oligomerization) and Na,K-β itself (homo-oligomerization).
Strikingly, the face of Na,K-β opposite the glycine zipper is strongly conserved, suggesting that this face may also play an important role in Na,K-ATPase structure or function (Fig. 1B). The conserved residues are spaced in accordance with the heptad repeat, which is another signature interaction motif found in TMD oligomerizations.16 Considering that the primary function of Na,K-β is to support the Na,K-ATPase enzyme activity, we hypothesized that this heptad repeat motif might be involved in the interaction with Na,K-α.
Recent studies from our laboratory and others have shown that Na,K-β has dual functions: in addition to its role in assisting Na,K-α , Na,K-β also functions as a cell adhesion molecule.4–6 As the sequence analyses suggest that there could be two separate packing faces on the Na,K-β TMD (see model in Fig. 1C), it is possible that the two faces are important for different functions of Na,K-β . To test the two-face hypothesis, we designed mutations that would presumably alter the TMD interactions of Na,K-β with Na,K-α (hetero-oligomerization), and Na,K-β itself (homo-oligomerization).
Analysis of heptad repeat mutants of Na,K-β
To study the significance of the highly conserved heptad repeat motif within the Na,K-β TMD, we generated two double mutants (Y39A/L46A and Y43A/F50A) targeting the conserved face of the Na,K-β TMD. These mutants were tagged with GFP and expressed in MSV-MDCK cells, which have very low levels of endogenous Na,K-β and Na,K-α .4 The Na,K-β-GFP fusion protein migrated at the expected molecular mass (~81 kDa). As shown in Fig. 2A, both the mutants were expressed at levels comparable to that of the wild-type (WT) GFP-tagged Na,K-β (middle panel, compare lane 1 with lanes 2 and 3). The levels of Na,K-α were comparable in all the three cell lines (top panel, compare lanes 1–3). It is known that Na,K-β interaction with Na,K-α is essential for the transport of Na,K-α to the plasma membrane.1,23 We therefore tested the levels of Na,K-α and Na,K-β on the plasma membrane by a cell surface biotinylation assay. We observed that there was about 60% reduction in the cell surface levels of Na,K-β as well as Na,K-α in cells expressing the Y43A/F50A mutant of Na,K-β compared to WT and the Y39A/L46A mutant (Fig. 2B). Consistent with the reduced plasma membrane expression of Na,K-α , we observed a 32 ± 5% reduction in the Na,K-ATPase enzyme activity as measured by the ouabain-sensitive rubidium flux (Fig. 2C). These results indicated that Na,K-β TMD mutants expressed similar total levels of Na,K-α , yet the cell surface expression of both Na,K-α and Na,K-β , and the Na,K-ATPase enzyme activity were significantly reduced in the Y43A/F50A mutant, suggesting that this mutation compromised Na,K-β interaction with Na,K-α.
Figure 2.
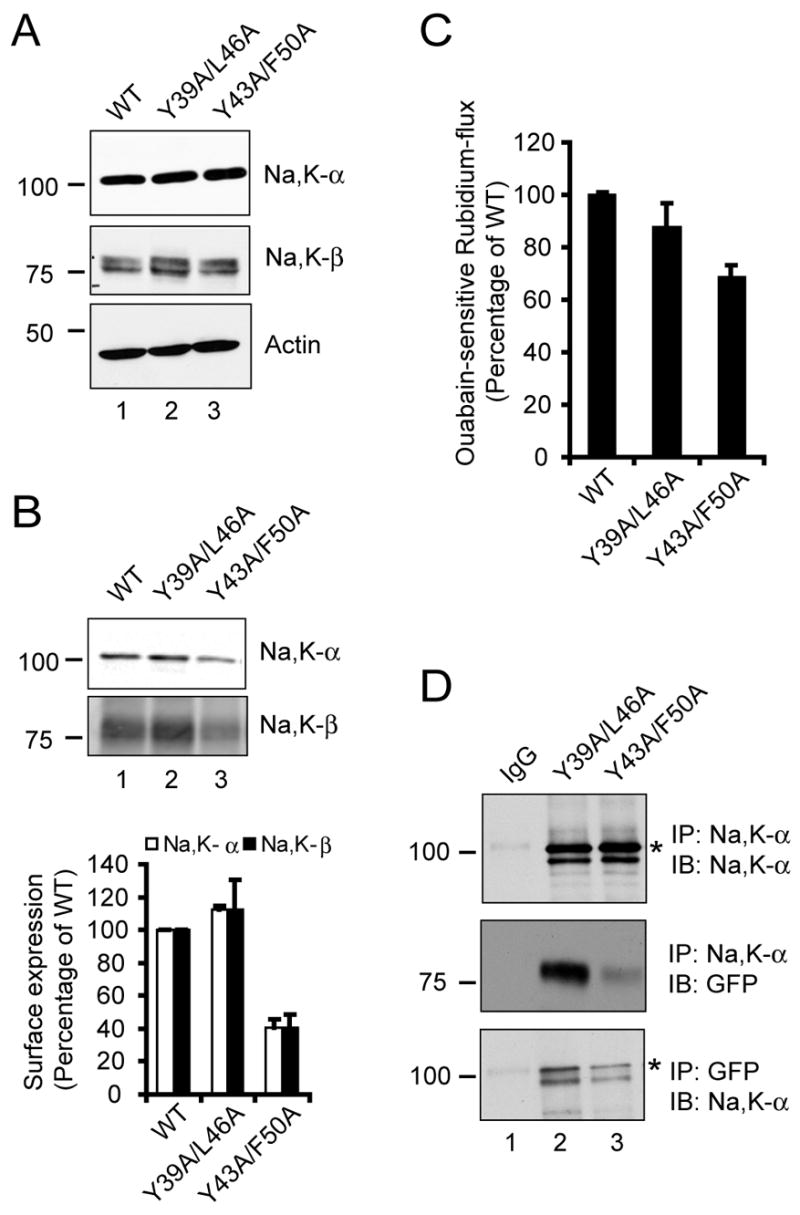
