Abstract
The human cytomegalovirus tegument protein pp71 localizes to the nucleus immediately upon infection, and functions to initiate viral gene expression. Analysis of a series of random insertion mutations revealed that sequences within the mid-region (MR) of pp71 are important for localization to the nucleus. Fusion of MR sequences with eGFP revealed that amino acids 94 to 300 were sufficient to target proteins to the nucleus. Random substitution mutagenesis within this domain resulted in two double-substitution mutants, pp71P203T/T223M and pp71T228M/L275Q, with a predominantly cytoplasmic localization. Disruption of nuclear targeting resulted in relocalization of the fusion proteins to a distinct perinuclear region. Using tandem mass spectrometry, we determined that threonine 223 can be phosphorylated. Mutation of this residue to a phosphomimetic amino acid resulted in abrogation of nuclear targeting. These results strongly suggest that the intracellular trafficking of pp71 is regulated by phosphorylation.
Keywords: HCMV, nuclear trafficking, phosphorylation, tegument, mass spectrometry
Introduction
Human cytomegalovirus (HCMV), a member of the beta subgroup of herpesviruses, is a ubiquitous human pathogen, which infects 40 to 60% of the human population in developed countries (Mocarski, 2007). HCMV infection is usually asymptomatic and maintains a latent state for the lifetime of the individual. However, primary infection or reactivation of a latent infection can cause severe morbidity and mortality in immunocompromised individuals such as AIDS patients and organ transplant patients (Griffiths, 2006; Snydman, 2006). The HCMV genome is a linear double-stranded DNA of approximately 235kb, which is enclosed in an icosahedral capsid surrounded by a tegument layer, and finally enveloped in a lipid bilayer embedded with a number of virus-encoded glycoproteins (Mocarski, 2007). HCMV gene expression is separated into three temporal phases termed immediate early (IE), early and late (Fortunato and Spector, 1999; Mocarski, 2007). Expression of immediately early genes begins immediately after virus entry, and encodes critical viral proteins, which regulate viral early and late gene expression (Anders, Kerry, and Pari, 2007; Mocarski, 2007).
The tegument layer of HCMV consists of at least 25 viral proteins, and the majority of the tegument proteins are phosphoproteins (Baldick and Shenk, 1996; Mocarski, 2007; Varnum et al., 2004). As might be predicted, tegument proteins can play critical roles in virus assembly and particle stability (AuCoin et al., 2006; Britt et al., 2004; Jones and Lee, 2004; Lorz et al., 2006; Munger, Yu, and Shenk, 2006; Silva et al., 2003). In addition, a number of the HCMV tegument proteins have key functions in the initiation of viral infection. For example, the tegument protein pp65 is the most abundant protein made during replication (Gibson, 1983) and is thought to be involved in immune evasion by modulating interferon signaling (Abate, Watanabe, and Mocarski, 2004; Browne and Shenk, 2003). The 559 amino acid pp71 protein, which is encoded by the UL82 gene, was the first HCMV tegument protein to be characterized as a transactivator, activating the major immediate early promoter and initiating viral gene expression (Bresnahan and Shenk, 2000; Chau, Vanson, and Kerry, 1999; Liu and Stinski, 1992). More recently, a number of other tegument proteins including pIRS1/pTRS1 (Iskenderian et al., 1996; Kerry et al., 1996; Romanowski and Shenk, 1997), pUL26 (Stamminger et al., 2002), ppUL35 (Schierling et al., 2004) and ppUL69 (Winkler, Rice, and Stamminger, 1994) have been added to the list of tegument transactivators of viral gene expression. The pIRS1/pTRS1 proteins have also been shown to assist in the evasion of the interferon response by relocalizing PKR to the nucleus (Hakki et al., 2006).
Upon fusion of the viral envelope with the cell membrane, the viral capsid rapidly localizes to the nucleus (Ogawa-Goto et al., 2003). In addition, some viral tegument proteins including pp71 are targeted to the nucleus (Hensel et al., 1995; Hensel et al., 1996), consistent with their role in initiating viral gene expression. The pp71 protein can also act with the immediate early gene products to activate early promoters of HCMV (Chau, Vanson, and Kerry, 1999), and interacts with the cellular protein hDaxx to facilitate viral transcription at nuclear domains 10 (ND10) (Cantrell and Bresnahan, 2005; Cantrell and Bresnahan, 2006; Hofmann, Sindre, and Stamminger, 2002; Ishov, Vladimirova, and Maul, 2002; Preston and Nicholl, 2006; Saffert and Kalejta, 2006). Furthermore, pp71 has been shown to enhance the infectivity of viral DNA and thereby accelerate the process of virus infection (Baldick et al., 1997). In addition, pp71 can bind to the retinoblastoma protein (Rb), resulting in its degradation and enhanced progression through the cell cycle (Kalejta, Bechtel, and Shenk, 2003; Kalejta and Shenk, 2003a; Kalejta and Shenk, 2003b), although this interaction is not important for the transcriptional activity of pp71 (Cantrell and Bresnahan, 2005).
Recent studies show that the nuclear import of pp71 is inhibited when assessed in nonpermissive cell lines such as undifferentiated THP-1 cells, suggesting that the nuclear localization of this protein plays a key role in the initiation of viral infection and that nuclear import is a regulated process (Saffert and Kalejta, 2007). However, little is known about the mechanism by which pp71 localizes to the nucleus. In addition, at the late phase of viral infection, pp71 relocates to the cytoplasm in dense perinuclear structures (Hensel et al., 1996), presumably sites of virion tegumentation. These data suggest that pp71 contains multiple subcellular targeting sequences. In the current study, we investigated the regions of pp71 responsible for nuclear accumulation of this protein. Our studies reveal that sequences within the mid region (MR) of pp71 are responsible for nuclear localization, even though this region contains no sequences matching known NLS motifs (Christophe, Christophe-Hobertus, and Pichon, 2000; Dingwall and Laskey, 1991; Macara, 2001). The disruption of sequences required for nuclear targeting within the MR domain results in protein localization to a distinct perinuclear region that may correspond to tegumentation sites. Further, our studies suggest that the phosphorylation status of the threonine residue at amino acid position 223 plays a critical role in the regulation of pp71 subcellular trafficking.
Results
Identification of pp71 sequences involved in nuclear trafficking
Based on the fact that pp71 localizes predominantly to the nucleus upon viral infection and in transfected cells (Hensel et al., 1996; Hofmann, Sindre, and Stamminger, 2002; Marshall et al., 2002), we reasoned that pp71 contained an intrinsic nuclear localization signal, allowing it to be actively imported into the nucleus. Initial sequence analysis of the pp71 559 amino acid open reading frame revealed a novel basic region in the C-terminus from amino acids 362 to 383 that had some similarity to nuclear localization signals (Christophe, Christophe-Hobertus, and Pichon, 2000; Dingwall and Laskey, 1991; Macara, 2001). However, preliminary experiments showed that this region is not involved in nuclear targeting (data not shown). Other than this basic region, pp71 contains no sequences with homology to known nuclear localization signals. Thus, in order to map the protein domain(s) responsible for nuclear localization of pp71, random insertion mutagenesis was performed. This approach has been used successfully to map functional domains within the major immediate early proteins (Stenberg et al., 1990) and was selected to minimize the potential impact of the mutations on protein conformation. The mutants were generated by randomly inserting a 12bp oligonucleotide linker into the pp71 open reading frame, resulting in four amino acid insertions within the amino- terminal region of pp71 at amino acids 66, 154, 191, 215, 217, and 284. The pp71 mutants were then transfected into HFF cells along with a construct expressing the HCMV IE86 protein as a marker for nuclear staining, and examined by immunofluorescence and confocal microscopy. The IE86 protein localizes exclusively to the nucleus, co-localizing with the wild type pp71 protein which exhibits predominantly nuclear staining (Figure 1A & B). Proteins with insertions at amino acids 66, 154, and 191 retained a predominantly nuclear localization, similar to the wild type pp71 protein (Figure 1A & B). However insertions at amino acids 215, 217, or 284 resulted in a diffuse cytoplasmic staining (Figure 1A & B). These results indicate that sequences around amino acids 215–284, that we term the mid-region (MR), are important for the nuclear targeting of pp71.
FIGURE 1.
