Abstract
A limiting factor in many molecular dating studies is shortage of reliable calibrations. Current methods for choosing calibrations (e.g. cross-validation) treat them as either correct or incorrect, whereas calibrations probably lie on a continuum from highly accurate to very poor. Bayesian relaxed clock analysis permits inclusion of numerous candidate calibrations as priors: provided most calibrations are reliable, the model appropriate and the data informative, the accuracy of each calibration prior can be evaluated. If a calibration is accurate, then the analysis will support the prior so that the posterior estimate reflects the prior; if a calibration is poor, the posterior will be forced away from the prior. We use this approach to test two fossil dates recently proposed as standard calibrations within vertebrates. The proposed bird–crocodile calibration (approx. 247 Myr ago) appears to be accurate, but the proposed bird–lizard calibration (approx. 255 Myr ago) is substantially too recent.
Keywords: molecular dating, Bayesian analysis, calibration, fossil, reptile
1. Introduction
Recent advances in molecular clock methods and expanding genetic databases provide an unprecedented opportunity to infer time-scales of organismal evolution. However, progress is lagging on one vital component of molecular divergence dating—calibration. Methods for choosing calibrations are being developed (e.g. van Tuinen & Dyke 2003; Near et al. 2005) and point calibrations are being replaced by distributions that better represent palaeontological uncertainty (e.g. Yang & Rannala 2005; Drummond et al. 2006). Much of the debate over calibration issues has involved amniotes (mammals, birds and reptiles). The mammal–bird split is probably the most commonly used calibration for these organisms, but its accuracy has been questioned due to a long preceding fossil gap (Reisz & Müller 2004; Müller & Reisz 2005). Instead, Reisz & Müller (2004) proposed using two other splits with tightly bounded divergence times as standard calibrations. They suggested a range of 252–257 Myr ago for the split between Archosauromorpha (birds, crocodiles and relatives) and Lepidosauromorpha (lizards, snakes, tuatara and relatives), and a range of 243–251 Myr ago for the split between Ornithodira (birds and relatives) and Crurotarsi (crocodiles and relatives). These dates, if correct, imply that both divergences occurred in close proximity (less than 14 Myr). More recently, Benton & Donoghue (2007), as part of a review across all metazoans, proposed a much broader interval (259.7–299.8 Myr ago) for the Archosauromorph–Lepidosauromorph (bird–lizard) split, but a similar interval (246.5–250.4 Myr ago) for the bird–crocodile split.
If the calibrations proposed by Reisz & Müller (2004) are accurate: (i) the molecular branches between the bird–lizard and the bird–crocodile divergences should be very short and (ii) these calibrations should be broadly consistent with other well-corroborated calibrations. Here, we use Bayesian relaxed clock analyses with soft and hard calibration bounds to demonstrate that the bird–lizard and bird–crocodile divergences were relatively broadly spaced in time; in particular, the bird–lizard divergence occurred much earlier than proposed and should be used with a much deeper maximum bound. The Bayesian approach used here does not treat calibrations as either correct or incorrect, but recognizes that calibrations lie on a continuum between highly accurate (narrow bounds straddling the actual divergence) and very inaccurate (wide bounds that still do not encompass the real date), and places greater weight on the former.
2. Material and methods
Taxa were selected to span the major amniote clades and a range of well-corroborated calibrations. Two nuclear loci were selected because they provide independent, large single-copy sequences uninterrupted by introns, and evolve at a suitable rate: approximately 3160 bp composing most of the recombination activating gene-1 (RAG-1) and approximately 1100 bp of c-mos (oocyte maturation factor). Sequences were obtained from GenBank, with additional sequences generated to improve sampling for birds and squamates. The tree was rooted with an amphibian (Xenopus). Primers and GenBank accession numbers are provided in electronic supplementary material.