Characterization of heptad repeat mutants of Na,K-β . (A) Immunoblots showing levels of Na,K-α , Na,K-β and actin (loading control) from 100 μg of total cell lysates of MSV-MDCK cells expressing Na,K-β-GFP (WT), Y39A/L46A or Y43A/F50A mutants. (B) Cell surface biotinylation assay of WT, Y39A/Y46A and Y43A/F50A cells. Lysates from cells labeled with biotin were immunoprecipitated using streptavidin-conjugated agarose beads and immunoblotted with anti-Na,K-α or anti-Na,K-β antibodies. The graph represents the mean from three independent experiments. The error bars denote SE of the mean. (C) Ouabain-sensitive rubidium flux of the above-mentioned cell lines expressed as a percentage of Na,K-β-GFP (WT), which was considered to be 100%. The error bars represent the SE of the mean. (D) MSV-MDCK cells expressing Y39A/L46A or Y43A/F50A mutants of Na,K-β were lysed and immunoprecipitated with either anti-Na,K-α or anti-GFP antibodies and immunoblotted with the indicated antibodies. IgG represents control immunoprecipitation with isotype matched irrelevant IgG from lysates of Y39A/L46A cells blotted similarly with indicated antibodies. The specific Na,K-α band is denoted by an asterisk.
Since Na,K-α and Na,K-β interactions are very stable and have been well documented by co-immunoprecipitation analysis,2,24 we performed co-immunoprecipitation to determine if the interaction between Na,K-α and the heptad repeat mutants of Na,K-β was affected. The total amount of Na,K-α immunoprecipitated in cell lines expressing either Y39A/L46A or Y43A/F50A mutants of Na,K-β was similar (Fig. 2D, top panel). However, there was approximately 78% reduction in the amount of GFP-tagged Y43A/F50A mutant of Na,K-β co-immunoprecipitated with Na,K-α (Fig. 2D, middle panel). This was further confirmed by a reverse co-immunoprecipitation experiment in which Na,K-β-GFP fusion protein was immunoprecipitated with anti-GFP antibody and immunoblotted for Na,K-α . Distinctly the amount of Na,K-α associated with Y43A/F50A mutant was reduced by nearly 57% in this analysis (Fig. 2D, bottom panel). Taken together, these results demonstrated that Tyr43 and Phe50 together, were critical for Na,K-β interaction with Na,K-α , and for Na,K-ATPase enzyme activity. These results also suggested that mutations of these residues reduced transport of Na,K-β as well as Na,K-α to the cell surface, and that Na,K-β also needed to bind to Na,K-α to be expressed on the plasma membrane.
Analysis of a glycine zipper mutant of Na,K-β
Na,K-β TMD has a glycine zipper motif comprised of G44xxxG48xxxG52. We altered the central G48 to disrupt the glycine zipper motif.20 The mutant, G48L, was expressed in MSV-MDCK cells as a GFP fusion protein. The expression level of the G48L mutant of Na,K-β was similar to that of WT Na,K-β (Fig. 3A). MSV-MDCK cells expressing either WT Na,K-β or the G48L mutant of Na,K-β also had similar levels of Na,K-α . Na,K-β G48L was not defective in the interaction between Na,K-α as determined by co-immunoprecipitation analysis (Fig. 3B). Cell surface biotinylation assay (Fig. 3C) demonstrated that the surface expression of the mutant Na,K-β was comparable to the levels of the WT. The cell surface levels of Na,K-α were also similar in cells expressing either WT or mutant Na,K-β (Fig. 3C). The rubidium uptake was not affected for this mutant, indicating that the pump was functioning at levels similar to WT (Fig. 3D). These results confirmed that the glycine zipper is neither involved in the hetero-oligomerization with Na,K-α nor in the modulation of Na,K-α function in the enzyme activity.
Figure 3.