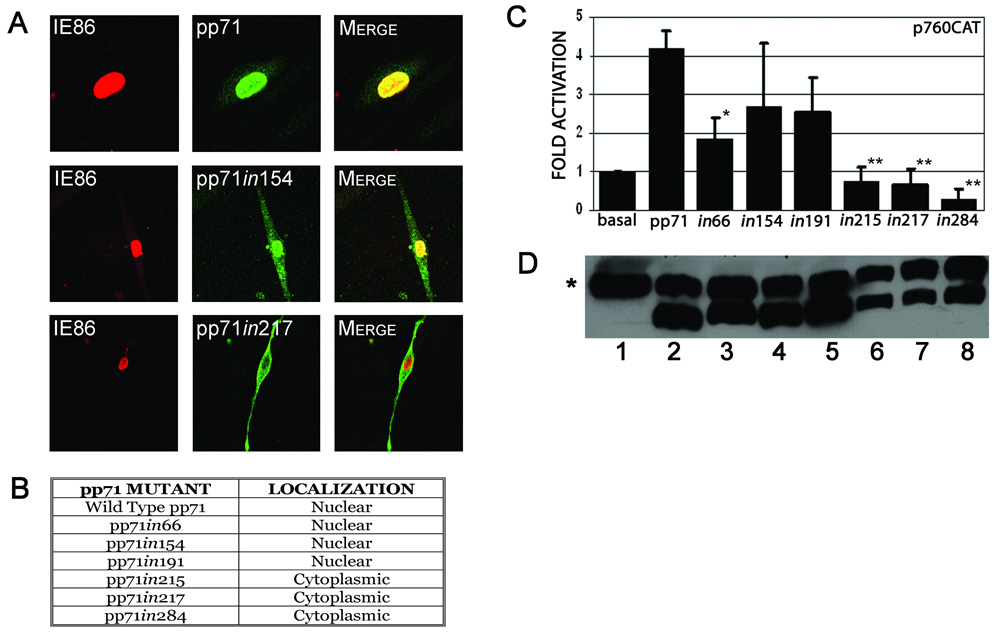
A & B. Subcellular localization of pp71 insertion mutants. HFF cells were co-transfected with constructs expressing the indicated pp71 insertion mutants and pMCRS86, and processed for immunofluorescence and confocal microscopy 48 hours after transfection using primary antibodies to the HA epitope tag of pp71 and IE86, with goat anti-mouse Alexa Fluor® 488 and anti-rabbit Alexa Fluor® 546 as secondary antibodies. The pp71 proteins are shown as green fluorescence, and IE86 shown as red fluorescence. The pp71in154 and pp71in217 proteins are shown as representatives of the insertion mutant phenotypes as indicated in B.
C. Transcriptional activity of pp71 insertion mutants. HFF cells were co-transfected with p760-CAT and pCMV71HAX or the indicated insertion mutants. Cells were harvested 48 hours post-transfection and assessed for CAT activity. Data is the average and standard deviation of a minimum of three experiments, normalized to promoter activity in the absence of pp71. Symbols: * significantly different from wild type, p<0.01; ** significantly different from wild type, p<0.001. D. Western blot analysis of the pp71 insertion mutants. HeLa cells were transfected with the following plasmids: pSI (lane 1), pSI71HAX (lane 2), pSI71HAXin66 (lane 3), pSI71HAXin154 (lane 4), pSI71HAXin191 (lane 5), pSI71HAXin215 (lane 6), pSI71HAXin217 (lane 7) or pSI71HAXin284 (lane 8). Cells were harvested at 48 hours post-transfection and the extracts subjected to western blot analysis using an antibody to the HA-epitope tag. Symbol: * non-specific band used as a loading control.
As pp71 activates the major immediate early promoter (MIEP) and initiates viral infection (Baldick et al., 1997; Liu and Stinski, 1992), we investigated the transcriptional activity of these pp71 mutants using a promoter-reporter assay. HFF cells were co-transfected with a CAT reporter plasmid containing the immediately early promoter region and expression plasmids containing either full-length pp71 or the random insertion mutants. Cells were assessed for CAT activity 48 hours after transfection. This study showed that the insertion at aa 66 caused an approximately 50% loss of pp71 activity (Figure 1C). In contrast, the insertions at aa's 154 and 191 resulted in a slight but not significant (p>0.05) reduction in activity compared with the wild type protein. However insertion at aa’s 215, 217, or 284 completely abrogated the activity of pp71 (p<0.001). Similar results were obtained when pp71 enhancement of IE protein activation of the HCMV UL54 DNA polymerase promoter was assessed (data not shown). These analyses provide a functional assessment of pp71 nuclear targeting and indicate that the insertions at aa’s 215, 217, and 284 disrupt nuclear targeting to the extent that transactivation is abolished. One possible explanation for the reduced activity of these pp71 mutants is that the mutations affect protein stability. To assess this, we subcloned the pp71 mutants into the pSI vector which expresses proteins under the control of the SV40 promoter. This promoter is less responsive to auto-activation by pp71 than the MIEP (data not shown) and therefore provides a more accurate measure of steady-state pp71 protein levels. Western blot analysis demonstrated that the pp71 mutants were expressed at similar levels to the wild type protein (Figure 1D). This analysis confirms that the decrease in activity that we observe is most likely the result of the change in subcellular localization, and is not related to major effects on the stability of the protein.
Analysis of the random insertion mutants suggested that a relatively large domain was involved in the nuclear targeting of pp71. We therefore decided to utilize a random site-directed domain mutagenesis approach in order to further map the sequences within the MR responsible for nuclear targeting of pp71 as described in the Materials and Methods. This strategy was used to generate pp71 mutants between aa 200–300 in the pp71 open reading frame cloned into the pSI vector. Using the random mutagenesis approach, we identified five mutants that resulted in amino acid changes: three single substitution mutants, M238I, H269Q, I287S, and two double substitution mutants, P206T/T223M, T228M/L275Q. Localization of these mutants was assessed by immunofluorescence and confocal microscopy (Figure 2A). The three single substitution mutants did not affect the nuclear targeting of pp71. In contrast, the two double substitution mutants exhibited a reduction in the nuclear accumulation of pp71, displaying a diffuse cytoplasmic fluorescence with some punctate staining, similar to that of the pp71 random insertion mutants. We then investigated the transcriptional activity of these pp71 mutants as a functional assessment of pp71 nuclear targeting using a construct containing the MIEP upstream of the luciferase reporter gene which provides for increased sensitivity over the CAT reporter. As expected, the three single substitution mutants did not significantly affect pp71 transactivation, whereas the two double substitution mutants significantly reduced the activity of pp71 (p<0.05, Figure 2B). Again, similar results were obtained when activation of the UL54 promoter by the pp71 mutants was assessed (data not shown). These data demonstrate that nuclear localization is important for the activity of pp71. In addition, these results clearly indicate that the sequences required for nuclear localization of pp71 do not correspond to a conventional nuclear localization signal. One possibility is that interactions with a cellular protein via the mid-region of pp71 may be required for nuclear transport. Preliminary studies showed that interaction with the Daxx protein was unaffected by these mutations (data not shown), suggesting that this protein is not involved in the nuclear transport of pp71.
FIGURE 2.
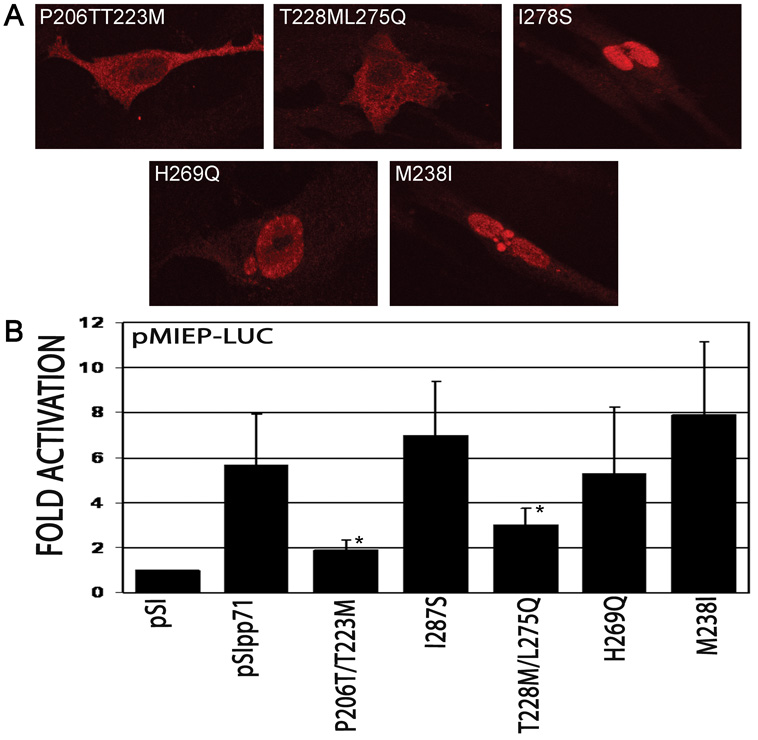
A. Subcellular localization of pp71 mutants generated by random mutagenesis. HFF cells were transfected with constructs expressing the indicated pp71 mutants, and processed for immunofluorescence and confocal microscopy 48 hours after transfection using antibodies to the HA epitope tag. B. Transcriptional activity of pp71 mutants. HFF cells were transfected with pMIEP-LUC and pSI71HAX or the indicated mutants. Cells were harvested 48 hours post-transfection and assessed for luciferase activity. Data is the average and standard deviation of a minimum of three experiments, normalized to promoter activity in the absence of pp71. Symbol: * significantly different from wild type, p<0.05.