Divergence dating used Beast v. 1.3 (Drummond & Rambaut 2003), which employs a Bayesian Markov chain Monte Carlo (MCMC) to co-estimate topology, substitution rates and node ages. Posterior probability distributions of node ages were obtained for each gene separately and the concatenated 2-gene alignment. The data were partitioned by gene and by codon (first+second versus third). Best-fit substitution models were identified for each data partition using MrModeltest v. 2 (Nylander 2004); the general time-reversible model with rate variation (six gamma categories) was implemented for all partitions, with an invariant site parameter added for one partition (RAG first+second codon). In the combined analysis, model parameters were unlinked across partitions. Each analysis implemented a Yule branching rate prior, with rate variation across branches assumed to be uncorrelated and lognormally distributed (Drummond et al. 2006). Each final MCMC chain was run for 4 000 000 generations (burnin 20%), with parameters sampled every 100 steps. Examination of MCMC samples using Tracer v. 1.2 (Rambaut & Drummond 2003) suggested that the independent chains were each adequately sampling the same probability distribution; effective sample sizes for all parameters of interest were greater than 500.
Four direct fossil calibrations were used in addition to the bird–lizard and bird–crocodile calibrations of Reisz & Müller (2004; table 1). Divergences were estimated using the above MCMC analyses and these calibrations in four ways. (i) First, a relaxed clock was used, with calibrations treated as having a translated lognormal distribution. This yields a skewed distribution consistent with the bias in the fossil record: there is a hard minimum bound meaning zero probability of dates much younger than the oldest known fossil (allowing for error in dating), peak probability is the mean age assigned to the oldest fossil, and there is a soft maximum bound meaning an indefinitely long (but increasingly unlikely) tail of older dates (to allow for non-preservation). Although lognormal priors were assigned wide minimum bounds (approx. 20 Myr younger than the oldest known fossils), they still bias against the possibility that the real dates for calibration nodes are much younger than the proposed fossil dates (e.g. owing to taxonomic error). (ii) Thus, a relaxed clock analysis was performed with calibrations assumed to have a normal distribution, which allows divergence dates to vary symmetrically (and with soft bounds). (iii) An analysis assuming a globally constant molecular clock, and calibrations with lognormal distributions, was performed in case the complex relaxed clock analyses were returning anomalous results due to overparametrization. (iv) Finally, a relaxed clock analysis was performed with all calibrations ‘fixed’ using hard bounds on narrow uniform distributions. This allows one to estimate branch-specific substitution rates implied if all six calibrations are assumed to be accurate. If inferred rates for some branches are implausible (e.g. substantially higher than other known tetrapods), this would suggest that some adjacent calibrations are incompatible (analyses 1–3 do not allow this test as the soft bounds on calibration nodes will allow their posterior distributions to shift significantly to avoid drastic implied substitution rates). In all analyses, a wide uniform prior constraint of 320–380 Myr (e.g. Benton & Donoghue 2007) was placed on the root (amphibian–amniote split) to prevent the chain from becoming fixed on unrealistic inflated values.
Table 1.
Prior probability distributions for divergence times enforced for the six calibration nodes based on fossil evidence.
| prior distribution | calibration node | |||||
|---|---|---|---|---|---|---|
| bird–crocodile | bird–lizard | scincomorph–anguimorph | pleurodire–cryptodire | Rattus–Mus | bird–mammal | |
| normal, mean (95% CI) | 247 (227, 267) | 254.5 (234.5, 274.5) | 168 (148, 188) | 210 (185, 235) | 12 (6, 18) | 315 (270, 360) |
| lognormal: mean (zero-offset, 95% CI) | 247 (228, 266) | 254.5 (237, 272) | 168 (155, 180) | 210 (190, 250) | 12 (9, 18) | 315 (295, 360) |
| uniform: hard min–max bounds | 243–251 | 252–257 | 158–178 | 195–225 | 10–15 | 295–370 |
| references | Reisz & Müller (2004) and Müller & Reisz (2005) | Reisz & Müller (2004) and Müller & Reisz (2005) | Evans et al. (2002) | Near et al. (2005) | Jacobs & Downs (1994) | Reisz & Müller (2004) and Benton & Donoghue (2007) |
Certain combinations of priors can interact to generate unexpected effective joint priors (e.g. a younger calibration for a large clade plus an older calibration for a smaller, included clade). Thus, analyses without data should be performed to (i) check that the effective priors are similar to the original priors and (ii) assess the informativeness of the data, by comparing these effective priors with posteriors obtained when data are added (Drummond et al. 2006). Here, these analyses indicated that the effective priors were similar to the original priors, with the posteriors obtained with data departing from both (indicating informative data).