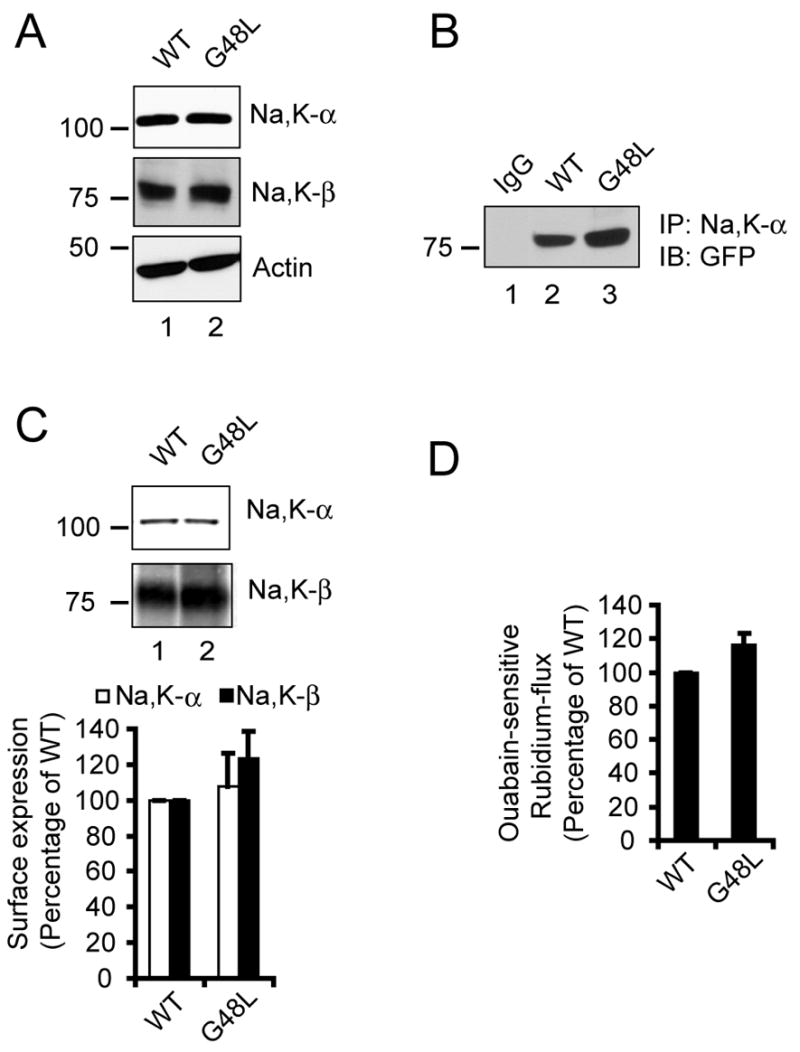
Characterization of G48L mutant of Na,K-β . (A) Immunoblots showing the levels of Na,K-α , Na,K-β and actin (loading control) in WT and G48L cells. (B) Co-immunoprecipitation assay showing the amount of GFP-tagged Na,K-β pulled down by Na,K-α in cells expressing either Na,K-β WT or the G48L mutant. IgG represents control immunoprecipitation with isotype matched irrelevant IgG from lysates of WT cells and blotted with anti-GFP antibody. (C) Cell surface biotinylation assay showing the levels of Na,K-α and Na,K-β on the plasma membrane in WT and G48L cells. The graph represents the quantifications of the blot from three independent experiments. The error bars denote the SE of the mean. (D) Ouabain-sensitive rubidium uptake of WT and G48L cells expressed as a percentage of Na,K-β-GFP (WT).
Na,K-β TMD oligomerizes in biological membranes
Since Na,K-β has cell-cell adhesion function, we reasoned that the glycine zipper motif is likely involved in the TMD interactions of Na,K-β itself. To directly test for homo-oligomerization, we used the TOXCAT assay, which investigates the association of transmembrane helices in the context of the biological membrane.25 The TOXCAT system utilizes a chimeric protein composed of the ToxR N-terminal transcriptional activation domain (ToxR’) fused to a transmembrane segment of interest and a C-terminal periplasmic domain of Escherichia coli maltose binding protein (MBP). The TMD mediated dimerization of the chimera in the E. coli inner membrane causes ToxR’ dimerization, thereby driving transcriptional activation of a reporter gene, chloramphenicol acetyltransferase (CAT) making the bacteria resistant to chloramphenicol. The efficiency of the TMD interaction can be determined in two ways: either by estimating the extent of acquired resistance to the antibiotic chloramphenicol in vivo or by the direct quantification of chloramphenicol acetylation by CAT in vitro.
We generated chimeric constructs as described above, consisting of either WT Na,K-β TMD or Na,K-β-G48L TMD and compared their oligomerization efficiencies. The disc diffusion assay showed a two-fold increase in the diameter of the clear zone around the chloramphenicol disc in G48L chimera compared to WT chimera (Fig. 4A, arrows), indicating that the mutant reduces oligomerization. The diameter of the clear zone observed in WT and G48L mutant of Na,K-β was similar to the diameter of the clear zone in WT and mutant TMD of Glycophorin A (GpA) respectively. GpA is a well-characterized oligomer used in TOXCAT assays19. In addition, this result was confirmed by a quantitative in vitro CAT assay (Fig. 4B). The CAT activity of the TMD of Glycophorin A WT chimera was considered 100%. The GpA mutant (G83I) showed negligible CAT activity. Na,K-β WT TM showed CAT activity higher than GpA WT (108.0 ± 19.5%), whereas the CAT activity of the G48L Na,K-β was only 38.0 ± 9.8%. These results demonstrated that the TMD of Na,K-β required G48 for homo-oligomerization within the bacterial membrane.
Figure 4.