To determine if the mid region (MR) is sufficient for nuclear targeting, a region containing aa’s 188–300 was cloned in frame with the amino-terminus of eGFP (188MR- eGFP) and the localization of the fusion protein examined (Figure 3). As expected, eGFP alone can be detected throughout the cell (Figure 3A). Surprisingly, the 188MR-eGFP fusion protein did not localize in the nucleus, but was distributed in a distinct punctate perinuclear pattern (Figure 3B) suggesting that while sequences within the MR are important for nuclear localization, these sequences are not sufficient to independently direct nuclear targeting. To assess if sequences adjacent to the MR domain participated in the nuclear localization of pp71, we generated additional eGFP fusion protein constructs containing sequences from aa 94–300 (94MR-eGFP) and 62–300 (62MR-eGFP). Confocal microscopy analysis revealed that amino acids 94 to 300 of pp71 were sufficient to mediate nuclear targeting (Figure 3C), suggesting that sequences adjacent to the MR influence recognition of the pp71 nuclear localization signal. To identify the location of the perinuclear fluorescence observed with the 188MR-eGFP fusion protein, HFF cells were transfected with this construct and co-stained with a series of subcellular markers including a panel of Golgi-related antibodies. These studies showed that the perinuclear fluorescence associated with 188MR-GFP colocalized with syntaxin 6 (data not shown) and syntaxin 11 (Figure 4A), SNAREs that are localized to the TGN-late endosomal compartment (Teng, Wang, and Tang, 2001). We also observed partial co-localization with other late TGN markers, including GS27 and Vti1a (data not shown). This staining pattern is reminiscent of that observed for pp71 at the late times of viral infection (Figure 3E & 7A (Hensel et al., 1996)) and may correspond to the site of cytoplasmic tegumentation (Mettenleiter, 2004).
FIGURE 3. Subcellular localization of MR-eGFP fusion proteins.
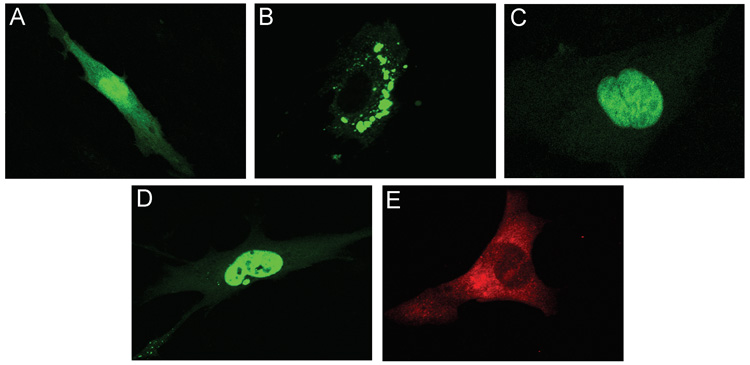
HFF cells were transfected with peGFP-N2 (A), p188MR-eGFP (B), p94MR-eGFP (C), or p62MR-eGFP (D) and processed for confocal microscopy 48 hours after transfection. HFF cells infected with HCMV were processed for immunofluoresence at 72 hours post-infection and subjected to immunofluorescence using a monoclonal antibody to pp71 (E).
FIGURE 4. Characterization of pp71 cytoplasmic localization.
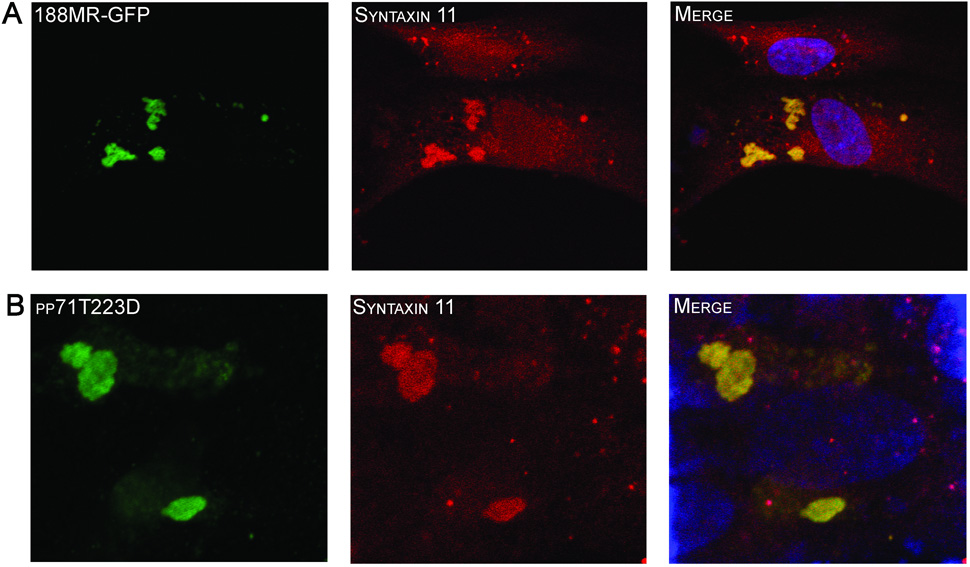
HFF cells were transfected constructs expressing p188MR-eGFP (A) or pp71T223D (B) and processed for confocal microscopy 48 hours after transfection. Cells transfected with the construct expressing pp71T223D were stained using and antibody to the HA-epitope tag (green) and both were costained with an antibody to syntaxin 11 (red). Nuclei were stained with TOTO3 dye (blue).
FIGURE 7. Subcellular localization of pp71 at the late stages of virus infection.
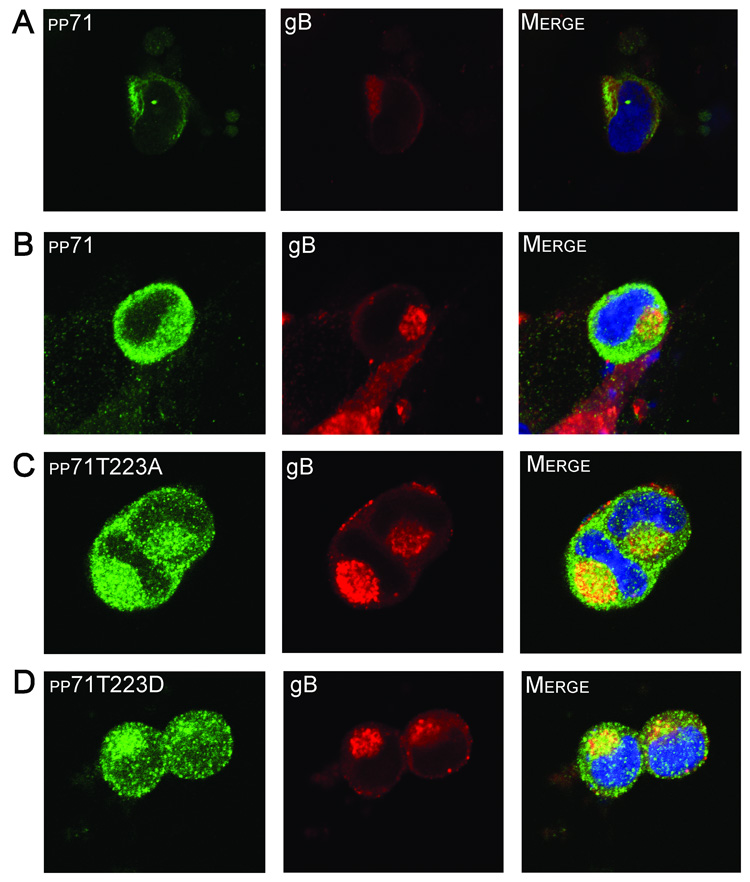
HFF cells were infected with HCMV strain AD169 (A) and fixed at 96 hours post-infection. The cells were then subjected to immunofluorescence using a monoclonal antibody to pp71 to or a polyclonal antibody to gB. Alternatively, HFF cells were transfected with constructs expressing pp71 (B), pp71T223A (C), or pp71T223D (D) and subsequently infected with HCMV strain AD169. Cells were fixed at 96 hours post-infection and subjected to immunofluorescence using an antibody to the HA epitope tag of pp71 (green) and the gB protein (red). Nuclei were stained with TOTO3 dye (blue).
