Abstract
Cell-free particulate fractions of extracts from the obligate methylotroph Methylococcus capsulatus catalyze the reduced nicotinamide adenine dinucleotide (NADH) and O2-dependent oxidation of methane (methane hydroxylase). The only oxidation product detected was formate. These preparations also catalyze the oxidation of methanol and formaldehyde to formate in the presence or absence of phenazine methosulphate with oxygen as the terminal electron acceptor. Methane hydroxylase activity cannot be reproducibly obtained from disintegrated cell suspensions even though the whole cells actively respired when methane was presented as a substrate. Varying the disintegration method or extraction medium had no significant effect on the activities obtained. When active particles were obtained, hydroxylase activity was stable at 0 C for days. Methane hydroxylase assays were made by measuring the methane-dependent oxidation of NADH by O2. In separate experiments, methane consumption and the accumulation of formate were also demonstrated. Formate is not oxidized by these particulate fractions. The effects of particle concentration, temperature, pH, and phosphate concentration on enzymic activity are described. Ethane is utilized in the presence of NADH and O2. The stoichiometric relationships of the reaction(s) with methane as substrate were not established since (i) the presumed initial product, methanol, is also oxidized to formate, and (ii) the contribution that NADH oxidase activity makes to the observed consumption of reactants could not be assessed in the presence of methane. Studies with known inhibitors of electron transport systems indicate that the path of electron flow from NADH to oxygen is different for the NADH oxidase, methane hydroxylase, and methanol oxidase activities.
Full text