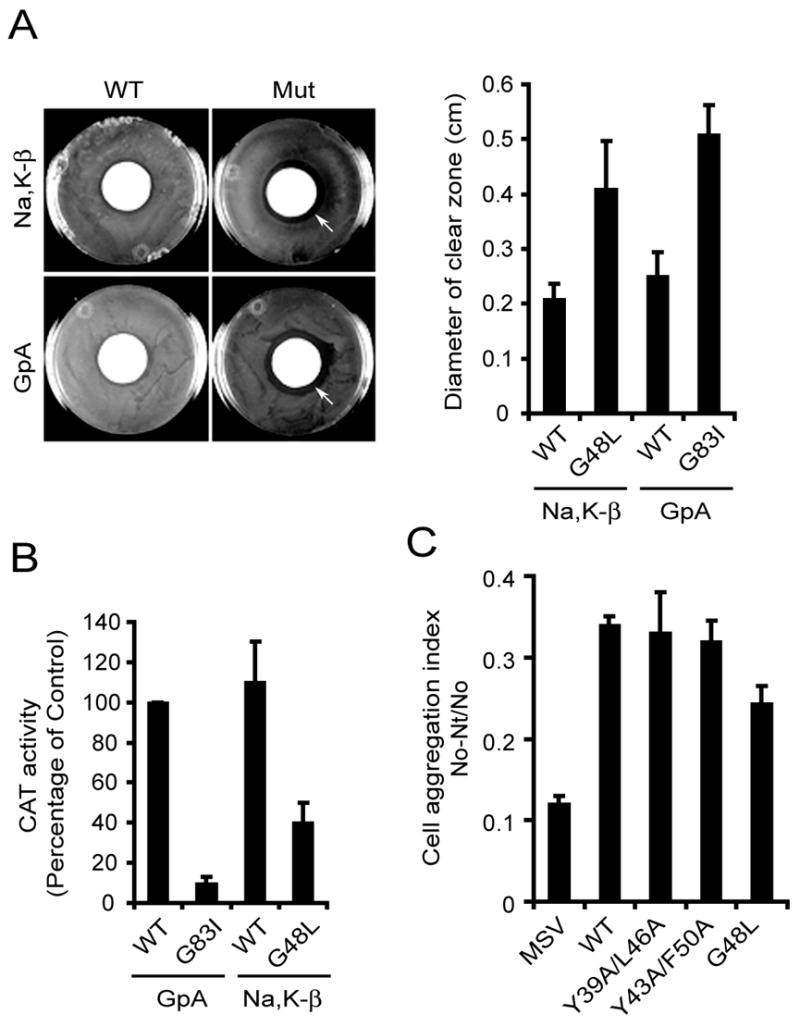
G48L mutant of Na,K-β does not self-associate and has reduced cell aggregation. (A) Disk diffusion assay of MM39 cells expressing chimeric constructs carrying either WT or mutant Na,K-β or Glycophorin A (GpA). Pictures of representative plates are shown for reference. The graph shows the diameter of the zone of cell inhibition in MM39 cells expressing various chimeras. The error bars denote SE of the mean from three independent experiments. (B) TOXCAT assay showing the amount of CAT activity from MM39 cells expressing different chimeras. The error bars denote SE of the mean from triplicates of two independent experiments. (C) Cell aggregation assay of the parental MSV-MDCK cells and those expressing Na,K-β WT or mutants. The error bars denote the SE of the mean of three independent experiments done in triplicates.
Analysis of cell aggregation for Na,K-β G48L mutant
The cell-cell adhesion function of epithelial cadherin (E-cadherin), a Ca2+-dependent cell adhesion molecule, has been well characterized.26 E-cadherin, not only requires trans-homodimerization but also requires cis-homodimerization for its cell adhesion function.27,28 MSV-MDCK cells expressing WT Na,K-β aggregate due to cell-cell adhesion mediated by Na,K-β ,4 whereas parental MSV-MDCK cells which express very low levels of Na,K-β do not aggregate in vitro (Fig. 4C). Therefore, we tested whether the G48L mutant of Na,K-β that failed to oligomerize within the bacterial plasma membrane, also show reduced cell-cell aggregation in MSV-MDCK cells. We observed a 29.0 ± 6.5% decrease in the cell-cell aggregation index of MSV-MDCK cells expressing G48L mutant compared to WT cells (Fig. 4C). The heptad repeat mutants of Na,K-β (Y39A/L46A and Y43A/F50A) had a minimal effect on the cell-cell aggregation (Fig. 4C). Taken together, these results demonstrated that Na,K-β laterally oligomerized within the membrane, and this interaction was involved in the cell-cell adhesion function of this protein.
Discussion
In this study, we have identified heptad repeat motif and glycine zipper motif in the TMD of Na,K-β by structure prediction, motif identification and evolutionary analysis. We validated our predictions by mutagenesis of critical residues within the motifs, and by analyzing the mutants in specific functional assays. We demonstrated that the heptad repeat motif and the glycine zipper motif are involved in two different interactions. Na,K-β interacts with Na,K-α via the heptad repeat motif, and mutagenesis of two amino acids within the heptad repeat motif, resulted in reduced binding to Na,K-α , and affected membrane targeting and consequently the ion transport function of Na,K-ATPase. The glycine zipper motif is involved in the self-association of Na,K-β , and the disruption of this motif by mutagenesis affected homo-oligomerization in biological membranes as revealed by the TOXCAT assay, and Na,K-β induced cell-cell aggregation. These results suggested that the TMD of Na,K-β not only facilitates the ion transport function of Na,K-α, but is also involved in its role as a cell-cell adhesion molecule.