Phosphorylation regulates pp71 trafficking
Using the random mutagenesis strategy, we identified two threonine residues that play a role in the subcellular trafficking of pp71 (Figure 2). Based on these findings, we speculated that phosphorylation events may be involved in regulating the subcellular localization of pp71. To address this hypothesis, we purified pp71 protein from transfected HeLa cells using S-tag affinity chromatography. The resultant protein was subjected to phosphopeptide mapping using LC-MS/MS (Durkin et al., 2006). Using this approach, we detected phosphorylation of nine residues within the pp71 open reading frame (Figure 5A). Within the MR, a single residue, the threonine residue at amino acid position 223 (T223) was found to be phosphorylated. A rapid semi-quantitative assessment of the total tryptic peptides derived from the wild type pp71 protein analyzed by mass spectrometry revealed that 25% of the peptides were phosphorylated at position T223, indicating that the majority of pp71 in transfected cells was not phosphorylated at this site. To assess the effect of T223 phosphorylation on pp71 trafficking, we mutated this residue to either alanine in order to block phosphorylation, or an aspartic acid to mimic phosphorylation within the context of the full-length pp71 protein. Assessment of the T223 mutants in transfection assays showed that mutation of amino acid 223 to an alanine (T223A) resulted in a reduction of the activity of pp71 (Figure 5B). However, mutation of this residue to aspartic acid (T223D) reduced transcriptional enhancement to a level not significantly different to the promoter activity observed in the absence of pp71 (p = 0.67). We next assessed the effect of the T223 mutations on the localization of pp71 (Figure 5C). The T223A mutant displayed an identical localization to the wild type pp71 protein, colocalizing with ND10 domains within the nucleus. In contrast, substitution of T223 with the phosphomimetic amino acid abrogated nuclear import of pp71, resulting in a punctate cytoplasmic localization. Analysis of the cytoplasmic localization of the pp71T223D protein revealed colocalization with syntaxin 11 (Figure 4B), consistent with the results obtained with the 188MR-GFP fusion protein (Figure 4A).
FIGURE 5.
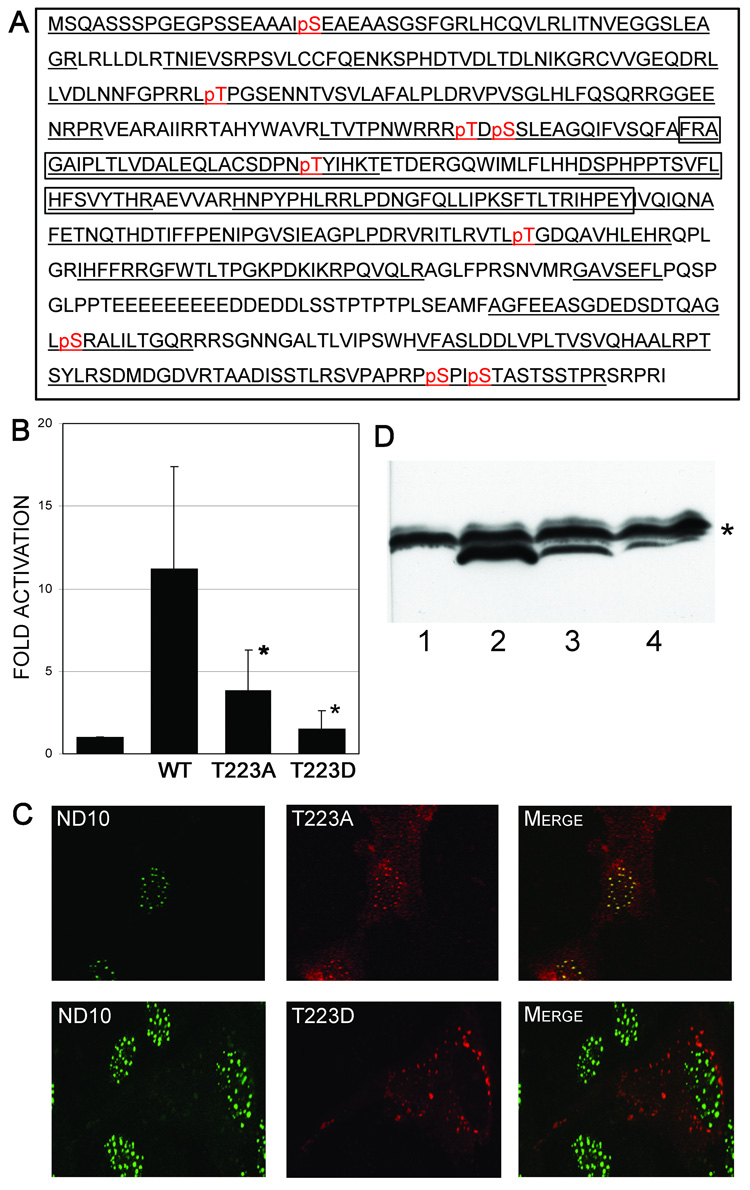
A. Phosphopeptide map of pp71. A compilation of the results obtained with MS/MS analysis of phosphorylation sites of the pp71 tegument protein. The total analyzed sequence is underlined, and the phosphorylated residues are indicated in red. The region corresponding to the Mid-Region (MR, aa 200–300) is boxed. B. Transcriptional activity of pp71 phosphorylation site mutants. HFF cells were transfected with pMIEP-LUC and pSI71HAX or the indicated mutants. Cells were harvested 48 hours post-transfection and assessed for luciferase activity. Data is the average and standard deviation of a minimum of three experiments and is expressed relative to the activity of the wild type (WT) pp71 protein. Symbol: * significantly different from wild type, p<0.05. C. Subcellular localization of pp71 phosphorylation site mutants. HFF cells were transfected with constructs expressing the indicated pp71 mutants, and processed for immunofluorescence and confocal microscopy 48 hours after transfection using antibodies to the HA epitope tag and nuclear ND10 domains. The pp71 proteins are shown as red fluorescence, and ND10 domains shown as green fluorescence. D. Western Blot analysis of pp71 phosphorylation site mutants. HeLa cells were transfected with constructs expressing eGFP (lane 1), pp71 (lane 2), pp71T223A (lane 3), or pp71T223D (lane 4). Cells were harvested at 48 hours post-transfection and the extracts subjected to western blot analysis using an antibody to the HA-epitope tag. Symbol: * non-specific band used as a loading control.
Analysis of the transfected cells by western blot using an antibody to the HA-tag showed lower levels of protein in cells transfected with pp71 containing either mutation of threonine 223 (Figure 5D). The decrease in protein levels could arise from the reduction in activity of the mutants, resulting in diminished auto-regulation of the SV40 promoter construct used to express pp71. However, the SV40 promoter displays minimal (approximately than 2-fold) activation by pp71 which would not account for the marked decrease in protein levels observed. Alternatively, this could indicate a decrease in the stability of the protein due to a conformational change resulting from the mutation of this residue. To assess this, we inserted the SV40 nuclear localization signal (NLS) into the amino terminus of both the wild type and pp71T223D open reading frames in order to independently target the protein to the nucleus. This analysis showed that localization of the pp71T223D mutant to the nucleus via the SV40 NLS did not result in restoration of pp71 activity (Figure 6A). This finding is consistent with the phosphomimetic mutation causing a conformational change in pp71. Surprisingly, insertion of the SV40 NLS into the wild type protein significantly enhanced the activity of pp71 (Figure 6A). Thus, it is possible that the addition of the SV40 NLS enhances nuclear import of pp71, or blocks nuclear export of this protein. To confirm the nuclear localization of the pp71 proteins containing the SV40 NLS, we performed immunofluorescence studies (Figure 6B). This analysis showed that both the wild type pp71 and the pp71 T223D mutant proteins containing the SV40 NLS localized to the nucleus of transfected cells. However, both proteins displayed only partial colocalization with ND10 domains, suggesting that the SV40 NLS influences the subnuclear localization of pp71.
FIGURE 6.
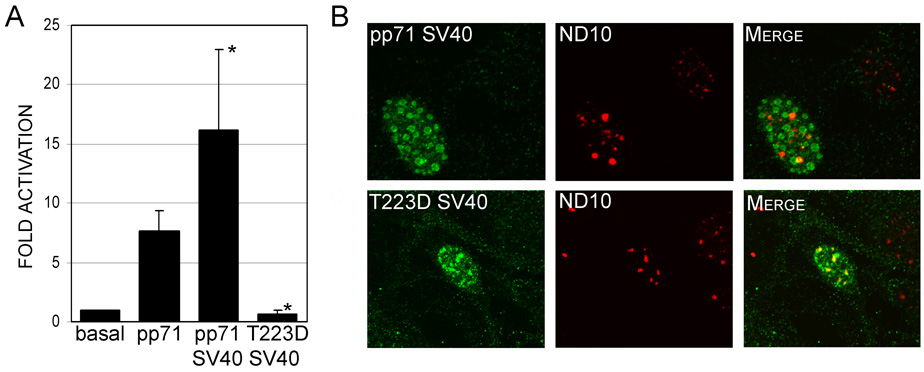
A. Transcriptional activity of pp71 mutants containing the SV40 NLS. HFF cells were transfected with pMIEP-LUC and pSI71HAX or the indicated mutants. Cells were harvested 48 hours post-transfection and assessed for luciferase activity. Data is the average and standard deviation of a minimum of three experiments and is expressed relative to the activity of the wild type (WT) pp71 protein. Symbol: * significantly different from wild type, p<0.01. B. Subcellular localization of pp71 mutants containing the SV40 NLS. HFF cells were transfected with constructs expressing the indicated pp71 mutants, and processed for immunofluorescence and confocal microscopy 48 hours after transfection using antibodies to the HA epitope tag and nuclear ND10 domains. The pp71 proteins are shown as green fluorescence, and ND10 domains shown as red fluorescence.