PDF

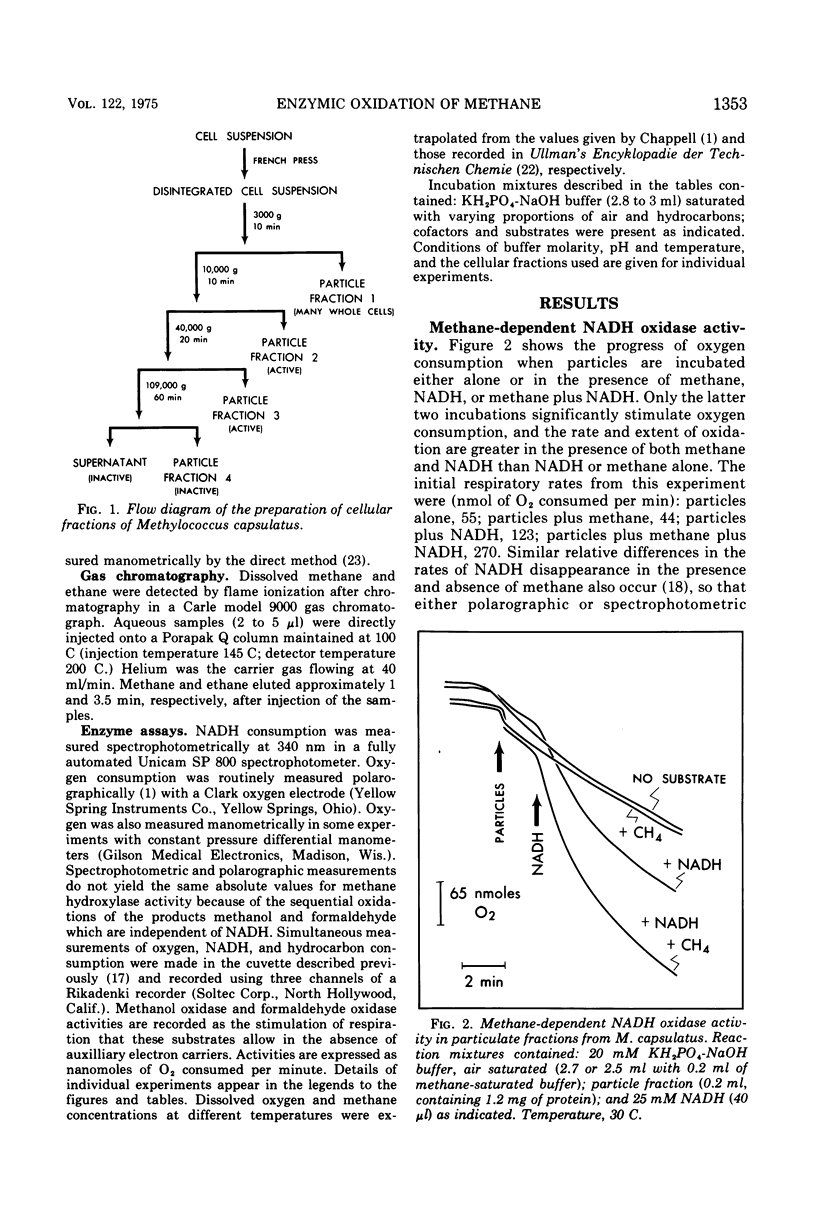

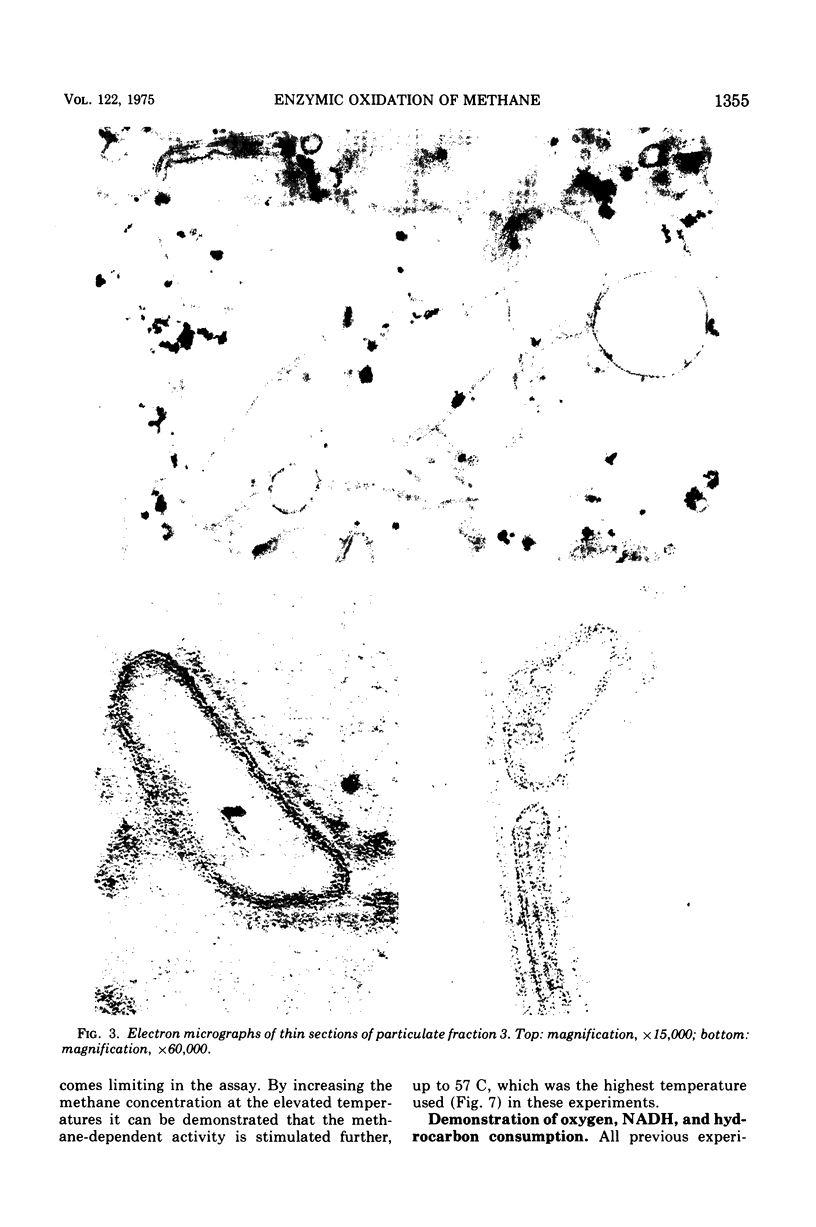
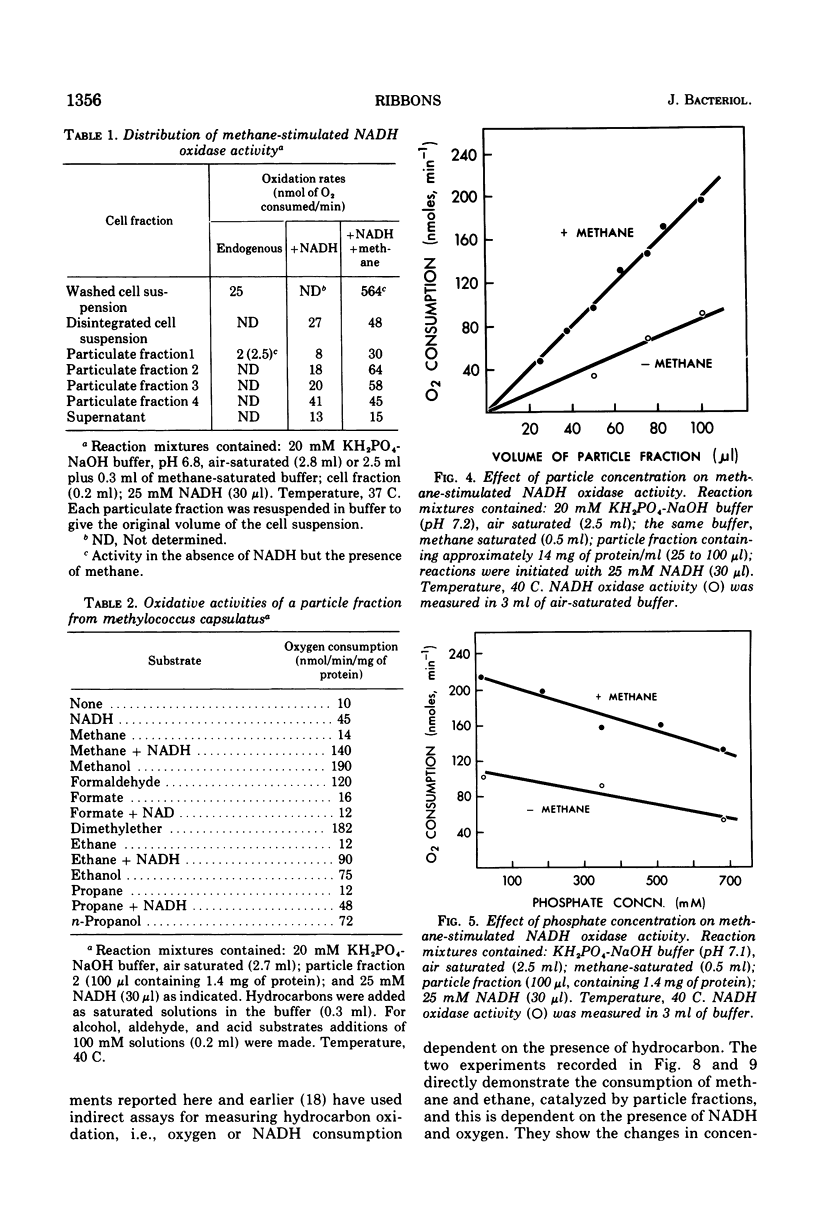
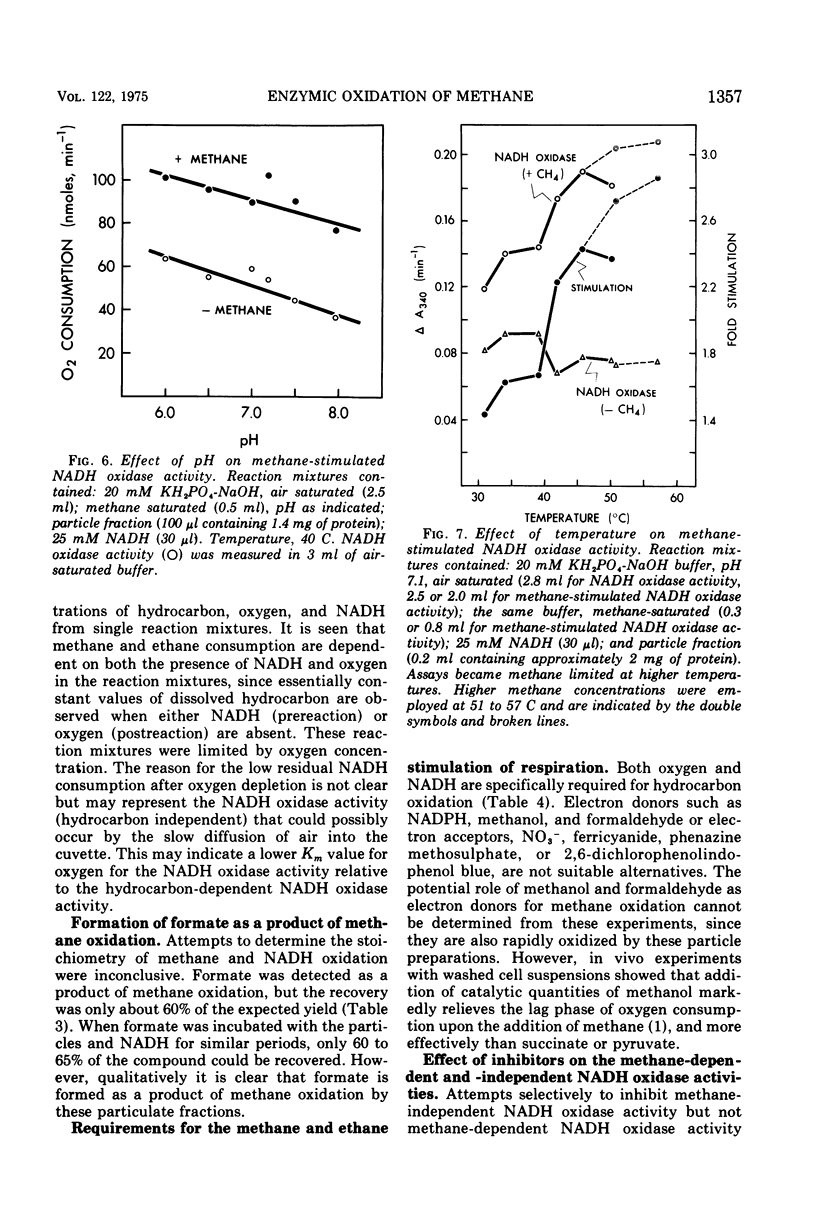
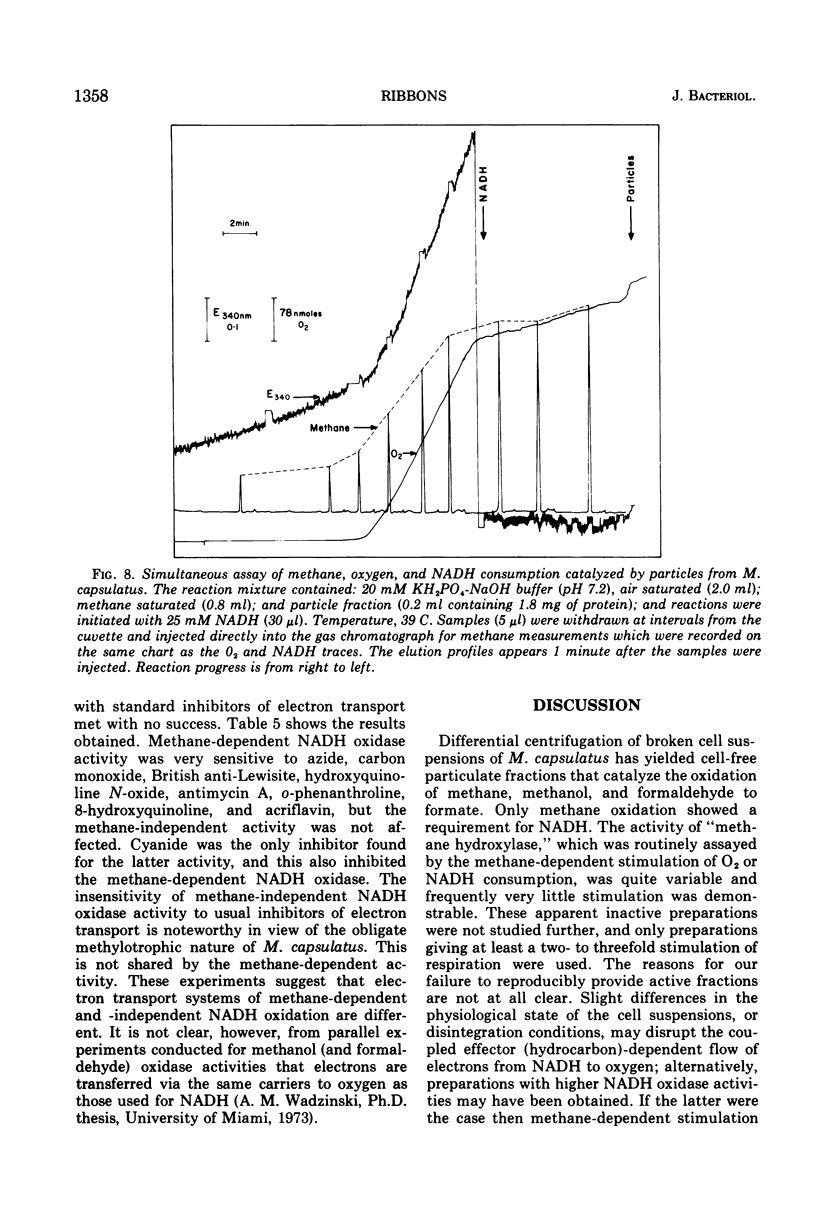
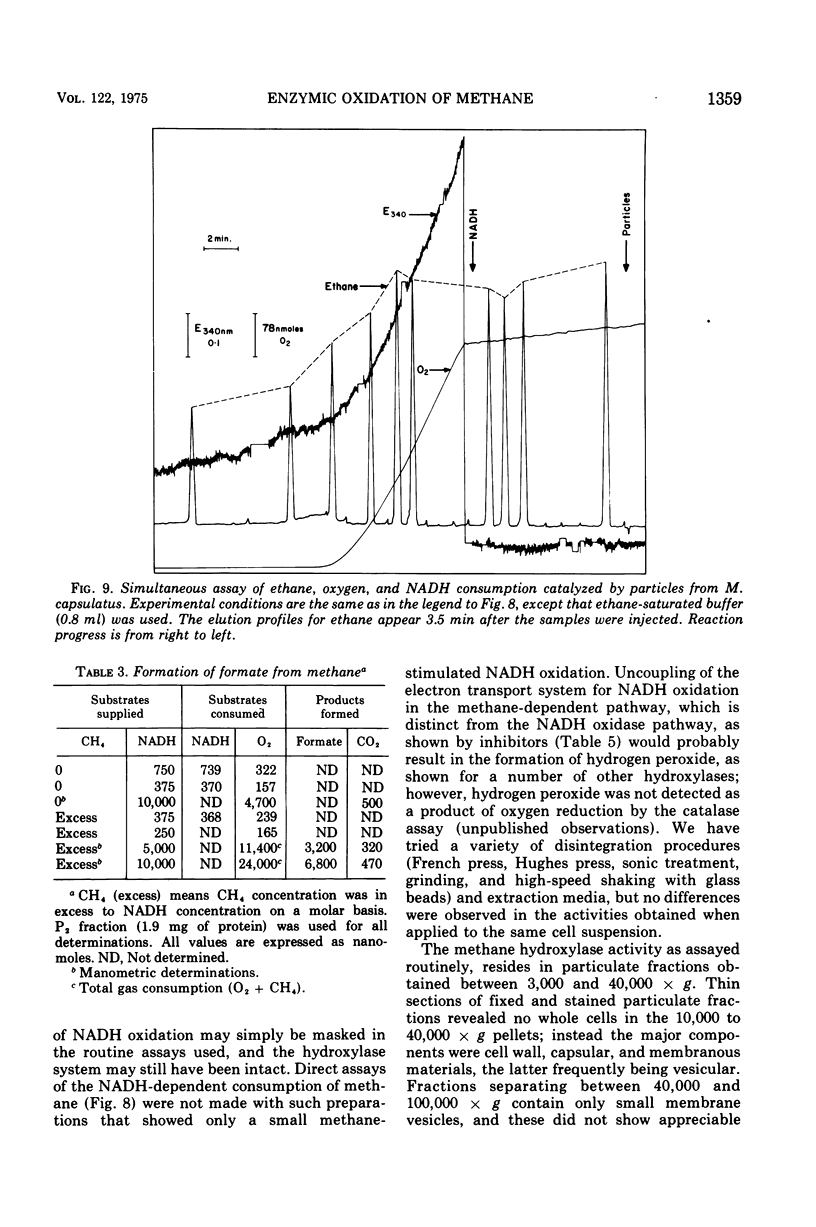
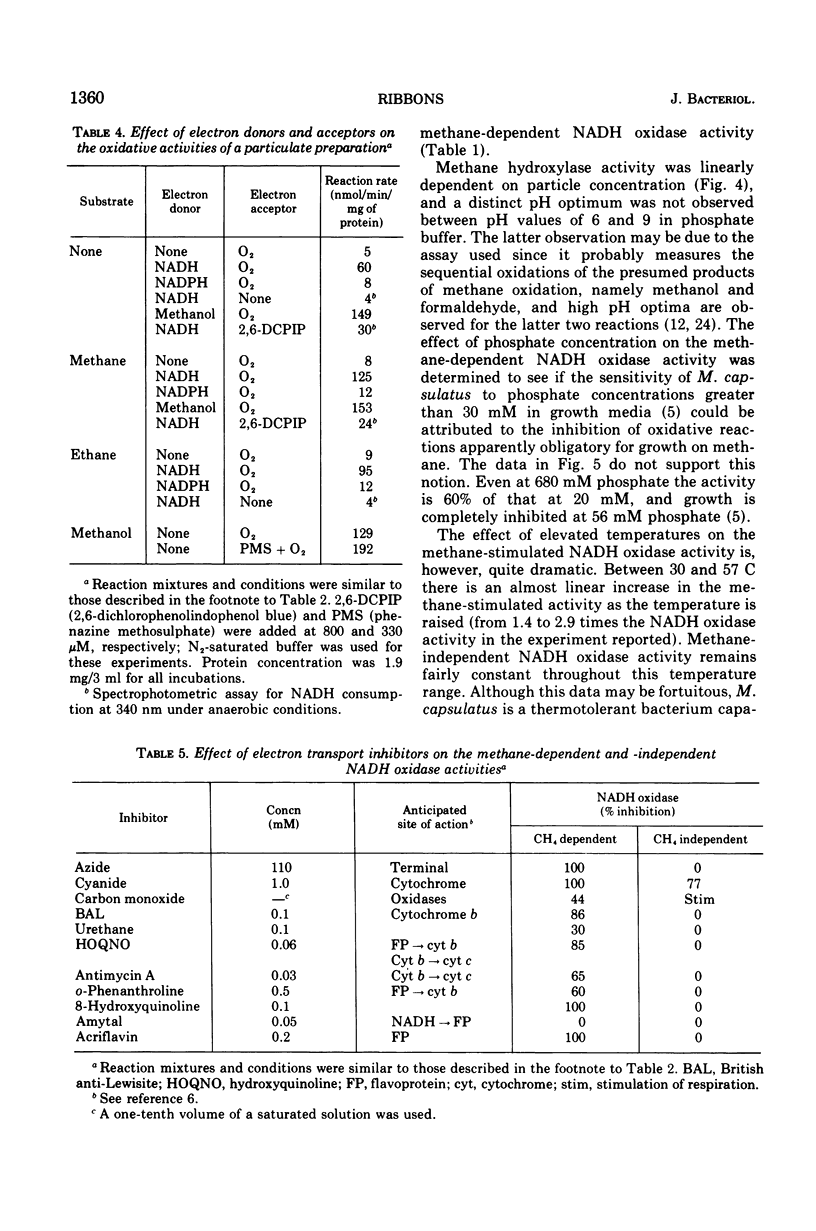



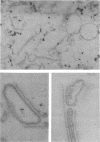
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Davies S. L., Whittenbury R. Fine structure of methane and other hydrocarbon-utilizing bacteria. J Gen Microbiol. 1970 May;61(2):227–232. doi: 10.1099/00221287-61-2-227. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Carbon monoxide-stimulated respiration in methane-utilizing bacteria. FEBS Lett. 1974 Apr 15;41(1):94–98. doi: 10.1016/0014-5793(74)80962-2. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Davis R. H. A methane-dependent coccus, with notes on classification and nomenclature of obligate, methane-utilizing bacteria. J Bacteriol. 