3. Results and discussion
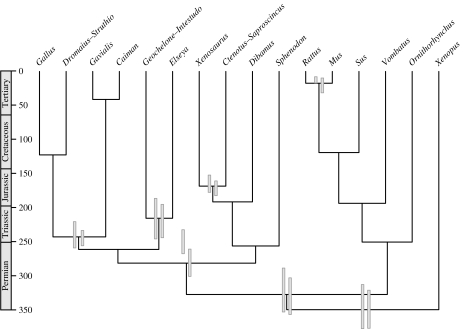
All MCMC searches converged on a topology highly congruent with recently published molecular phylogenies (e.g. Townsend et al. 2004; Iwabe et al. 2005; van Rheede et al. 2005). Both genes showed high levels of rate heterogeneity (coefficients of branch rate variation greater than 0.8) and low autocorrelation of rates between adjacent branches (rate covariance less than 0.1). Mean and highest posterior density (HPD) node age estimates inferred from individual and combined gene datasets were very similar (table 2) and the discussion below focuses on the combined analyses. Age estimates from the relaxed clock analysis with lognormal calibration priors produced narrow 95% HPD intervals that are mostly consistent with the fossil dates (figure 1, table 2). A notable feature of all these trees is the relatively long branches between the bird–lizard and bird–crocodile divergences, indicating that they are widely spaced in time (contra, Reisz & Müller 2004). Thus, either the bird–lizard divergence is older than proposed or the bird–crocodilian divergence is younger; our analyses indicate the former. The combined gene analysis dates the most recent common ancestor of the bird–crocodile clade at 247.3 Myr ago (95% HPD: 237.7–259.0). The posterior estimate is therefore very similar to the prior (mode=247; figure 1). However, the bird–lizard split is dated at 282.8 Myr ago (95% HPD: 263.0–303.9) and greatly exceeds the prior (mode=254.5). This result suggests that a more liberal lower bound is required for this calibration and is consistent with the older dates proposed by Benton & Donoghue (259.7–299.8 Myr ago).
Table 2.
Posterior probability distributions for divergence times for the bird–crocodile and bird–lizard nodes, obtained from the combined and single gene data analysed using a relaxed clock with normal, lognormal and uniform hard-bounded calibration priors (table 1), and a globally constant molecular clock with lognormal calibrations.
| posterior—mean (95% HPD)—millions of years before present | ||
|---|---|---|
| bird–crocodile | bird–lizard | |
| RAG-1+C-MOS | ||
| normal | 232.7 (213.2, 250.9) | 269.2 (250.8, 287.8) |
| lognormal | 247.3 (237.7, 259.0) | 282.8 (263.0, 303.9) |
| uniform | 244.8 (243.0, 248.1) | 258.2 (254.8, 260.1) |
| global clock | 235.8 (232.6, 240.0) | 261.1 (249.5, 269.1) |
| RAG-1 | ||
| normal | 231.9 (214.1, 250.9) | 269.1 (250.7, 287.9) |
| lognormal | 247.7 (237.1, 259.1) | 279.8 (275.9, 302.1) |
| uniform | 244.2 (243.0, 236.4) | 255.8 (253.7, 257.0) |
| C-MOS | ||
| normal | 232.4 (212.0, 253.9) | 264.2 (245.9, 282.4) |
| lognormal | 247.6 (237.5, 259.7) | 273.6 (254.0, 293.9) |
| uniform | 248.2 (243.0, 251.0) | 256.2 (252.6, 257.0) |
Figure 1.
