Abstract
Temperate phage Mu-1, which is able to integrate at random in its host chromosome, is also able to mediate integration of other circular deoxyribonucleic acid, as a lambda-gal mutant unable to integrate by itself. After mixed infection with lambda-gal and Mucplus, galplus transductants are recovered that have the lambda-gal integrated in any circular permutation, sandwiched between two complete Mu genomes in the same orientation, the whole Mu-lambda-gal-Mu structure being found at any location in the bacterial chromosome. Here we show that such a lambda-gal can integrate in an induced Mu lysogen. In this case the lambda-gal is again in any circular permutation, between two Mu in the same orientation, but it is always located at the site of the original Mu prophage, and the two surrounding Mu have always the same genotype as the original Mu prophage. Active Mu replication functions are not essential for that process to occur. This suggests that bacterial replication may generate two Mu copies that in some way can regenerate a Mu attachment site that recombines with the lambda-gal. A model is presented that accounts for these observations, may be helpful for understanding some complex features of Mu development, and may possibly offer a basis for explaining spontaneous duplications.
Full text
PDF




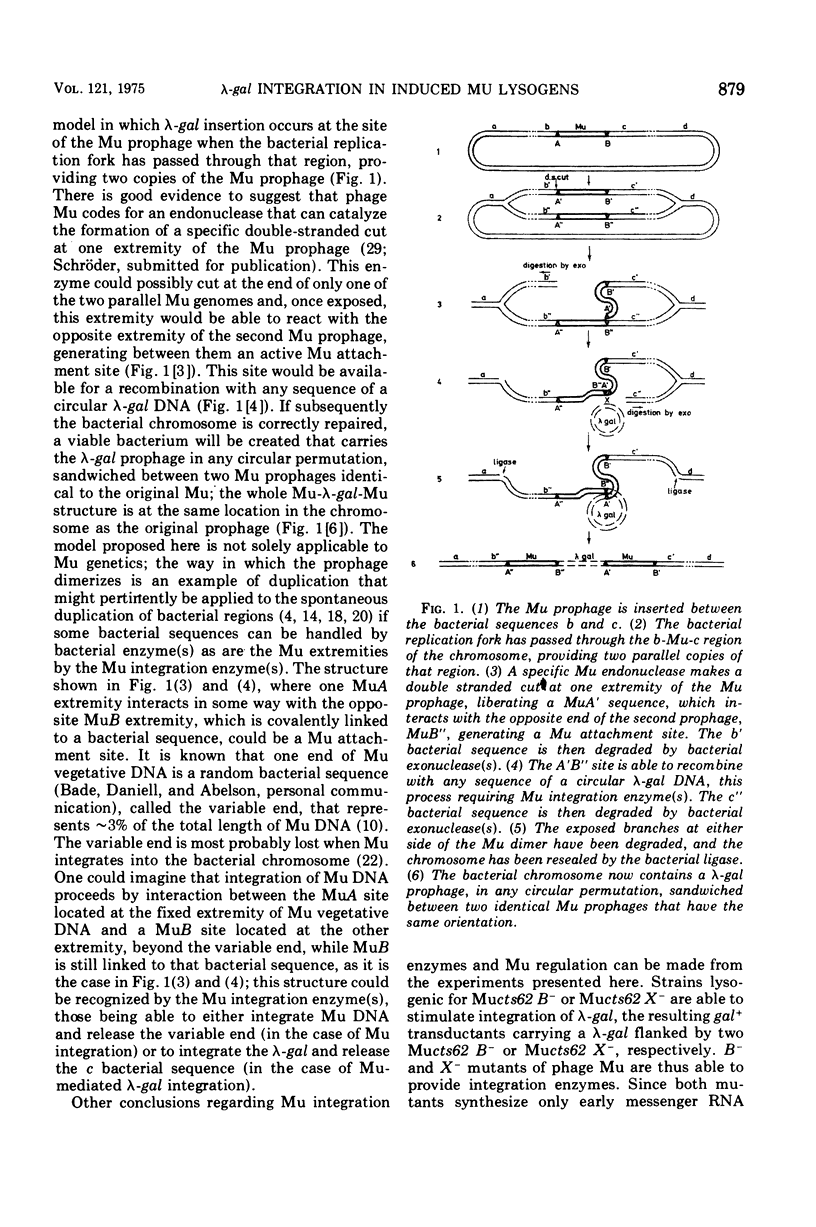
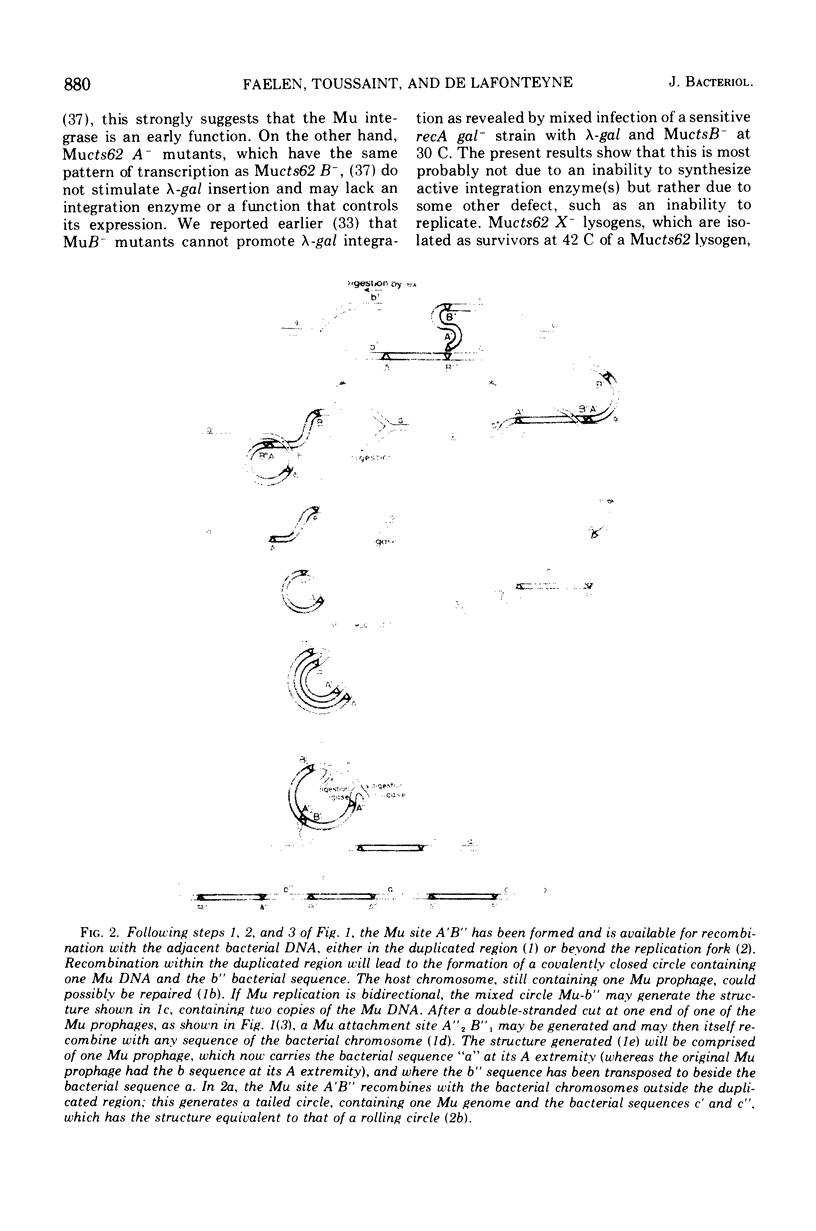


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J., Boram W., Bukhari A. I., Faelen M., Howe M., Metlay M., Taylor A. L., Toussaint A., Van de Putte P., Westmaas G. C. Summary of the genetic mapping of prophage Mu. Virology. 1973 Jul;54(1):90–92. doi: 10.1016/0042-6822(73)90117-7. [DOI] [PubMed] [Google Scholar]
- Appleyard R K. Segregation of Lambda Lysogenicity during Bacterial Recombination in Escherichia Coli K12. Genetics. 1954 Jul;39(4):429–439. doi: 10.1093/genetics/39.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeftinck F., Cunin R., Glansdorff N. Arginine gene duplications in recombination proficient and deficient strains of Escherichia coli K 12. Mol Gen Genet. 1974;132(3):241–253. doi: 10.1007/BF00269397. [DOI] [PubMed] [Google Scholar]
- Boram W., Abelson J. Bacteriophage Mu integration: on the orientation of the prophage. Virology. 1973 Jul;54(1):102–108. doi: 10.1016/0042-6822(73)90119-0. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Metlay M. Genetic mapping of prophage Mu. Virology. 1973 Jul;54(1):109–116. doi: 10.1016/0042-6822(73)90120-7. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. Sensitive mutants of bacteriophage lambda. Virology. 1961 May;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- Couturier M., Van Vliet F. Vegetative recombination in bacteriophage Mu-1. Virology. 1974 Jul;60(1):1–8. doi: 10.1016/0042-6822(74)90359-6. [DOI] [PubMed] [Google Scholar]