The trafficking of the pp71 tegument protein to the TGN-endosomal compartment when nuclear import is inhibited may be related to the finding that pp71 localizes to a punctate perinuclear region at the late stages of virus infection (Hensel et al., 1996), thought to be sites of tegumentation. To investigate this further we performed transfection/superinfection experiments to assess the localization of these pp71 mutants at the late stages of virus infection when virion morphogenesis is occurring. In our initial experiments, we confirmed that during infection, the transfected pp71 protein colocalized with endogenous pp71 (data not shown), and that endogenous pp71 localizes adjacent to gB (Figure 7A), a marker for virus assembly compartments (Homman-Loudiyi et al., 2003; Jarvis et al., 2004; Sanchez et al., 2000). Analysis of the co-transfected cells showed that both the wild type pp71 and the pp71T223D protein localized to the cytoplasm with some evidence of colocalization with gB (Figure 7B & D), suggesting that cytoplasmic pp71 localization partially overlaps the virus assembly compartments (Homman-Loudiyi et al., 2003; Krzyzaniak, Mach, and Britt, 2007; Sanchez et al., 2000). Surprisingly, the pp71 protein containing the T223A substitution that is unable to be phosphorylated displayed a similar pattern of localization (Figure 7C), indicating that this mutant protein is capable of cytoplasmic trafficking at the late stages of virus infection. These studies indicate that viral and/or cellular proteins that may be modified by infection are important for pp71 nuclear export at the late stages of infection and suggest that nuclear import and export are controlled via different mechanisms.
Discussion
Transport of proteins into the nucleus occurs via two primary mechanisms, either via passive diffusion or by active transport through nuclear pore complexes (Macara, 2001). Based on its molecular weight, pp71 most likely contains a nuclear targeting signal allowing it to be actively transported into nucleus. In the current study, we determined that mutations within the mid-region (MR) of pp71, consisting of sequences from aa's 200–300 disrupted nuclear accumulation of pp71, suggesting that the MR domain is important for the nuclear import of this protein. This finding is consistent with studies using a transposon mutagenesis approach demonstrating that insertions within the central domain of pp71 disrupted nuclear trafficking (Willey, S. J. & Preston, C. M., personal communication). We further show that nuclear localization of pp71 is necessary, although not sufficient, for its transcriptional activities, consistent with the mechanism of pp71 transactivation via Daxx degradation (Cantrell and Bresnahan, 2005; Cantrell and Bresnahan, 2006; Hwang and Kalejta, 2007; Saffert and Kalejta, 2006; Saffert and Kalejta, 2007). It should be noted that we observed some variability in the assessment of pp71 activity that reflects the nature of these transient transfection studies, although the mutants that displayed disruption of pp71 nuclear trafficking all exhibited significantly lower activity than the wild type protein. In addition, our studies show that the threonine residue at position 223 within the MR plays a critical role in regulating the subcellular trafficking of pp71. Mass spectrometric analysis demonstrated that T223 can be phosphorylated, and our data strongly suggests that phosphorylation at this site influences nuclear import. Other studies have reported the effects of mutations within the MR domain of pp71, as this region contains one of two Daxx interaction domains, as well as the Rb-interaction motif (Cantrell and Bresnahan, 2005; Hofmann, Sindre, and Stamminger, 2002; Kalejta, Bechtel, and Shenk, 2003). These analyses demonstrated that mutation of aa 219 from cysteine to glycine that disrupts the Rb-interaction motif (Kalejta, Bechtel, and Shenk, 2003) had no affect on virus replication, suggesting that nuclear import was unaffected by this mutation (Cantrell and Bresnahan, 2005). Likewise, deletion of the Daxx interaction domain from aa 206–213, while disrupting pp71 transcriptional activation, had no affect on nuclear import (Cantrell and Bresnahan, 2005; Hofmann, Sindre, and Stamminger, 2002). Taken together, these studies support the notion that the effect of the pp71T223D mutation on nuclear import is specific to this residue and not a generic effect of mutations within the MR domain.
Analysis of the mid region fusion proteins demonstrated that sequences from aa 188–300 were unable to independently target proteins to the nucleus. These findings suggest that while the mid region of pp71 is necessary for the nuclear trafficking of pp71, it is not sufficient to target proteins to the nucleus. Further, these findings suggest that the higher-ordered structure of the pp71 protein or amino acids outside this essential region also play a role in subcellular targeting. Together, our data suggest that the pp71 mid region contains an unconventional, non-sequential, long nuclear targeting sequence that may be conformation sensitive. However, the mechanism by which the T223 phosphomimetic mutation affects nuclear import is currently unclear. Our current model is that phosphorylation of T223 alters the conformation of pp71, such that recognition of the nuclear import sequence is blocked. This effect is likely to be specific as other mutations and deletions within this region had no effect on pp71 nuclear import (Cantrell and Bresnahan, 2005; Hofmann, Sindre, and Stamminger, 2002). An alternative possibility is that phosphorylation of this residue affects the interaction with a cellular protein required for nuclear transport. Our preliminary experiments suggest that Daxx- interaction is not sufficient for nuclear import (data not shown). The identification of additional cellular pp71-interacting proteins would help to address this question.
A recent study has shown that nuclear import of pp71 can be inhibited upon infection of in vitro models of quiescent or "latent-like" infections (Saffert and Kalejta, 2007). Specifically, this study showed that nuclear import of pp71 was blocked in non-permissive THP-1 cells, and that this block contributes, at least in part, to the defect in virus replication. In contrast, pp71 nuclear import was unaffected when these cells are induced to their permissive states, suggesting that the block in pp71 nuclear import is a cell specific effect (Saffert and Kalejta, 2007). However, the precise mechanism by which pp71 nuclear import is regulated in these cell types is currently unclear. Our findings suggest that the phosphorylation of the threonine residue at position 223 may be important in this phenomenon. Specifically, our studies revealed that T223 can be phosphorylated by a cellular kinase, consistent with a cell specific effect. Analysis of the sequence surrounding T223 using Scansite software did not result in any matches to known cellular kinase recognition motifs, even when the search was conducted at a low stringency (Obenauer, Cantley, and Yaffe, 2003). Thus, it is unclear at this point which cellular kinase is involved in phosphorylating pp71 at this site. However, our findings are consistent with other studies that show both cellular and viral kinases associate with HCMV tegument proteins, and phosphorylation events modulate their function and/or localization. For example, the HCMV pp65 protein interacts with a cellular polo-like kinase, although the phosphorylation targets of this complex have yet to be defined (Gallina et al., 1999). The HCMV UL97 kinase may also be a component of the virion tegument (van Zeijl et al., 1997) and UL97 kinase activity is important for pp65 subcellular localization and virion morphogenesis (Prichard et al., 2005). Interestingly, the cellular phosphatase PP2A is also a component of HCMV particles (Michelson et al., 1996), suggesting the possibility of finely-tuned regulation of virion protein phosphorylation. Other studies demonstrated that of three differentially phosphorylated forms of the HCMV UL69 tegument protein, only one form is actually incorporated into the tegument of virion particles (Winkler and Stamminger, 1996). In addition, cyclin-dependent kinases can modulate the phosphorylation state and localization of UL69 and pp65 (Sanchez et al., 2007; Sanchez and Spector, 2006). Together, these studies demonstrate that differential phosphorylation can influence both subcellular localization and virion incorporation of the HCMV tegument proteins. In addition, HCMV infection can influence both the localization and activity of a number of cellular kinases (Hakki et al., 2006; Kudchodkar et al., 2004; Sanchez et al., 2004; Tamrakar, Kapasi, and Spector, 2005), suggesting the possibility of differential regulation of virion protein phosphorylation during the course of infection. Our findings are therefore consistent with a model whereby phosphorylation of pp71 modulates the subcellular localization of this tegument protein to enable efficient virus replication.