The heptad repeat motif belongs to the leucine zipper type of side-chain packing similar to certain soluble proteins, and is a commonly found TMD sequence motif that governs interactions between two transmembrane proteins.16 The interacting residues form a repeated heptad (abcdefg) pattern, wherein residues at a- and d- positions constitute the core of the interfaces.29 Such heptad repeat motifs have been shown to form the interfaces of phospholamban30 and cadherins.16 Breaking the heptad repeat interaction in a TMD is expected to be difficult due to strong interactions among side chains and the high effective concentration possible when restricted to a bilayer, so a single point mutation may be insufficient to abolish binding. It was shown earlier in Xenopus oocytes that single or double mutations in Y39 and Y43 residues within Na,K-β TMD did not alter the binding of Na,K-β to Na,K-α, but these mutations had an additive effect on the transport properties of the Na,K-α .31 We therefore introduced mutations in two different residues in a heptad repeat such that they were separated by seven amino acids. Mutations closer to the bilayer core (Y43A/F50A) disrupted the interactions between Na,K-α and Na,K-β . Mutations closer to the membrane interface (Y39A/L46A) had a minimal effect on the interaction, however, suggesting that the core region is more important in Na,K-β interaction with Na,K-α . Alternatively, the ‘a’ residues (Y43A/F50A) had an effect on the oligomerization, whereas the ‘d’ residues (Y39A/L46A) had minimal effect, suggesting that the ‘a’ residues are more critical for the helix-helix interface than the ‘d’ residues.
Na,K-β is known to interact with Na,K-α via different sites present throughout the protein. These sites include, the cytoplasmic domain as shown by controlled proteolysis assays of Na,K-β ,32 the membrane proximal region consisting of 63 amino acids as revealed by yeast two-hybrid assays,33 and also the TMD as shown by oxidative cross-linking studies of fragments of proteolyzed Na,K-ATPase.34,35 Although Na,K-β TMD was implicated in binding to Na,K-α , our results for the first time show that disruption of the TMD interaction site within Na,K-β dramatically affects its interaction with Na,K-α . The finding that the association of Na,K-α to Na,K-β is drastically reduced by mutagenesis of Y43 and F50 indicates that the Na,K-β TMD is critical for Na,K-α association, and disruption of this interaction site destabilizes other potential associations between the two subunits.
Some Na,K-β TMDs have tandem GxxxG motifs (glycine zipper) and others have a mixture of small residues on the face opposite from the heptad repeat motif. The GxxxG sequence motif was the most commonly isolated motif in a screen of a randomized sequence library to identify sequence patterns that can mediate high-affinity inter-helical associations.25 The GxxxG motif is also found in a number of self-associating transmembrane proteins including glycophorin A,36 receptor tyrosine kinases like ErbB,37 and integrins.38 The G48L mutation had no effect on the association of Na,K-β with Na,K-α which is consistent with the data obtained from a triple glycine mutant of Na,K-β expressed in Xenopus oocytes.31 We found that the G48L mutant disrupted homo-oligomerization of the TMD, and showed a reduced cell aggregation index as compared to WT Na,K-β . The sequence conservation scoring data (Fig. 1B) indicated that G48 is likely to be the most important of the three glycine residues in the zipper as only a G or A was found at this position. It is interesting to note that the sequence variations of GxxxG motifs among homologs are also found in other cell signaling receptors such as erbB and integrins. This might explain the variations of binding affinities or changing interaction partners, which are a characteristic of single pass cell membrane signaling receptor proteins. Likewise, other functions of Na,K-β , such as cell adhesion, may be differentiated in the different families, leading to consequent sequence divergence in the Na,K-β-β homo-oligomeric interface.
Na,K-ATPase oligomer has been shown to consist of two Na,K-α and two Na,K-β molecules based on molecular distance measurements.39 Whether the Na,K-β molecules involved in cell adhesion are complexed with Na,K-α remains to be determined. However, the observation that the heptad repeat mutant of Na,K-β (Y43A/F50A), that has reduced interaction with Na,K-α , exhibits cell aggregation index similar to WT Na,K-β , suggests that Na,K-β oligomerization with Na,K-α is less likely involved in Na,K-β mediated cell adhesion.
Cell-cell adhesion involves interaction of the extracellular domains. E-cadherin, a well-defined Ca2+-dependent cell adhesion molecule undergoes trans-homophilic interactions between two molecules on neighboring cells. However, it is also known to form cis-dimers by lateral interactions within the same cell membrane. It has been shown that the cis-dimerization is required for the adhesive function of E-cadherin.27,28 Although E-cadherin TMD does not have a GxxxG, it interacts via its TMD, and this interaction was found to be essential for E-cadherin mediated adhesiveness.40 Thus, it is clear from studies with E-cadherin that both trans-dimerization (mediated by the extracellular domain) and cis-dimerization (mediated by its TMD) are essential for the cell adhesion function. Similarly, disruption of the lateral oligomers of Na,K-β by G48L mutation reduced the cell-cell aggregation index but did not completely abolish the aggregation, perhaps because of trans-dimerization mediated by the extracellular domain of the Na,K-β . The mechanism of cell adhesion mediated by Na,K-β still needs to be characterized, however.
Materials and Methods
Sequence conservation scoring
The program ConSeq (http://bioinfo.tau.ac.il/ConSeq,41) was used to map the sequence conservation reflected in the multiple sequence alignment of human Na,K-β (Swiss-Prot accession number P05026) with 34 representative sequence homologs. This server compares the sequence of a reference protein with the proteins deposited in Swiss-Prot42 and finds the ones that are homologous to the sequence. The number of PSI-BLAST iterations and the E-value cutoff used were 1 and 0.001, respectively. All the sequences that were found to be evolutionarily related with the human Na,K-β in the data set were used in the subsequent conservation scoring. Briefly, the ConSeq server assigns relative conservation scores to each residue, taking into account the evolutionary relationships among the family of homologs. The scores are normalized such that the average score is zero, and negative and positive deviations represent the degrees of conservation and variation, respectively.