- Daniell E., Abelson J., Kim J. S., Davidson N. Heteroduplex structures of bacteriophage Mu DNA. Virology. 1973 Jan;51(1):237–239. doi: 10.1016/0042-6822(73)90385-1. [DOI] [PubMed] [Google Scholar]
- Daniell E., Roberts R., Abelson J. Mutations in the lactose operon caused by bacteriophage Mu. J Mol Biol. 1972 Aug 14;69(1):1–8. doi: 10.1016/0022-2836(72)90019-8. [DOI] [PubMed] [Google Scholar]
- FRY B. A. Conditions for the infection of Escherichia coli with lambda phage and for the establishment of lysogeny. J Gen Microbiol. 1959 Dec;21:676–684. doi: 10.1099/00221287-21-3-676. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A., Couturier M. Mu-1 promoted integration of a -gal phage in the chromosome of E. coli. Mol Gen Genet. 1971;113(4):367–370. doi: 10.1007/BF00272338. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A. Isolation of conditional defective mutants of temperate phage Mu-1 and deletion mapping of the Mu-1 prophage. Virology. 1973 Jul;54(1):117–124. doi: 10.1016/0042-6822(73)90121-9. [DOI] [PubMed] [Google Scholar]
- Folk W. R., Berg P. Duplication of the structural gene for glycyl-transfer RNA synthetase in Escherichia coli. J Mol Biol. 1971 Jun 14;58(2):595–610. doi: 10.1016/0022-2836(71)90374-3. [DOI] [PubMed] [Google Scholar]
- GLANSDORFF N. TOPOGRAPHY OF COTRANSDUCIBLE ARGININE MUTATIONS IN ESCHERICHIA COLI K-12. Genetics. 1965 Feb;51:167–179. doi: 10.1093/genetics/51.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. The integration and excision of the bacteriophage lambda genome. Cold Spring Harb Symp Quant Biol. 1968;33:735–747. doi: 10.1101/sqb.1968.033.01.084. [DOI] [PubMed] [Google Scholar]
- HORIUCHI T., HORIUCHI S., NOVICK A. The genetic basis of hyper-synthesis of beta-galactosidase. Genetics. 1963 Feb;48:157–169. doi: 10.1093/genetics/48.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. W., Foulds J., Soll L., Berg P. Instability of a missense suppressor resulting from a duplication of genetic material. J Mol Biol. 1969 Feb 14;39(3):563–581. doi: 10.1016/0022-2836(69)90146-6. [DOI] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Davidson N. Electron microscope heteroduplex study of the heterogeneity of Mu phage and prophage DNA. Virology. 1974 Mar;58(1):229–239. doi: 10.1016/0042-6822(74)90157-3. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Davidson N. Structure of inserted bacteriophage Mu-1 DNA and physical mapping of bacterial genes by Mu-1 DNA insertion. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2823–2827. doi: 10.1073/pnas.69.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOW B., WOOD T. H. A QUICK AND EFFICIENT METHOD FOR INTERRUPTION OF BACTERIAL CONJUGATION. Genet Res. 1965 Jul;6:300–303. doi: 10.1017/s001667230000416x. [DOI] [PubMed] [Google Scholar]