While our studies suggest that phosphorylation of T223 regulates the nuclear import of pp71, the mechanism that controls nuclear export at the late stages of infection is less clear. Recent studies suggest that cyclin-dependent kinases play a role in pp65 nuclear egress at the late stages of infection (Sanchez et al., 2007), and a similar mechanism may be involved in regulating pp71 cytoplasmic localization. Analysis of the 188MR-eGFP protein and the pp71T223D mutant revealed that pp71 also contains a sequence that targets proteins to a distinct perinuclear region that colocalizes with syntaxin 6 and syntaxin 11, markers of the TGN-late endosomal compartment (Teng, Wang, and Tang, 2001). Trafficking to this region is consistent with findings that pp71 is localized to a punctate perinuclear region at late times of infection (Hensel et al., 1996), that may corresponds to sites of viral tegumentation (Mettenleiter, 2004). Also of note are findings that in non-permissive cell lines where nuclear import of pp71 is blocked, the protein localizes to punctate perinuclear region (Saffert and Kalejta, 2007). The presence of a domain that enables perinuclear targeting within pp71 suggests that relocalization at the late stages of infection is an inherent property of this protein to allow for complete tegumentation during virion morphogenesis. Two models have been proposed by which nuclear tegument proteins become incorporated into the virion (Mettenleiter, 2004). Firstly, nuclear tegument proteins may associate with nascent nucleocapsids and traffic to the cytoplasm together with the nucleocapsids. Second, the viral tegument proteins may independently traffic to the cytoplasm to specific tegumentation sites where they then associate with the newly formed capsids. Our studies showing that the pp71T223D mutant protein can independently traffic to a specific cytoplasmic site that partially overlaps previously characterized virus assembly compartments would be consistent with this latter model. However, analysis of the location of the pp71T223A mutant at the late stages of infection demonstrating that viral and/or cellular proteins modified by virus infection can regulate the nuclear export of pp71 could be consistent with either model. Further analysis of these mutant proteins in the context of virus infection will be necessary to distinguish between these two models.
In summary, we have identified an unconventional non-sequential NLS sequence in the mid region of pp71 which is sufficient for targeting pp71 to the nucleus. This NLS appears to be conformational sensitive, and does not fit any known consensus motifs. In addition, sequences targeting pp71 to a distinct perinuclear region were identified. Further, our studies strongly suggest that differential phosphorylation events modulate the subcellular localization of pp71. Our observations provide novel insight into the mechanisms that regulate the trafficking of this key viral protein, and offer useful tools for a full understanding of the functions of pp71 in HCMV infection, assembly and egress.
Materials and Methods
Plasmids
The plasmids pSVH and pSV0d have been previously described (Stenberg et al., 1990). The plasmids pCMV71 and p760CAT were obtained from Dr. Mark Stinski. The pMIEPLuc plasmid was constructed by inserting the major immediate early promoter region from p760CAT into pGL2Basic (Promega). The plasmid pCMV71 was modified by the insertion of the influenza hemagglutinin (HA) epitope tag immediately downstream of the initiating AUG codon by overlapping PCR mutagenesis using primer HA-1 (Table 1) to generate plasmid pCMV71HA. An additional Xba I site was also introduced immediately upstream of the pp71 ORF by PCR using primer 71X-1 to generate pCMV71HAX. The resultant Xba I fragment was then cloned into pBSKS(+) (Stratagene) modified by the deletion of the Kpn I site (pBSKN71HAX). Random insertion mutants within the pp71 open reading frame were generated as previously described (Stenberg et al., 1990). Briefly, the pCMV71 plasmid was partially digested with either Hae III or Pvu II and the linear DNA isolated. A 12 nt linker containing a unique Bst BI site (5′ CGCTTCGAAGCG) was ligated to the linear DNA and the plasmid recircularized. Clones that contained Bst BI sites were identified by restriction enzyme digestion and the site of insertion confirmed by complete sequencing of the pp71 open reading frame. Some insertion mutants that contained multiple copies of the linker were further digested with Bst BI, and recircularized, resulting in a single copy of the linker. Mutants were subcloned by inserting the Not I and Bsm I fragments from pCMV71 into pBSKN71HAX. The resultant Xba I fragments containing the pp71 ORF and its mutants forms were then subcloned into the final expression vectors pSI (Promega) and pRc/RSV (Invitrogen).
Table 1.
Primers used for mutagenesis. Mutated sequences are underlined. For primers used in PCR mutagenesis procedures, only the forward primer sequence is shown.
| Primer Name | Primer Sequence | Purpose |
|---|---|---|
| HA-1 | 5’-CCGCCTTCGCAATGTACCCATACGATGTTCCAGATTACGCTTCTCAGGCATCGTCC | Introduce HA-Tag sequence |
| 71X-1 | 5’-TTGCCGGAACCCTTCTAGACCTCCCACGAAGAC | Introduce Xba I site 5’ of pp71 ORF |
| MRS | 5’-GCAGATCTTTGTCAGCCAGT | Amplify MR for random mutagenesis |
| MRAS | 5’-TGACTCGGGATGTATGCGC | Amplify MR for random mutagenesis |
| pp7162MR-For | 5’-CTCGAGTCTCTGCTGTTTTC | Introduce Xho I site for eGFP-62MR cloning |
| pp7162MR-N | 5’-CTCGAGGAACTTCGTGCCACCATGGAGGTG | N-terminal primer for 62MR-eGFP cloning |
| pp7194MR-N | 5′-CTAAACATCAAGGAGCTCACCATGGTGGGCG | N-terminal primer for 94MR-eGFP cloning |
| pp71188MR-N | 5′-GCAGAACCGAGCTCACCATGGAGG | N-terminal primer for 188MR-eGFP cloning |
| pp71MR-Rev | 5’-GTACCCGGGATGTATGCG | Introduce Xma I site for MR-eGFP cloning |
| S-tag1 | 5′-CGCCTTCGCAATGAAAGAAACCGCTGCTGCGAAATTTTCTCAGGCATCGTCC | Introduce 8 aa of the S-tag sequence |
| S-tag2 | 5′-GCTGCTAAATTTGAACGCCAGCACATGGACTCGTCTCAGGCATCGTCC | Introduce 7 aa of the S-tag sequence |
| SV40 | 5′-GATGTTCCAGATTACGCTCCAAAGAAGAAGAGAAAGGTGTCTCAGGCATCGTCCTCG | Introduce the SV40 NLS |
Random mutagenesis of the MR domain (amino acids 198–294) was performed using the GeneMorph® II EZClone Domain Mutagenesis kit (Stratagene) using the MRS and MRAS primers (Table 1). Mid region (MR)-eGFP fusion protein constructs were generated using PCR-based mutagenesis and standard cloning. Briefly, the pp71 mid region sequences from amino acid 62, 94, and 188 to 300 were amplified using the appropriate forward primers with a common reverse primer (Table 2). The PCR products were cloned into pCR4-TOPO, sequenced and then subcloned into peGFP-N2 to generate p62MR-eGFP, p94MR-eGFP and p188MR-eGFP. Insertion of the SV40NLS into the pp71 open reading frame immediately downstream of the HA-epitope tag was performed using the Quikchange mutagenesis procedure (Stratagene) with the appropriate primers (Table 1).
Cells and transient transfection
Human foreskin fibroblasts (HFF) were grown in minimal essential medium supplemented with 10% newborn calf serum and 1% penicillin, streptomycin and glutamine. HFFs were transfected using the DEAE-dextran method as previously described (Stenberg et al., 1989) or using Lipofectamine 2000 according to the manufacturer's directions (Invitrogen Corporation). HeLa cells (ATCC) were maintained in Iscove's media supplemented with 10% fetal calf serum and 1% penicillin and streptomycin and transfected using standard calcium-phosphate procedures.
Reporter Assays and Western Blot analysis
HFF cells were co-transfected with either p760CAT or pMIEPLuc and the corresponding expression vectors containing either wild type or mutant pp71. Cells were harvested 48 hours post-transfection and assessed for either CAT activity or luciferase activity using the luciferase assay kit (Promega) and a TD-20/20 luminometer (Turner designs). Western Blot analysis was performed as previously described (Ciocco-Schmitt et al., 2002) using a monoclonal antibody to the HA-tag (12CA5, Roche Applied Science).