1966 May;91(5):1924–1931. doi: 10.1128/jb.91.5.1924-1931.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood J. H., Pirt S. J. Quantitative aspects of growth of the methane oxidizing bacterium Methylococcus capsulatus on methane in shake flask and continuous chemostat culture. J Appl Bacteriol. 1972 Dec;35(4):597–607. doi: 10.1111/j.1365-2672.1972.tb03741.x. [DOI] [PubMed] [Google Scholar]
- Higgins I. J., Quayle J. R. Oxygenation of methane by methane-grown Pseudomonas methanica and Methanomonas methanooxidans. Biochem J. 1970 Jun;118(2):201–208. doi: 10.1042/bj1180201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp M. B. Hexose phosphate synthase from Methylcoccus capsulatus makes D-arabino-3-hexulose phosphate. Biochem J. 1974 Apr;139(1):129–134. doi: 10.1042/bj1390129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp M. B., Quayle J. R. Microbial growth on C1 compounds. Uptake of [14C]formaldehyde and [14C]formate by methane-grown Pseudomonas methanica and determination of the hexose labelling pattern after brief incubation with [14C]methanol. Biochem J. 1967 Jan;102(1):94–102. doi: 10.1042/bj1020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEADBETTER E. R., FOSTER J. W. Incorporation of molecular oxygen in bacterial cells utilizing hydrocarbons for growth. Nature. 1959 Oct 31;184(Suppl 18):1428–1429. doi: 10.1038/1841428a0. [DOI] [PubMed] [Google Scholar]
- Lawrence A. J., Kemp M. B., Quayle J. R. Synthesis of cell constituents by methane-grown Methylococcus capsulatus and Methanomonas methanooxidans. Biochem J. 1970 Feb;116(4):631–639. doi: 10.1042/bj1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Bose H. R., Mandy W. J., Hoare D. S. Physiological studies of methane- and methanol-oxidizing bacteria: comparison of a primary alcohol dehydrogenase from Methylococcus capsulatus (Texas strain) and Pseudomonas species M27. J Bacteriol. 