- Lederberg J. Gene Recombination and Linked Segregations in Escherichia Coli. Genetics. 1947 Sep;32(5):505–525. doi: 10.1093/genetics/32.5.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambach A., Brachet P. Sélection de mutants du bactériophage lambda incapables de se répliquer. C R Acad Sci Hebd Seances Acad Sci D. 1971 Jan 4;272(1):149–152. [PubMed] [Google Scholar]
- Schroeder W., Bade E. G., Delius H. Participation of Escherichia coli DNA in the replication of temperate bacteriophage Mu1. Virology. 1974 Aug;60(2):534–542. doi: 10.1016/0042-6822(74)90347-x. [DOI] [PubMed] [Google Scholar]
- Schröder W., van de Putte P. Genetic study of prophage excision with a temperature inducible mutant of Mu-1. Mol Gen Genet. 1974 May 21;130(2):99–104. doi: 10.1007/BF00269081. [DOI] [PubMed] [Google Scholar]
- TAYLOR A. L. BACTERIOPHAGE-INDUCED MUTATION IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1963 Dec;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R., Leurs C., Dambly C., Parmentier D., Lambert L., Brachet P., Lefebvre N., Mousset S., Porcheret J., Szpirer J. Isolation and characterization of new sus (amber) mutants of bacteriophage lambda. Mutat Res. 1967 Nov-Dec;4(6):735–741. doi: 10.1016/0027-5107(67)90082-6. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Faelen M. Connecting two unrelated DNA sequences with a Mu dimer. Nat New Biol. 1973 Mar 7;242(114):1–4. doi: 10.1038/newbio242001a0. [DOI] [PubMed] [Google Scholar]
- Waggoner B. T., González N. S., Taylor A. L. Isolation of heterogeneous circular DNA from induced lysogens of bacteriophage Mu-1. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1255–1259. doi: 10.1073/pnas.71.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffelman C. A., Westmaas G. C., van de Putte P. Similarity of vegetative map and prophage map of bacteriophage Mu-1. Virology. 1973 Jul;54(1):125–134. doi: 10.1016/0042-6822(73)90122-0. [DOI] [PubMed] [Google Scholar]
- Wijffelman C. A., Westmaas G. C., van de Putte P. Vegetative recombination of bacteriophage Mu-1 in Escherichia coli. Mol Gen Genet. 1972;116(1):40–46. doi: 10.1007/BF00334258. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Zeldis J. B., Bukhari A. I., Zipser D. Orientation of prophage Mu. Virology. 1973 Sep;55(1):289–294. doi: 10.1016/s0042-6822(73)81033-5. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Gruijthuijsen M. Chromosome mobilization and integration of F-factors in the chromosome of RecA strains of E. coli under the influence of bacteriophage Mu-1. Mol Gen Genet. 1972;118(2):173–183. doi: 10.1007/BF00267086. [DOI] [PubMed] [Google Scholar]