Cell culture and plasmid construction
MSV-MDCK cells (DoCl1) from American Type Culture Collection (Manassas, VA) were cultured in DMEM supplemented with 10% fetal bovine serum, 1 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Canine Na,K-β cDNA obtained from Dr. Robert Farley (University of Southern California, Los Angeles, CA) was sub-cloned into pEGFP vector (Clontech, Palo Alto, CA) as described previously,7 to yield a fusion protein with GFP at the C-terminus (Na,K-β-GFP). Point mutations were introduced in Na,K-β-GFP by using a QuikChange mutagenesis kit (Stratagene, La Jolla, CA) following manufacturer’s protocol. The sequences of the mutants were confirmed by DNA sequencing. These GFP-fusion constructs were stably transfected into MSV-MDCK cells followed by the sorting of GFP-positive cells by using fluorescence activated cell sorting.
The vector for TOXCAT assay (pccKAN), glycophorin A (GpA) WT and G83I mutant were kindly provided by Dr. Donald M. Engelman (Yale University, New Haven, CT). Na,K-β TMD (WT- consisting of amino acids 35–60) was cloned into pccKAN using specific oligonucleotides to generate a chimera consisting of an N-terminal DNA binding domain of ToxR, Na,K-β TMD, and the periplasmic maltose binding protein (MBP). The presence of MBP in the periplasm was confirmed by growth on minimal maltose media. The G48L mutation was introduced within the Na,K-β TMD using the QuikChange mutagenesis kit.
Antibodies
Mouse monoclonal antibodies (mAbs) raised against Na,K-α (M7-PB-E9 for immunoblotting and M8- for immunoprecipitation) and Na,K-β (M17-P5-F11) have been described previously.4 Anti-actin mAb, anti-GFP mAb, and horseradish peroxidase (HRP)-anti-mouse antibody was purchased from Sigma-Aldrich (St. Louis, MO), Zymed Laboratories (San Francisco, CA), and Transduction Laboratories (Lexington, KY) respectively.
Immunoblotting and immunoprecipitation
Cells were lysed in a lysis buffer containing 20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 1 mM sodium glycerolphosphate, 1 mM sodium orthovanadate, 1 mM PMSF, and 5 μg/ml each of antipain, leupeptin, and pepstatin. The lysates were sonicated, and clarified by centrifugation at 13,000 rpm for 10 min at 4°C. Total protein was estimated from the supernatants using the Bio-Rad DC reagent (Bio-Rad, Hercules, CA) as per manufacturer's instructions. Lysates corresponding to equal amounts of total protein were used. For immunoprecipitation, lysates were incubated on a rotator overnight with antibody bound to protein A agarose beads at 4°C. The proteins bound to the beads were collected by centrifugation and washed thrice with lysis buffer, separated by SDS-PAGE and transferred to nitrocellulose membrane (Schleicher & Schuell, Keene, NH). Immunoblotting was performed using a primary antibody, and HRP-conjugated secondary antibody diluted in tris-buffered saline containing 5% non-fat dried milk and 0.1% Tween20. The proteins were detected by using the enhanced chemiluminescence lighting system according to the manufacturer's recommendations (PerkinElmer Life and Analytical Sciences, Boston, MA).
Cell surface biotinylation
Cell surface biotinylation was performed as described previously,4,43 by using 0.5 μg/ml membrane-impermeable EZ-Link Sulfo-NHS-Biotin (Pierce, Rockford, IL). The cells were lysed in 150 mM NaCl, 20 mM Tris pH 8, 5 mM EDTA, 1% Triton X-100, 0.1% bovine serum albumin (BSA), 1 mM PMSF, 5 μg/ml each of antipain, leupeptin, pepstatin. Lysate precipitated for 16 h at 4°C with 30 μl of Ultralink streptavidin beads (Pierce). The precipitates were washed, loaded on a gel, and immunoblotted as described above).
Rubidium transport assay
The ouabain-sensitive ion transport was measured by determining the uptake of86Rb+ as described earlier.4,44 To measure the ouabain-sensitive86Rb+ transport, the cells were incubated with 50 μM ouabain (Sigma-Aldrich) for 30 min at 37°C before the experiment. The samples were normalized to the protein content and the ouabain-sensitive Rb flux was calculated. The data are presented as mean ± SE from three independent experiments done in triplicates.
Cell aggregation assay
Cells were washed briefly in PBS and trypsinized with 0.05% trypsin/1 mM CaCl2 to obtain single cells. Cells (200,000) in culture medium were plated on 60-mm Petri dishes coated with 1% agarose and incubated on a gyratory shaker at 37°C, 5% CO2. After 16 h single cells were counted. The cell aggregation data are represented by the index (N0 − Nt)/N0 where Nt is the single cell number after the incubation time t and N0 is the cell number at the initiation of incubation.
Disk diffusion assay
Disk diffusion assay was performed as described previously.19 Briefly, a Whatman 3MM filter paper disk (2.4 cm diameter) was soaked with 60 μl of 90 mg/ml chloramphenicol in ethanol, air-dried and was placed in the center of a LB/Amp agar plate. MM39 cells were electroporated with Mal-Na,K-β (WT-TM)-ToxR or Mal-NamK-β (G48L-TM)-ToxR chimeric constructs and grown to an OD600 of 0.6, diluted 6× with LB medium, and plated on 20 ml of LB/Amp agar plates. The plates were incubated O/N at 37 ° C and the diameter of the clear zone of inhibition of growth around the disk was measured.