Consensus ultrametric tree obtained using combined data and lognormal priors. Left hand node bars indicate prior distributions; right hand node bars, posterior distributions (95% HPD). Time-scale in millions of years.
The relaxed clock analysis with normal calibration priors again produced posterior estimates for the bird–crocodile and bird–lizard divergences that are relatively broadly spaced (table 2). However, as expected (see above), posterior ages for most calibration nodes are younger than the minimum plausible dates implied by fossils, including the bird–crocodile posterior estimate (mean=232.7 Myr ago; 95% HPD=213.2–250.9). Only the bird–lizard posterior is older than the prior mode (mean=269.2 Myr ago; 95% HPD=250.8–287.8). Similarly, in the global clock analysis, the two divergences of interest were widely spaced: posterior estimates for the bird–crocodile split are slightly shallower than the prior (mean=235.8 Myr ago; 95% HPD=232.6–240.0); whereas the bird–lizard posterior exceeds the prior (mean=261.1 Myr ago; 95% HPD=249.5–269.1).
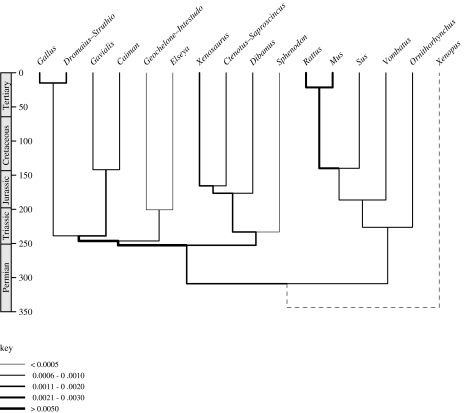
The above data suggest that the bird–crocodile and bird–lizard calibrations cannot be as closely spaced in time as proposed: the long intervening molecular branches cause the posterior estimates for the bird–lizard split to move towards older dates. However, we also investigated whether these long branches can be reconciled with the proposed closely spaced divergences by assuming very rapid molecular evolution. Fixing all proposed calibrations (table 1) with hard, narrow bounds implies extremely high branch-specific substitution rates on the branches between the two calibrations of interest. Results for the two-gene analysis are shown (table 2, figure 2). The mean rate of molecular evolution for the branches between the bird–lizard and bird–crocodile nodes is 0.0055 substitutions per site per Myr: this is by far the highest anywhere on the tree, and more than double that of rodents (0.0026), which have the highest rate known in amniotes (e.g. Douzery et al. 2003). There is nothing about the inferred ecology of early archosauromorphs (e.g. generation time, metabolic rate) that would suggest such an exceptionally high rate of molecular evolution.
Figure 2.
Consensus ultrametric tree obtained using combined data with hard uniform bounds for all calibrations. Time-scale in millions of years. Thickness of each branch proportional to rate of evolution in substitutions per site per Myr (based on 10 000 post-burnin trees filtered for topological concordance).
The present study shows how Bayesian analysis of molecular data with soft and hard bounds can be used to evaluate the accuracy of proposed calibrations. However, for this approach to work, the phylogenetic model must be reasonably accurate, the molecular data informative and most calibrations reliable. All potential calibration information is included in the analysis: the concordant, reliable calibrations contribute most to the final date estimates (priors consistent with posteriors), while less reliable calibrations have less influence (priors inconsistent with posteriors). This is a promising alternative to the existing methods of choosing calibration points (e.g. cross-validation: Near et al. 2005), which often use an arbitrary cut-off to retain a subset of calibrations, and thus treat calibrations in an all-or-nothing fashion.