Immunofluorescence
HFF cells were transfected and seeded onto 2-well permanox™ chamber slides or 8-chamber cover glasses pretreated with 2% gelatin (Nalge Nunc International) 24 hours post-transfection. After an additional 24 hours, HFF cells were first fixed with 4% paraformaldehyde for 10 minutes at room temperature, washed twice with PBS, and then fixed with 100% methanol for 2 minutes and washed four times with PBS. HFF cells were permeabilized with 0.1% Triton X-100 in PBS for 10 minutes at room temperature and blocked with 5% normal goat serum (NGS) in PBS for 45 minutes at 37°C. Primary antibodies were diluted in PBS containing 5% NGS, and added to HFF cells for 45 minutes including 1218 peptide antibody to IE86 (Stenberg et al., 1989), anti-HA (12CA5, Roche Applied Science) or HA-probe (Y-11, Santa Cruz Biotechnology, Inc.) to pp71 and anti-PML (PG-M3, Santa Cruz Biotechnology, Inc.). Cells were washed 3 times with PBS containing 0.1% BSA and incubated with fluorochrome-conjugated secondary antibodies (goat anti-rabbit Alexa Fluor® 546, anti-mouse Alexa Fluor® 488, Molecular Probes) diluted in 5% NGS in PBS as indicated by the manufacturers for 45 minutes at 37°C. Following three washes with 0.1% BSA in PBS, the cells were washed 2 times with sterile water, dried in a fume hood, and finally mounted with cover glass using vecta-shield mounting medium (Vector Laboratories, Inc.) and assessed by confocal microscopy using a Zeiss LSM 510 confocal microscope and the images analyzed using Metamorph. Identification of the cytoplasmic localization of pp71 was performed by co-staining with a panel of antibodies to Golgi-related markers (BD Biosciences).Nuclei were stained using the TOTO3 dye (Invitrogen Corporation) according to the manufacturer's directions. Immunofluorescence of infected cells was performed at 72 or 96 hours post-infection by the AD169 strain using a monoclonal antibody to gB kindly provided by Dr. W. J. Britt (University of Alabama School of Medicine, Birmingham), a polyclonal antibody to the gB protein provided by Dr. T. Compton (Novartis Institutes for BioMedical Research) or an antibody to pp71 provided by Dr. R. F. Kalejta (University of Madison-Wisconsin).
S-tag protein purification and Mass Spectrometry
The pBSKN71HAX plasmid was subjected to a two-step mutagenesis procedure (Quikchange, Stratagene) using the S-tag1 and S-tag2 primers (Table 1) to introduce the 15 amino acid sequence corresponding to the S-tag (Hackbarth et al., 2004). The resultant Xba I fragment was then subcloned into pSI to generate pSI71S. This plasmid was then transfected into HeLa cells using a standard calcium-phosphate procedure. Approximately 1.5×107 transfected cells were harvested and the cell nuclei isolated as previously described (Kerry, Priddy, and Stenberg, 1994), except for the addition of 1 mM sodium vanadate in the buffers. The resultant nuclear extracts were then subjected to S-tag affinity chromatography essentially as described by the manufacturer (Novagen). Bound proteins were eluted with 1 × SDS-PAGE Gel Loading Buffer and treated for 20 min at 95°C to reverse the formaldehyde cross-links. Proteins were then separated on a 4–20% SDS-PAGE gel and visualized with a Bio-Safe Coomassie staining kit (Bio-Rad).
Protein bands were excised from 1-D polyacrylamide gels. Gel slices were cut into 1–2 mm cubes; washed 3X with 500 µl Ultra-pure water and incubated in 100% acetonitrile for 45 minutes. Samples were reduced with 50 mM DTT at 56°C for 45 minutes then alkylated with 55 mM iodoacetamide for 1 hour at room temperature. The material was dried in a speed-vac, rehydrated in a 12.5 ng/µl modified sequencing grade trypsin solution (Promega, Madison, WI) and incubated in an ice bath for 40–45 min. The excess trypsin solution was then removed and replaced with 40–50 µl of 50mM ammonium bicarbonate, 10% acetonitrile, pH 8.0 and the mixture was incubated overnight at 37°C. Peptides were extracted 2X with 25 µl 50% acetonitrile, 5% formic acid and dried in a speed-vac. Digests were resuspended in 20 µl Buffer A (5% Acetonitrile, 0.1% Formic Acid, 0.005% heptafluorobutyric acid) and 3–6 µl were loaded onto a 12-cm × 0.075 mm fused silica capillary column packed with 5µM diameter C-18 beads (The Nest Group, Southboro, MA) using a N2 pressure vessel at 1100 psi. Peptides were eluted over 55 minutes, by applying a 0–80% linear gradient of Buffer B (95% Acetonitrile, 0.1% Formic Acid, 0.005% HFBA) at a flow rate of 150 µl/min with a pre-column flow splitter resulting in a final flow rate of ~200 nl/min directly into the source. In some cases, the gradient was extended to 150 minutes to acquire more MS/MS spectra. A LTQ™ Linear Ion Trap (ThermoFinnigan, San Jose, CA) was run in an automated collection mode with an instrument method composed of a single segment and 5 data-dependent scan events with a full MS scan followed by 4 MS/MS scans of the highest intensity ions. Normalized collision energy was set at 35, activation Q was 0.250 with minimum full scan signal intensity at 1 × 105 with no minimum MS2 intensity specified. Dynamic exclusion was turned on utilizing a three minute repeat count of 2 with the mass width set at 1.0 m/z. Sequence analysis was performed with TurboSEQUEST™ (ThermoFinnigan, San Jose, CA) or MASCOT (Matrix Sciences, London GB) using an indexed viral subset database of the non-redundant protein database from National Center for Biotechnology Information (NCBI) web site (http://www.ncbi.nlm.nih.gov/).
Acknowledgements
This work was supported in part by PHS Grant AI038372 to JAK. The authors would like to thank Dr. Laura Hanson for her assistance with the confocal microscopy and Dr. Rob Kalejta, Dr. Theresa Compton and Dr. Bill Britt for the provision of the pp71 and gB antibodies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abate DA, Watanabe S, Mocarski ES. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol. 2004;78(20):10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders DG, Kerry JA, Pari GS. Betaherpesviruses: DNA synthesis and late viral gene expression. In: Arvin A, Campadielli-Fume G, Mocarski ES, Moore P, Roizman B, Whitley R, Yamanishi K, editors. "Human Herpesviruses: Biology, Therapy and Immunoprophylaxis". Cambridge: Cambridge University Press; 2007. pp. 295–310. [Google Scholar]
- AuCoin DP, Smith GB, Meiering CD, Mocarski ES. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J Virol. 2006;80(16):8199–8210. doi: 10.1128/JVI.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldick CJ, Jr, Marchini A, Patterson CE, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71(6):4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldick CJ, Jr, Shenk T. Proteins associated with purified human cytomegalovirus particles. J Virol. 1996;70(9):6097–6105. doi: 10.1128/jvi.70.9.6097-6105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnahan WA, Shenk TE. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc Natl Acad Sci U S A. 2000;97(26):14506–14511. doi: 10.1073/pnas.97.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt WJ, Jarvis M, Seo JY, Drummond D, Nelson J. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J Virol. 2004;78(1):539–543. doi: 10.1128/JVI.78.1.539-543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Shenk T. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci U S A. 2003;100(20):11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell SR, Bresnahan WA. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J Virol. 2005;79(12):7792–7802. doi: 10.1128/JVI.79.12.7792-7802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell SR, Bresnahan WA. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J Virol. 2006;80(12):6188–6191. doi: 10.1128/JVI.02676-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau NH, Vanson CD, Kerry JA. Transcriptional regulation of the human cytomegalovirus US11 early gene. J Virol. 1999;73(2):863–870. doi: 10.1128/jvi.73.2.863-870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe D, Christophe-Hobertus C, Pichon B. Nuclear targeting of proteins: how many different signals? Cell Signal. 2000;12(5):337–341. doi: 10.1016/s0898-6568(00)00077-2. [DOI] [PubMed] [Google Scholar]
- Ciocco-Schmitt GM, Karabekian Z, Godfrey EW, Stenberg RM, Campbell AE, Kerry JA. Identification and Characterization of Novel Murine Cytomegalovirus M112--113 (e1) Gene Products. Virology. 2002;294(1):199–208. doi: 10.1006/viro.2001.1311. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991;16(12):478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Durkin SS, Ward MD, Fryrear KA, Semmes OJ. Site-specific phosphorylation differentiates active from inactive forms of the human T-cell leukemia virus type 1 Tax oncoprotein. J Biol Chem. 2006;281(42):31705–31712. doi: 10.1074/jbc.M607011200. [DOI] [PubMed] [Google Scholar]
- Fortunato EA, Spector DH. Regulation of human cytomegalovirus gene expression. Adv Virus Res. 1999;54:61–128. doi: 10.1016/s0065-3527(08)60366-8. [DOI] [PubMed] [Google Scholar]
- Gallina A, Simoncini L, Garbelli S, Percivalle E, Pedrali-Noy G, Lee KS, Erikson RL, Plachter B, Gerna G, Milanesi G. Polo-like kinase 1 as a target for human cytomegalovirus pp65 lower matrix protein. J Virol. 1999;73(2):1468–1478. doi: 10.1128/jvi.73.2.1468-1478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. Protein counterparts of human and simian cytomegaloviruses. Virology. 1983;128(2):391–406. doi: 10.1016/0042-6822(83)90265-9. [DOI] [PubMed] [Google Scholar]
- Griffiths PD. CMV as a cofactor enhancing progression of AIDS. J Clin Virol. 2006;35(4):489–492. doi: 10.1016/j.jcv.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Hackbarth JS, Lee SH, Meng XW, Vroman BT, Kaufmann SH, Karnitz LM. S-peptide epitope tagging for protein purification, expression monitoring, and localization in mammalian cells. Biotechniques. 2004;37(5):835–839. [PubMed] [Google Scholar]