CAT assays
MM39 cells electroporated with the two chimeric constructs described above were grown to OD600 of 0.6. 200 μl of this culture was pelleted, resuspended in 0.5 ml of 100 mM Tris·HCl, pH 8.0, and lysed by addition of 20 μl of 100 mM EDTA, 100 mM DTT, 50 mM Tris·HCl, pH 8.0, and a small drop of toluene at 30°C for 30 min. CAT assays were performed on the cell-free extracts by using Promega CAT Enzyme Assay System.
Supplementary Material
Supplemental Figure 1. Multiple sequence alignment of Na,K-β homologs as detected by a BLAST search.
Acknowledgments
Supported by NIH DK56216. We thank Dr. Donald M. Engelman (Yale University, New Haven, CT), for providing us the reagents for the TOXCAT assay, and Drs. William James Ball, Jr. and Robert Farley for Na,K-ATPase antibodies and canine Na,K-β cDNA, respectively.
Abbreviations used
- CAT
Chloramphenicol acetyltransferase
- GpA
Glycophorin A
- MBP
maltose binding protein, Na,K-α, Na,K-ATPase α-subunit
- Na
K-β, Na,K-ATPase β-subunit
- TMD
transmembrane domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaplan JH. Ion movements through the sodium pump. Annu Rev Physiol. 1985;47:535–544. doi: 10.1146/annurev.ph.47.030185.002535. [DOI] [PubMed] [Google Scholar]
- 2.Rajasekaran SA, Gopal J, Willis D, Espineda C, Twiss JL, Rajasekaran AK. Na,K-ATPase beta1-subunit increases the translation efficiency of the alpha1-subunit in MSV-MDCK cells. Mol Biol Cell. 2004;15:3224–3232. doi: 10.1091/mbc.E04-03-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geering K. The functional role of beta subunits in oligomeric P-type ATPases. J Bioenerg Biomembr. 2001;33:425–438. doi: 10.1023/a:1010623724749. [DOI] [PubMed] [Google Scholar]
- 4.Rajasekaran SA, Palmer LG, Quan K, Harper JF, Ball WJ, Jr, Bander NH, Peralta Soler A, Rajasekaran AK. Na,K-ATPase beta-subunit is required for epithelial polarization, suppression of invasion, and cell motility. Mol Biol Cell. 2001;12:279–295. doi: 10.1091/mbc.12.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoshani L, Contreras RG, Roldan ML, Moreno J, Lazaro A, Balda MS, Matter K, Cereijido M. The Polarized Expression of Na+,K+-ATPase in Epithelia Depends on the Association between {beta}-Subunits Located in Neighboring Cells. Mol Biol Cell. 2005;16:1071–1081. doi: 10.1091/mbc.E04-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitamura N, Ikekita M, Sato T, Akimoto Y, Hatanaka Y, Kawakami H, Inomata M, Furukawa K. Mouse Na+/K+-ATPase beta1-subunit has a K+-dependent cell adhesion activity for beta-GlcNAc-terminating glycans. Proc Natl Acad Sci U S A. 2005;102:2796–2801. doi: 10.1073/pnas.0409344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barwe SP, Anilkumar G, Moon SY, Zheng Y, Whitelegge JP, Rajasekaran SA, Rajasekaran AK. Novel Role for Na,K-ATPase in Phosphatidylinositol 3-Kinase Signaling and Suppression of Cell Motility. Mol Biol Cell. 2005;16:1082–1094. doi: 10.1091/mbc.E04-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 9.Lemmon MA, Engelman DM. Specificity and promiscuity in membrane helix interactions. Q Rev Biophys. 1994;27:157–218. doi: 10.1017/s0033583500004522. [DOI] [PubMed] [Google Scholar]
- 10.Dieckmann GR, DeGrado WF. Modeling transmembrane helical oligomers. Curr Opin Struct Biol. 1997;7:486–494. doi: 10.1016/s0959-440x(97)80111-x. [DOI] [PubMed] [Google Scholar]
- 11.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 12.Shai Y. Molecular recognition within the membrane milieu: implications for the structure and function of membrane Pproteins. J Membr Biol. 2001;182:91–104. doi: 10.1007/s00232-001-0034-a. [DOI] [PubMed] [Google Scholar]
- 13.DeGrado WF, Gratkowski H, Lear JD. How do helix-helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 2003;12:647–665. doi: 10.1110/ps.0236503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemmon MA, Treutlein HR, Adams PD, Brunger AT, Engelman DM. A dimerization motif for transmembrane alpha-helices. Nat Struct Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- 15.Sternberg MJ, Gullick WJ. A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. Protein Eng. 1990;3:245–248. doi: 10.1093/protein/3.4.245. [DOI] [PubMed] [Google Scholar]
- 16.Gurezka R, Laage R, Brosig B, Langosch D. A heptad motif of leucine residues found in membrane proteins can drive self-assembly of artificial transmembrane segments. J Biol Chem. 1999;274:9265–9270. doi: 10.1074/jbc.274.14.9265. [DOI] [PubMed] [Google Scholar]
- 17.Dawson JP, Weinger JS, Engelman DM. Motifs of serine and threonine can drive association of transmembrane helices. J Mol Biol. 2002;316:799–805. doi: 10.1006/jmbi.2001.5353. [DOI] [PubMed] [Google Scholar]