1972 May;110(2):570–577. doi: 10.1128/jb.110.2.570-577.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Hoare D. S. Physiological studies of methane and methanol-oxidizing bacteria: oxidation of C-1 compounds by Methylococcus capsulatus. J Bacteriol. 1971 Jul;107(1):187–192. doi: 10.1128/jb.107.1.187-192.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbons D. W., Harrison J. E., Wadzinski A. M. Metabolism of single carbon compounds. Annu Rev Microbiol. 1970;24:135–158. doi: 10.1146/annurev.mi.24.100170.001031. [DOI] [PubMed] [Google Scholar]
- Ribbons D. W., Michalover J. L. Methane oxidation by cell-free extracts of Methylococcus capsulatus. FEBS Lett. 1970 Nov 9;11(1):41–44. doi: 10.1016/0014-5793(70)80487-2. [DOI] [PubMed] [Google Scholar]
- Suzuki I., Kwok S. C. Cell-free ammonia oxidation by Nitrosomonas europaea extracts: effects of polyamines, Mg2+ and albumin. Biochem Biophys Res Commun. 1970 Jun 5;39(5):950–955. doi: 10.1016/0006-291x(70)90416-x. [DOI] [PubMed] [Google Scholar]
- Wadzinski A. M., Ribbons D. W. Oxidation of C1 compounds by particulate fractions from Methylococcus capsulatus: properties of methanol oxidase and methanol dehydrogenase. J Bacteriol. 1975 Jun;122(3):1364–1374. doi: 10.1128/jb.122.3.1364-1374.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S. W., Asbell M. A., Valois F. W. Ammonia oxidation by cell-free extracts of Nitrosocystis oceanus. Biochem Biophys Res Commun. 1970 Mar 27;38(6):1113–1119. doi: 10.1016/0006-291x(70)90354-2. [DOI] [PubMed] [Google Scholar]
- Whittenbury R., Phillips K. C., Wilkinson J. F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970 May;61(2):205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]