- Hakki M, Marshall EE, De Niro KL, Geballe AP. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J Virol. 2006;80(23):11817–11826. doi: 10.1128/JVI.00957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel G, Meyer H, Gartner S, Brand G, Kern HF. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32) J Gen Virol. 1995;76(Pt 7):1591–1601. doi: 10.1099/0022-1317-76-7-1591. [DOI] [PubMed] [Google Scholar]
- Hensel GM, Meyer HH, Buchmann I, Pommerehne D, Schmolke S, Plachter B, Radsak K, Kern HF. Intracellular localization and expression of the human cytomegalovirus matrix phosphoprotein pp71 (ppUL82): evidence for its translocation into the nucleus. J Gen Virol. 1996;77(Pt 12):3087–3097. doi: 10.1099/0022-1317-77-12-3087. [DOI] [PubMed] [Google Scholar]
- Hofmann H, Sindre H, Stamminger T. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J Virol. 2002;76(11):5769–5783. doi: 10.1128/JVI.76.11.5769-5783.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homman-Loudiyi M, Hultenby K, Britt W, Soderberg-Naucler C. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-golgi network 46, and mannosidase II. J Virol. 2003;77(5):3191–3203. doi: 10.1128/JVI.77.5.3191-3203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Kalejta RF. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology. 2007 doi: 10.1016/j.virol.2007.05.037. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Vladimirova OV, Maul GG. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J Virol. 2002;76(15):7705–7712. doi: 10.1128/JVI.76.15.7705-7712.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskenderian AC, Huang L, Reilly A, Stenberg RM, Anders DG. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70(1):383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis MA, Jones TR, Drummond DD, Smith PP, Britt WJ, Nelson JA, Baldick CJ. Phosphorylation of human cytomegalovirus glycoprotein B (gB) at the acidic cluster casein kinase 2 site (Ser900) is required for localization of gB to the trans-Golgi network and efficient virus replication. J Virol. 2004;78(1):285–293. doi: 10.1128/JVI.78.1.285-293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Lee SW. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J Virol. 2004;78(3):1488–1502. doi: 10.1128/JVI.78.3.1488-1502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta RF, Bechtel JT, Shenk T. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol Cell Biol. 2003;23(6):1885–1895. doi: 10.1128/MCB.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta RF, Shenk T. The human cytomegalovirus UL82 gene product (pp71) accelerates progression through the G1 phase of the cell cycle. J Virol. 2003a;77(6):3451–3459. doi: 10.1128/JVI.77.6.3451-3459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta RF, Shenk T. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc Natl Acad Sci U S A. 2003b;100(6):3263–3268. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry JA, Priddy MA, Jervey TY, Kohler CP, Staley TL, Vanson CD, Jones TR, Iskenderian AC, Anders DG, Stenberg RM. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J Virol. 1996;70(1):373–382. doi: 10.1128/jvi.70.1.373-382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry JA, Priddy MA, Stenberg RM. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J Virol. 1994;68(7):4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzaniak M, Mach M, Britt WJ. The Cytoplasmic Tail of Glycoprotein M (gpUL100) Expresses Trafficking Signals Required for Human Cytomegalovirus Assembly and Replication. J Virol. 2007;81(19):10316–10328. doi: 10.1128/JVI.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol. 2004;78(20):11030–11039. doi: 10.1128/JVI.78.20.11030-11039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Stinski MF. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66(7):4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorz K, Hofmann H, Berndt A, Tavalai N, Mueller R, Schlotzer-Schrehardt U, Stamminger T. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J Virol. 2006;80(11):5423–5434. doi: 10.1128/JVI.02585-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65(4):570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall KR, Rowley KV, Rinaldi A, Nicholson IP, Ishov AM, Maul GG, Preston CM. Activity and intracellular localization of the human cytomegalovirus protein pp71. J Gen Virol. 2002;83(Pt 7):1601–1612. doi: 10.1099/0022-1317-83-7-1601. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC. Budding events in herpesvirus morphogenesis. Virus Res. 2004;106(2):167–180. doi: 10.1016/j.virusres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Michelson S, Turowski P, Picard L, Goris J, Landini MP, Topilko A, Hemmings B, Bessia C, Garcia A, Virelizier JL. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and a host-cell derived PP2A. J Virol. 1996;70(3):1415–1423. doi: 10.1128/jvi.70.3.1415-1423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Jr, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. "Fields Virology". 5 ed. Vol. 2. Philadelphia: Lippincott, Williams and Wilkins; 2007. pp. 2701–2772. 2 vols. [Google Scholar]
- Munger J, Yu D, Shenk T. UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J Virol. 2006;80(7):3541–3548. doi: 10.1128/JVI.80.7.3541-3548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Goto K, Tanaka K, Gibson W, Moriishi E, Miura Y, Kurata T, Irie S, Sata T. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J Virol. 2003;77(15):8541–8547. doi: 10.1128/JVI.77.15.8541-8547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM, Nicholl MJ. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J Gen Virol. 2006;87(Pt 5):1113–1121. doi: 10.1099/vir.0.81566-0. [DOI] [PubMed] [Google Scholar]
- Prichard MN, Britt WJ, Daily SL, Hartline CB, Kern ER. Human cytomegalovirus UL97 Kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J Virol. 2005;79(24):15494–15502. doi: 10.1128/JVI.79.24.15494-15502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski MJ, Shenk T. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J Virol. 1997;71(2):1485–1496. doi: 10.1128/jvi.71.2.1485-1496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffert RT, Kalejta RF. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol. 2006;80(8):3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffert RT, Kalejta RF. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J Virol. 2007;81(17):9109–9120. doi: 10.1128/JVI.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Greis KD, Sztul E, Britt WJ. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol. 2000;74(2):975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Mahr JA, Orazio NI, Spector DH. Nuclear export of the human cytomegalovirus tegument protein pp65 requires cyclin-dependent kinase activity and the Crm1 exporter. J Virol. 2007;81(21):11730–11736. doi: 10.1128/JVI.02760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, McElroy AK, Yen J, Tamrakar S, Clark CL, Schwartz RA, Spector DH. Cyclin-dependent kinase activity is required at early times for accurate processing and accumulation of the human cytomegalovirus UL122-123 and UL37 immediate-early transcripts and at later times for virus production. J Virol. 2004;78(20):11219–11232. doi: 10.1128/JVI.78.20.11219-11232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Spector DH. Cyclin-dependent kinase activity is required for efficient expression and posttranslational modification of human cytomegalovirus proteins and for production of extracellular particles. J Virol. 2006;80(12):5886–5896. doi: 10.1128/JVI.02656-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierling K, Stamminger T, Mertens T, Winkler M. Human cytomegalovirus tegument proteins ppUL82 (pp71) and ppUL35 interact and cooperatively activate the major immediate-early enhancer. J Virol. 2004;78(17):9512–9523. doi: 10.1128/JVI.78.17.9512-9523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Yu QC, Enquist L, Shenk T. Human cytomegalovirus UL99- encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol. 2003;77(19):10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snydman DR. The case for cytomegalovirus prophylaxis in solid organ transplantation. Rev Med Virol. 2006 doi: 10.1002/rmv.514. [DOI] [PubMed] [Google Scholar]
- Stamminger T, Gstaiger M, Weinzierl K, Lorz K, Winkler M, Schaffner W. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. J Virol. 2002;76(10):4836–4847. doi: 10.1128/JVI.76.10.4836-4847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg RM, Depto AS, Fortney J, Nelson JA. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol. 1989;63(6):2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg RM, Fortney J, Barlow SW, Magrane BP, Nelson JA, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64(4):1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamrakar S, Kapasi AJ, Spector DH. Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7. J Virol. 2005;79(24):15477–15493. doi: 10.1128/JVI.79.24.15477-15493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng FY, Wang Y, Tang BL. The syntaxins. Genome Biol. 2001;2(11) doi: 10.1186/gb-2001-2-11-reviews3012. REVIEWS3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl M, Fairhurst J, Baum EZ, Sun L, Jones TR. The human cytomegalovirus UL97 protein is phosphorylated and a component of virions. Virology. 1997;231(1):72–80. doi: 10.1006/viro.1997.8523. [DOI] [PubMed] [Google Scholar]
- Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG, 2nd, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004;78(20):10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M, Rice SA, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68(6):3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M, Stamminger T. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J Virol. 1996;70(12):8984–8987. doi: 10.1128/jvi.70.12.8984-8987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