- 18.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 19.Russ WP, Engelman DM. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci U S A. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc Natl Acad Sci U S A. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang S, Tovchigrechko A, Vakser IA. The role of geometric complementarity in secondary structure packing: a systematic docking study. Protein Sci. 2003;12:1646–1651. doi: 10.1110/ps.0304503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eilers M, Shekar SC, Shieh T, Smith SO, Fleming PJ. Internal packing of helical membrane proteins. Proc Natl Acad Sci U S A. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow DC, Forte JG. Functional significance of the beta-subunit for heterodimeric P-type ATPases. J Exp Biol. 1995;198:1–17. doi: 10.1242/jeb.198.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Caplan MJ, Forbush B, 3rd, Palade GE, Jamieson JD. Biosynthesis of the Na,K-ATPase in Madin-Darby canine kidney cells. Activation and cell surface delivery. J Biol Chem. 1990;265:3528–3534. [PubMed] [Google Scholar]
- 25.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 26.Ranscht B. Cadherins and catenins: interactions and functions in embryonic development. Curr Opin Cell Biol. 1994;6:740–746. doi: 10.1016/0955-0674(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 27.Ozawa M. Lateral dimerization of the E-cadherin extracellular domain is necessary but not sufficient for adhesive activity. J Biol Chem. 2002;277:19600–19608. doi: 10.1074/jbc.M202029200. [DOI] [PubMed] [Google Scholar]
- 28.Takeda H, Shimoyama Y, Nagafuchi A, Hirohashi S. E-cadherin functions as a cis-dimer at the cell-cell adhesive interface in vivo. Nat Struct Biol. 1999;6:310–312. doi: 10.1038/7542. [DOI] [PubMed] [Google Scholar]
- 29.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 30.Simmerman HK, Kobayashi YM, Autry JM, Jones LR. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J Biol Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 31.Hasler U, Crambert G, Horisberger JD, Geering K. Structural and functional features of the transmembrane domain of the Na,K-ATPase beta subunit revealed by tryptophan scanning. J Biol Chem. 2001;276:16356–16364. doi: 10.1074/jbc.M008778200. [DOI] [PubMed] [Google Scholar]
- 32.Geering K, Beggah A, Good P, Girardet S, Roy S, Schaer D, Jaunin P. Oligomerization and maturation of Na,K-ATPase: functional interaction of the cytoplasmic NH2 terminus of the beta subunit with the alpha subunit. J Cell Biol. 1996;133:1193–1204. doi: 10.1083/jcb.133.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colonna TE, Huynh L, Fambrough DM. Subunit interactions in the Na,K-ATPase explored with the yeast two-hybrid system. J Biol Chem. 1997;272:12366–12372. doi: 10.1074/jbc.272.19.12366. [DOI] [PubMed] [Google Scholar]
- 34.Or E, Goldshleger R, Karlish SJ. Characterization of disulfide cross-links between fragments of proteolyzed Na,K-ATPase. Implications for spatial organization of trans-membrane helices. J Biol Chem. 1999;274:2802–2809. doi: 10.1074/jbc.274.5.2802. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov A, Zhao H, Modyanov NN. Packing of the transmembrane helices of Na,K-ATPase: direct contact between beta-subunit and H8 segment of alpha-subunit revealed by oxidative cross-linking. Biochemistry. 2000;39:9778–9785. doi: 10.1021/bi001004j. [DOI] [PubMed] [Google Scholar]
- 36.Brosig B, Langosch D. The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 1998;7:1052–1056. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 38.Li R, Gorelik R, Nanda V, Law PB, Lear JD, DeGrado WF, Bennett JS. Dimerization of the transmembrane domain of Integrin alphaIIb subunit in cell membranes. J Biol Chem. 2004;279:26666–26673. doi: 10.1074/jbc.M314168200. [DOI] [PubMed] [Google Scholar]
- 39.Linnertz H, Urbanova P, Obsil T, Herman P, Amler E, Schoner W. Molecular distance measurements reveal an (alpha beta)2 dimeric structure of Na+/K+-ATPase. High affinity ATP binding site and K+-activated phosphatase reside on different alpha-subunits. J Biol Chem. 1998;273:28813–28821. doi: 10.1074/jbc.273.44.28813. [DOI] [PubMed] [Google Scholar]
- 40.Huber O, Kemler R, Langosch D. Mutations affecting transmembrane segment interactions impair adhesiveness of E-cadherin. J Cell Sci. 1999;112:4415–4423. doi: 10.1242/jcs.112.23.4415. [DOI] [PubMed] [Google Scholar]
- 41.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18:S71–77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 42.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anilkumar G, Barwe SP, Christiansen JJ, Rajasekaran SA, Kohn DB, Rajasekaran AK. Association of prostate-specific membrane antigen with caveolin-1 and its caveolae-dependent internalization in microvascular endothelial cells: Implications for targeting to tumor vasculature. Microvasc Res. 2006;72:54–61. doi: 10.1016/j.mvr.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Lambrecht N, Corbett Z, Bayle D, Karlish SJ, Sachs G. Identification of the site of inhibition by omeprazole of a alpha-beta fusion protein of the H,K-ATPase using site-directed mutagenesis. J Biol Chem. 1998;273:13719–13728. doi: 10.1074/jbc.273.22.13719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Multiple sequence alignment of Na,K-β homologs as detected by a BLAST search.