Supplementary Material
Information on primers and PCR protocols for sequence generation
Taxonomic information and Genbank accession numbers for all sequences
BEAST xml input: C-mos+RAG-1 combined data with lognormal calibration priors
References
- Benton M.J, Donoghue P.C.J. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. doi:10.1093/molbev/msl150 [DOI] [PubMed] [Google Scholar]
- Douzery E.J.P, Delsuc F, Stanhope M.J, Huchon D. Local molecular clocks in three nuclear genes: divergence times for rodents and other mammals and incompatibility among fossil calibrations. J. Mol. Evol. 2003;57:S201–S213. doi: 10.1007/s00239-003-0028-x. doi:10.1007/s00239-003-0028-x [DOI] [PubMed] [Google Scholar]
- Drummond, A. J. & Rambaut, A. 2003. Beast version 1.0. Available from http://evolve.zoo.ox.ac.uk/beast/
- Drummond A.J, Ho S.Y.W, Phillips M.J, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.E, Prasad G.V.R, Manhas B.K. Fossil lizards from the Jurassic Kota formation of India. J. Vertebr. Paleontol. 2002;22:299–312. doi:10.1671/0272-4634(2002)022[0299:FLFTJK]2.0.CO;2 [Google Scholar]
- Iwabe N, Hara Y, Kumazawa Y, Shibamoto K, Saito Y, Miyata T, Katoh K. Sister group relationship of turtles to the bird–crocodilian clade revealed by DNA-coded proteins. Mol. Biol. Evol. 2005;22:810–813. doi: 10.1093/molbev/msi075. doi:10.1093/molbev/msi075 [DOI] [PubMed] [Google Scholar]
- Jacobs L.L, Downs W.R. The evolution of murine rodents in Asia. In: Tomida Y, Li C.-K, Setoguchi T, editors. Rodent and lagomorph families of Asian origins and diversification. National Science Museum Monographs; Tokyo, Japan: 1994. pp. 149–156. [Google Scholar]
- Müller J, Reisz R.R. Four well-constrained calibration points from the vertebrate fossil record for molecular clock estimates. BioEssays. 2005;27:1069–1075. doi: 10.1002/bies.20286. [DOI] [PubMed] [Google Scholar]
- Near T.J, Meylan P.A, Shaffer H.B. Assessing concordance of fossil calibration points in molecular clock studies: an example using turtles. Am. Nat. 2005;165:137–146. doi: 10.1086/427734. doi:10.1086/427734 [DOI] [PubMed] [Google Scholar]
- Nylander, J. A. A. 2004 MrModeltest: program distributed by the author, v2. Evolutionary Biology Centre, Uppsala University.
- Rambaut A, Drummond A. University of Oxford; Oxford, UK: 2003. Tracer: MCMC trace analysis tool. [Google Scholar]
- Reisz R.R, Müller J. Molecular timescales and the fossil record: a paleontological perspective. Trends Genet. 2004;20:237–241. doi: 10.1016/j.tig.2004.03.007. doi:10.1016/j.tig.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Townsend T, Larson A, Louis E, Macey J.R. Molecular phylogenetics of Squamata: the position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 2004;53:735–757. doi: 10.1080/10635150490522340. doi:10.1080/10635150490522340 [DOI] [PubMed] [Google Scholar]
- van Rheede T, Bastiaans T, Boone D.N, Hedges S.B, de Jong W.W, Madsen O. The platypus is in its place: nuclear genes and indels confirm the sister group relationship of monotremes and therians. Mol. Biol. Evol. 2005;23:587–597. doi: 10.1093/molbev/msj064. doi:10.1093/molbev/msj064 [DOI] [PubMed] [Google Scholar]
- van Tuinen M, Dyke G.J. Calibration of galliform molecular clocks using multiple fossils and genetic partitions. Mol. Phylogenet. Evol. 2003;30:74–86. doi: 10.1016/s1055-7903(03)00164-7. doi:10.1016/S1055-7903(03)00164-7 [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Branch-length prior influences Bayesian posterior probability of phylogeny. Syst. Biol. 2005;54:455–470. doi: 10.1080/10635150590945313. doi:10.1080/10635150590945313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information on primers and PCR protocols for sequence generation
Taxonomic information and Genbank accession numbers for all sequences
BEAST xml input: C-mos+RAG-1 combined data with lognormal calibration priors